Abstract
Membrane-permeable compounds that reversibly inhibit a particular step in gene expression are highly useful tools for cell biological and biochemical/structural studies. In comparison with other gene expression steps where multiple small molecule effectors are available, very few compounds have been described that act as general inhibitors of pre-mRNA splicing. Here we report construction and validation of a set of mammalian cell lines suitable for the identification of small molecule inhibitors of pre-mRNA splicing. Using these cell lines, we identified the natural product isoginkgetin as a general inhibitor of both the major and minor spliceosomes. Isoginkgetin inhibits splicing both in vivo and in vitro at similar micromolar concentrations. It appears to do so by preventing stable recruitment of the U4/U5/U6 tri-small nuclear ribonucleoprotein, resulting in accumulation of the prespliceosomal A complex. Like two other recently reported general pre-mRNA splicing inhibitors, isoginkgetin has been previously described as an anti-tumor agent. Our results suggest that splicing inhibition is the mechanistic basis of the anti-tumor activity of isoginkgetin. Thus, pre-mRNA splicing inhibitors may represent a novel avenue for development of new anti-cancer agents.
The removal of introns from nascent transcripts by the process of pre-mRNA (precursor to messenger RNA) splicing is an essential step in eukaryotic gene expression. Splicing is mediated by the spliceosome, a highly dynamic, multimegadalton machine composed of five small stable nuclear RNAs (snRNAs)2 and more than 100 polypeptides (reviewed in Ref. 1). Within the spliceosome, intron excision occurs in two chemical steps: 1) 5′ splice site cleavage accompanied by lariat formation at the branch point adenosine and 2) 3′ splice site cleavage accompanied by exon ligation. Both of these steps are readily observable in in vitro reactions containing crude nuclear extract and ATP as an energy source. In such reactions, spliceosome assembly occurs in a distinctly stepwise fashion. First, the pre-mRNA substrate is coated with a heterogeneous mixture of RNA-binding proteins (referred to as H complex). Interaction of U1 snRNP (U1 snRNA and its associated proteins) with the 5′ splice site and recognition of the branch point adenosine by U2 snRNP generates an early commitment complex (E or CC complex). A subsequent ATP-dependent step stabilizes the U2 snRNP-branch point interaction, resulting in formation of the prespliceosome (A complex). Entry of the U4/U5/U6 tri-snRNP to form B complex is followed by multiple structural rearrangements, which produce the catalytically active C complex, wherein the two chemical steps of splicing occur. Finally, the ligated exon and lariat products are released, and the remaining spliceosome components are disassembled.
In the more than two decades since its initial description (2, 3), a wealth of information has been gleaned regarding the parts list of the spliceosome, its gross assembly/disassembly pathway, certain key local structural interactions, and the activities of individual components. However, in comparison with other macromolecular machines, such as the ribosome and RNA polymerase II, our understanding of the spliceosome's inner workings and its detailed structure is still in its infancy. Mechanistic understanding of other macromolecular complexes has been greatly enhanced by the availability of multiple small molecule inhibitors impeding their functional cycles at different points (4, 5). Such small molecules have been likened to wrenches that can be thrown into the works to freeze cellular machines in specific conformations, making them more amenable to biochemical and structural investigation (4). Although a set of such wrenches that inhibit splicing in vitro or inhibit specific or alternative splicing events in vivo is being generated (4, 6–10), there is currently a paucity of small molecules that affect general pre-mRNA splicing in vivo.
In this paper, we describe a cell-based assay to screen for general splicing inhibitors. This assay takes advantage of the observation that some amount of unspliced pre-mRNA can escape from the nucleus and become available for translation in the cytoplasm (11, 12). By screening for an increase in reporter protein expression from a mammalian pre-mRNA designed such that only the unspliced version generates active protein, we were able to identify a compound that acts as a general inhibitor of splicing both in vivo and in vitro. This inhibitor is the naturally occurring biflavonoid isoginkgetin. In in vitro reactions, isoginkgetin causes accumulation of the prespliceosomal A complex. Like two other compounds recently described as in vivo splicing inhibitors (13, 14), isoginkgetin is a known anti-tumor agent (15). Our results suggest that the mechanistic basis of the anti-tumor activity of isoginkgetin is its inhibition of pre-mRNA splicing.
EXPERIMENTAL PROCEDURES
Plasmids—Reporter construct I was created by replacing the Renilla luciferase gene in plasmid triose-phosphate isomerase (TPI)/Renilla luciferase 5′ intron (pSHM06T) (16) with the firefly luc2 gene from plasmid pGL4.10 (Promega). Construct II is identical to construct I except that site-directed mutagenesis was used to remove an in-frame stop codon in the intron and add a G at position 6 in TPI exon 7. Construct III was generated by site-directed mutagenesis of II to inactivate the 5′ splice site. Constructs I, II, and III were subcloned into vector pcDNA5/FRT for integration into the flp recombinase target (FRT) sequence in HEK293 cells (described below).
Cell Culture and Generation of Stable Cell Lines—Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics (Invitrogen), as indicated below. Trypan blue (Invitrogen) and Alamar blue (BIOSOURCE) assays were done according to standard protocols. To generate a host cell line containing an integrated FRT site, HEK293 cells (ATCC) were transfected with ScaI-linearized pFRT/lacZeo (Invitrogen) using FuGENE 6 (Roche Applied Science) as per the Flp-In™ system manual (Invitrogen). Stable integrants were selected with 100 μg/ml zeocin. Southern blotting and β-galactosidase assays identified cell clones with integrants expressing intermediate levels of the LacZ-Zeo fusion. Two such clones (293F1 and 293F2) were subsequently co-transfected with one of the three reporter plasmids and pOG44 (Invitrogen), which encodes Flp recombinase. Successful targeting of the reporter plasmid to the FRT site was selected by treatment with 100 μg/ml hygromycin.
Luciferase Assays and RNA Isolation—Cells were grown in 24-well, 6-well, or 6-cm dishes and treated with DMSO or DMSO plus isoginkgetin, as indicated in the figure legends. Luciferase assays (luciferase assay system; Promega) were performed using a TD-20/20 Luminometer (Turner Systems). Protein concentrations were determined for lysates using the DC protein assay (Bio-Rad). Total RNA was isolated using TRI Reagent (Molecular Research Corp.) and DNase-treated (Promega) according to the manufacturers' protocols. When a sample was to be used both for monitoring luciferase activity and for preparing RNA, cells were harvested in ice-cold 1× phosphate-buffered saline; 25% was diluted 2-fold with 2× passive lysis buffer (Promega) and used for luciferase assays, and the remainder was added to 5.5 volumes of TRI Reagent for RNA isolation.
Reverse Transcription (RT)-PCR—RT-PCR was carried out using the SuperScript III one-step RT-PCR kit (Invitrogen). For qualitative experiments, 60 ng of DNase-treated total RNA was used per 25-μl reaction, run on 2% agarose gels, and stained with ethidium bromide. For semiquantitative RT-PCR (using [α-32P]dATP), different amounts of DNase-treated total RNA (0.1–100 ng) were used, depending on the transcript being amplified. Reactions were performed with at least two concentrations of RNA to ensure that they were within the linear range. When the amplification product crossed an intron-exon boundary, primers amplifying U6 snRNA, an unspliced polymerase III transcript, were included as a control. Primer sequences are listed in supplemental Table 1.
High Throughput Screening—All high throughput screening was performed at the Broad Institute in association with the Chemical Biology Program.
Cells were plated in 384-well plates at cell densities ranging from 1500 to 9000 cells/well. Plating ∼4000 cells/well gave the least variability in luciferase activity while maximizing the signal difference between cell lines II and III. Cells were plated in a 30-μl total volume using a Multidrop 384 or a Multidrop Micro (Thermo Scientific) liquid handler. To test sensitivity to DMSO (the carrier for the compound libraries), cell lines II and III were incubated overnight in growth media containing 0–1% DMSO. Concentrations up to 1% DMSO did not cause significant changes in signal intensity or reproducibility for any of the cell lines.
For high throughput screening, cell lines II and III were plated on day 0 in growth medium in white, opaque bottom 384-well plates (Nunc) at a density of 3000–6000 cells/30 μl. On day 1, 100 nl of test compounds or DMSO was pin-transferred using a CyBio robot with a 384-well pin array to duplicate plates. On day 2, ∼24 h after the pin transfer, luciferase activities were read using the Steady-Glo Luciferase Assay System (Promega) according to the manufacturer's protocol. Briefly, 30 μl of the combined cell lysis and assay reagent buffer were added directly to the well using a Matrix Wellmate (Thermo Scientific). After agitating plates for 10 min, luciferase readings were collected using an EnVision plate reader (PerkinElmer Life Sciences). Data analysis was performed as described (17) to determine Z-scores and assess reproducibility of compound effects. For follow up experiments, cells were grown in 24-well, 6-well, or 6-cm dishes and treated with DMSO or DMSO plus isoginkgetin, as described above.
Isoginkgetin—Screening and initial follow up experiments were carried out with isoginkgetin obtained from MicroSource. Isoginkgetin for subsequent experiments was purchased from Gaia Corp. To validate their identity and purity, all samples were subjected to C18 reverse phase high pressure liquid chromatography-mass spectrometry analysis. One sample from Gaia that exhibited no activity in cellular assays proved <50% pure, whereas a second sample that proved >99% pure by high pressure liquid chromatography-mass spectrometry did exhibit the expected activity.
In Vitro Splicing Reactions—Uniformly 32P-labeled adenovirus major late (AdML), TPI (exon 6-intron 6-exon 7), and β-globin (exon 1-intron 1-exon 2) pre-mRNA splicing substrates were generated by T7 run-off transcription of plasmids pHMS81, pMJM542, and pMJM540 and gel-purified as previously described (18). HeLa cell nuclear extract was prepared as previously described (19–21). Splicing reactions containing ∼20 nm pre-mRNA, 30% nuclear extract, 20 mm additional potassium chloride, 2.5 mm magnesium chloride, 10 mm creatine phosphate, 0.5 mm dithiothreitol, 0.4 units/μl RNasin, 40 mm Tris-HCl (pH 8), 0.5 mm ATP, and 1% (v/v) DMSO or isoginkgetin dissolved in DMSO were incubated for the indicated times (0–90 min) at 30° C. Native splicing complexes were separated on 4% nondenaturing gels as described previously (21, 22). Splicing efficiencies were assessed by separating purified RNAs on 15% denaturing polyacrylamide gels. To estimate the IC50 of isoginkgetin, nonlinear regression analysis was performed using GraphPad Prism version 5.0a for Macintosh.
RESULTS
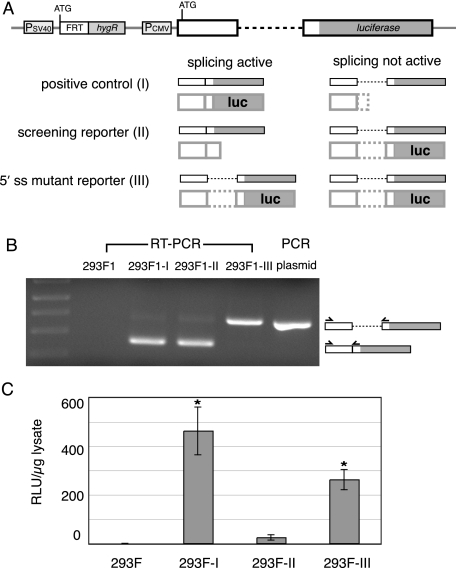
Development of Cell Lines for in Vivo Screening—To generate constructs appropriate for in vivo screening, we created a splicing reporter consisting of the human TPI gene exon 6-intron 6-exon 7 cassette (16) upstream of the firefly luciferase open reading frame (Fig. 1A, positive control; construct I). In this construct, the start codon is in the first exon, and active luciferase is generated only from spliced mRNA. In order to generate the screening reporter (construct II), we removed all in frame stop codons from the intron and added one base at the beginning of the second exon, such that the luciferase open reading frame is out of frame with the start codon in the spliced mRNA but is in frame when the intron is retained. We also created a mutant reporter (construct III) identical to the screening reporter except for a 5′ splice site point mutation predicted to abolish splicing (U to A at position 2 in the intron). All three reporters were subcloned downstream of the cytomegalovirus promoter in plasmid pcDNA5/FRT. This plasmid also contains an FRT site.
FIGURE 1.
A, schematic representations of reporter constructs (post genomic integration) and the transcripts and proteins generated from each construct with and without splicing. Heavy solid and dotted lines represent exon and intron sequences, respectively. B, RT-PCR analysis of total RNA isolated from the indicated cell lines plus PCR of plasmid I pcDNA5/FRT (size control for unspliced transcripts). C, luciferase activity/μg of protein for the same cell lines as in B (error bars, ±S.D., n ≥ 3 for each cell line; *, significant difference, p < 0.01; Student's t test (two-tailed with unequal variance)).
Stable cell lines expressing each reporter were generated using the Flp/FRT recombinase system. To do so, we clonally selected HEK293 cell lines stably transfected with a plasmid containing a FRT site at a single position in a transcriptionally active portion of the genome, as confirmed by LacZ expression (see “Experimental Procedures”). Independent co-transfection of two of these cell lines (293F1 and 293F2) with the reporter constructs and a plasmid encoding Flp recombinase generated the screening cell lines (293F1-I, -II, and -III and 293F2-I, -II, and -III). RT-PCR analysis of total RNA from each cell line confirmed that transcripts from constructs I and II were spliced, whereas those from construct III were not (see Fig. 1B for the 293F1-based lines; 293F2-based lines, data not shown). Consistent with this, luciferase activity levels were high in the positive control cells (I), low in the splicing reporter cells (II), and intermediate in the mutant reporter cells (III) (Fig. 1C). Since the three reporters behaved identically in both the 293F1 and 293F2 backgrounds, the two cell line sets were used interchangeably.
High Throughput Screening of Compound Libraries—Paired cell lines containing either reporter II or III were used to screen for small molecules that specifically inhibited splicing. We expected that such inhibitors would increase the luciferase activity from reporter II but would have little or no effect on reporter III. Comparison of compound effects on reporters II and III allowed us to discriminate between effects on splicing and other potential means of increased protein expression (e.g. increased cell proliferation, transcription, translation, etc.). The 6-fold difference between the luciferase activities of reporters II and III under the screening conditions (0.3% DMSO) provided a sufficient window for compound identification.
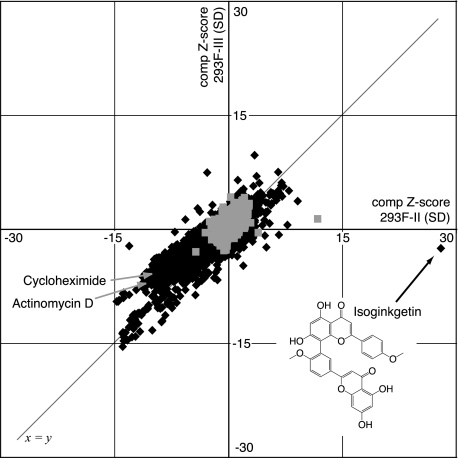
For high throughput screening, cells were grown in 384-well plates, exposed in duplicate to compounds dissolved in DMSO or an equivalent volume of DMSO alone for 24 h, after which luciferase activities were measured. Each luciferase value was expressed as the number of S.D. values from the mean (the Z-score) of control DMSO-treated cells (17). A composite Z-score was derived by combining the vectors of each replicate's Z-score for every given compound/cell line pair. This score allows easy comparison of effects upon two separate biological systems that have differently distributed spreads of average values (in this case, the average luciferase activity of cells expressing construct II versus those expressing construct III) (17). For each compound deemed to have a reproducible effect (i.e. its duplicates matched in direction and degree), the composite Z-score for reporter II was plotted against the composite Z-score for reporter III (Fig. 2).
FIGURE 2.
Plot of composite Z-scores for 293F-II versus 293F-III cells from high throughput screen. Light gray squares (n = 3172) represent values from control DMSO-treated wells. Dark gray diamonds (n = 9729) represent values from compound-treated wells. Only results with |reproducibility| ≥ 0.7 are shown. The structure of isoginkgetin is also indicated.
Approximately 8000 compounds were screened from natural product and synthetic libraries, some at multiple concentrations (15–100 μm). Of these, about half were known bioactives from commercially available NINDS, SpecPlus, and Biomol collections, and the remainder were the products of multiple diversity-oriented synthesis endeavors. A complete list of compounds screened and the scores each yielded in our assay can be found on the World Wide Web via ChemBank. The vast majority of compounds fell either within the noise range as defined by DMSO treatment (|composite Z-score| ≤ 2.5) or on the line defined by x = y (i.e. affecting reporters II and III similarly). As expected, compounds such as the transcription inhibitor actinomycin D and the translation inhibitor cycloheximide decreased luciferase activity of both reporters II and III, thus yielding negative composite Z-scores for both.
A limited number of compounds yielded a positive composite Z-score ≥3 for one reporter while having a negligible effect on the other (Table 1). Among this set, one compound stood out strikingly as having a very large positive effect on luciferase activity in cells expressing construct II (composite Z-score = 27.9) but no significant effect in cells expressing construct III (composite Z-score = –2.5). This compound is the biflavonoid isoginkgetin, a natural product found in a variety of plant species, including Ginkgo biloba.
TABLE 1.
Compounds with reproducible differential effects on reporters II and III
| Compound name | ChemBank IDa | Composite Z-scorebII | Composite Z-score III | ReproducibilitycII | Reproducibility III |
|---|---|---|---|---|---|
| Isoginkgetin | 2060300 | 27.9 | -2.5 | 0.9996 | -0.9978 |
| Piceatannol | 648 | 9.1 | 3.7 | 0.9996 | 0.9966 |
| Forskolin | 424 | 7.7 | 0.6 | 0.9965 | 0.9395 |
| SB-203580 | 1907775 | 6.6 | -0.7 | 0.9963 | -0.9580 |
| PK04_097119 | 2141419 | 5.4 | 0.8 | 0.9963 | 0.9781 |
| Resveratrol 4′-methyl ether | 2060112 | 3.4 | 9.8 | 0.9606 | 0.9481 |
| Cosmosiin | 3055391 | 2.0 | 5.3 | 0.9225 | 0.9978 |
| Bisacodyl | 982 | 1.8 | 5.1 | 0.9694 | 0.9998 |
| PK04_100001 | 3052787 | 1.6 | 5.0 | 1.0000 | 0.9976 |
| PK04_102320 | 3414053 | 1.2 | 6.5 | 0.8535 | 0.9973 |
| Genistein | 3103903 | -2.8 | 7.5 | -0.9412 | 0.9367 |
ChemBank ID is the unique identifier for screening compounds within the ChemBank data base.
The composite Z-score is a normalized value representing the number of S.D. values away from the empirically determined and mathematically defined mean of mock-treated cells.
Reproducibility is defined as the cosine of the angle between the vector (ZScoreA, ZScoreB) and the imaginary line defined by ZScoreA = ZScoreB; it ranges from -1 to +1.
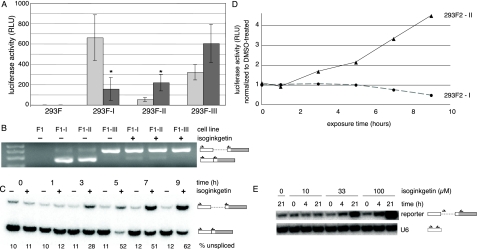
Validation and Characterization of Isoginkgetin as an in Vivo Splicing Inhibitor—To confirm the isoginkgetin results from the screen, cells containing reporters I, II, and III as well as the parental cell line without a reporter were treated with 33 μm isoginkgetin (the screening concentration) dissolved in DMSO or with DMSO alone. Luciferase activity of reporter II increased significantly (∼4-fold) with isoginkgetin treatment, whereas that of reporter III increased only modestly (∼1.8-fold) (Fig. 3A). As observed in the screen, the effect on reporter III was not statistically significant (p ≤ 0.05). Consistent with an inhibition of splicing, a marked decrease (∼5-fold) in luciferase activity was observed upon isoginkgetin treatment of cells expressing reporter I.
FIGURE 3.
Isoginkgetin inhibits splicing of reporter transcripts in vivo. A, luciferase activity of indicated cells treated with DMSO (light bars) or 33 μm isoginkgetin (dark bars) overnight (error bars, ±S.D., n ≥ 3; *, significant difference, p < 0.05; Student's t test (two-tailed with unequal variance)). B, RT-PCR of total RNA isolated from the indicated cell lines treated with DMSO or 33 μm isoginkgetin overnight. Sizes of unspliced and spliced products are indicated. C, semiquantitative RT-PCR of total RNA isolated from 293F-II cells treated with DMSO or 33 μm isoginkgetin for the length of time indicated. D, luciferase activity versus time for 293F-I (circles, dashed line) and 293F-II (triangles, solid line) cells treated with isoginkgetin relative to the luciferase activity of the same cells treated with DMSO alone for the same period of time. E, RT-PCR of total RNA isolated from 293F-II cells treated with DMSO and 0, 10, 33, or 100μm isoginkgetin for 0, 4, or 21 h. Multiplexed RT-PCR of U6 snRNA served as a nonspliced control. RLU, relative luciferase units.
To verify that the observed changes in luciferase activity were due to inhibition of splicing, we performed RT-PCR analysis of total RNA extracted from isoginkgetin- and DMSO-treated cells. This analysis confirmed that overnight exposure to 33 μm isoginkgetin was sufficient to shift the predominant species generated from constructs I and II from spliced mRNA to unspliced pre-mRNA (Fig. 3B). Semiquantitative RT-PCR revealed that this shift toward accumulation of unspliced pre-mRNA was detectable within 3 h of isoginkgetin addition (Fig. 3C). Consistent with this, luciferase activity from reporter I began to decrease, and that from reporter II began to increase within a similar time frame (Fig. 3D). The effect of isoginkgetin treatment was also concentration-dependent, since treatment with 100 μm isoginkgetin led to a greater increase in pre-mRNA accumulation within 4 h than did treatment with either 10 or 33 μm isoginkgetin (Fig. 3E).
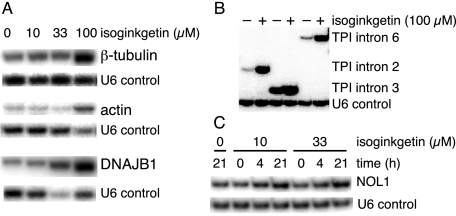
To test whether the effect of isoginkgetin on splicing was specific to our reporter system or more general, we analyzed several endogenous transcripts, including those encoding β-tubulin, actin, DNAJB1, TPI, glyceraldehyde-3-phosphate dehydrogenase, and RIOK3. All of these transcripts exhibited pre-mRNA accumulation (2–20-fold) upon treatment with isoginkgetin (Fig. 4, A and B, and data not shown). We also monitored NOL1 intron 7, a known substrate for the minor spliceosome (23). As observed for other endogenous transcripts, isoginkgetin treatment led to an increase in the amount of unspliced NOL1 intron 7 (Fig. 4C). Taken together, these results indicate that isoginkgetin is a general inhibitor of splicing that targets both the major and minor spliceosomes.
FIGURE 4.
Isoginkgetin treatment leads to the accumulation of endogenous pre-mRNAs. To determine pre-mRNA accumulation, intron inclusion was assessed using primers that crossed exon-intron boundaries (supplemental Table 1); multiplexed RT-PCR of U6 snRNA served as a nonspliced control. A, RT-PCR of total RNA isolated from 293F-II cells treated with DMSO and 0, 10, 33, or 100 μm isoginkgetin for 21 h to monitor single introns within the β-tubulin, actin, and DNAJB1 transcripts. B, RT-PCR of total RNA isolated from 293F-II cells treated with DMSO or 100 μm isoginkgetin for 21 h to monitor multiple introns within the endogenous TPI gene. C, RT-PCR of total RNA isolated from 293F-II cells treated with DMSO and 0, 10 or 33 μm isoginkgetin for 0, 4, or 21 h to monitor an intron that is a substrate for the minor spliceosome.
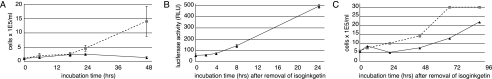
As expected for a general inhibitor of an essential step in gene expression, treatment of cells with isoginkgetin at concentrations inhibitory for splicing resulted in growth arrest (Fig. 5A). Trypan blue and Alamar blue analyses suggested that this growth arrest was not accompanied by a loss of cell viability (data not shown). Consistent with this, removal of isoginkgetin after 24 h restored both expression of luciferase from reporter I and cellular proliferation (Fig. 5, B and C). Thus, isoginkgetin appears to be a reversible inhibitor of pre-mRNA splicing.
FIGURE 5.
Isoginkgetin inhibits cell proliferation in a reversible fashion. A, cell growth upon treatment with DMSO (squares, dashed line) or 33 μm isoginkgetin (triangles, solid line). B, luciferase activity of 293F-I cells exposed to fresh medium after 24 h treatment with 33 μm isoginkgetin. C, cell growth in fresh medium after an 18-h exposure to DMSO (squares, dashed line) or 33 μm isoginkgetin (triangles, solid line).
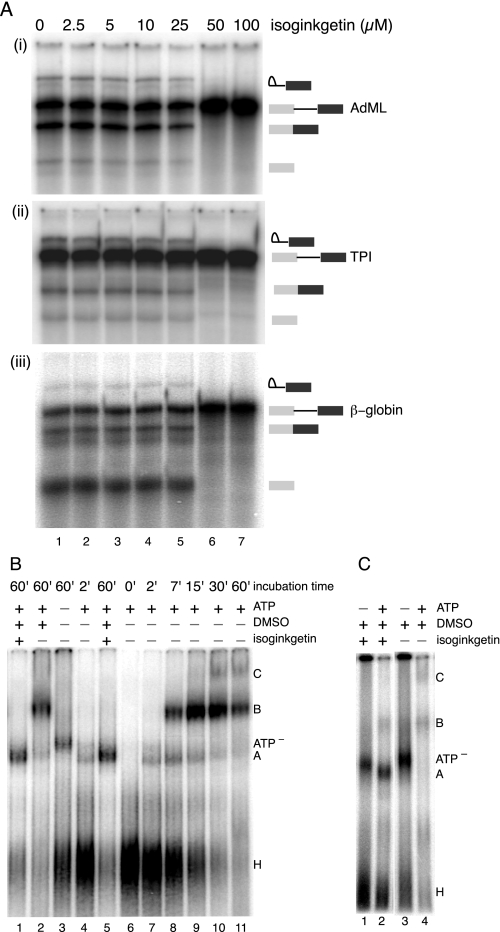
Isoginkgetin Inhibits Splicing in Vitro—To determine whether isoginkgetin could inhibit splicing in vitro, we performed splicing assays in HeLa cell nuclear extract using multiple RNA substrates. For these assays, isoginkgetin was diluted in DMSO, which itself did not affect splicing efficiency at concentrations of ≤2% (data not shown). Radiolabeled pre-mRNAs were spliced for 60 min in standard reactions supplemented with 0–100 μm isoginkgetin (≤1% v/v). Splicing of AdML, β-globin, and TPI (exon 6-intron 6-exon 7, equivalent to the in vivo splicing reporter) transcripts was inhibited by isoginkgetin in a dose-dependent manner. For all three constructs, splicing was completely inhibited by 50 μm isoginkgetin, with an IC50 of ∼30 μm (Fig. 6A). This in vitro splicing inhibition was reproducible with several different HeLa nuclear extract preparations, and preincubation of the extracts with the compound was not required for inhibition (data not shown). Thus, as observed in vivo, isoginkgetin is a general inhibitor of splicing in vitro.
FIGURE 6.
Isoginkgetin inhibits splicing of multiple substrates in vitro and stalls spliceosome assembly at A complex. A, denaturing gels showing concentration dependence of isoginkgetin-mediated inhibition of in vitro splicing of the indicated pre-mRNA substrates AdML (i), TPI (ii), and β-globin (iii). B, native gel of splicing reactions with or without 70 μm isoginkgetin, DMSO, and ATP. Lanes 6–11 show a time course of splicing complex formation; positions of complexes are indicated to the right. C, native gel of splicing reactions (60 min) with or without 70 μm isoginkgetin and ATP.
Isoginkgetin Arrests Spliceosome Assembly and Sequesters Pre-mRNA in Complex A—To ascertain the stage at which splicing is inhibited, splicing reactions containing AdML pre-mRNA ±70 μm isoginkgetin were subjected to native gel electrophoresis. In the absence of isoginkgetin, progression through H/E, A, B, and C complexes was readily detectable over a 60-min time course (Fig. 6B, lanes 6–11). Reactions lacking ATP instead accumulated a complex, probably E, having a slightly lower mobility than A complex (Fig. 6B, lane 3, ATP– complex) (22, 24). This same complex was observable in reactions containing isoginkgetin but lacking ATP (Fig. 6C). In reactions containing both isoginkgetin and ATP, little or no complex comigrating with B or C could be observed. Rather, these reactions accumulated a little or no complex with mobility identical to that of A complex (Fig. 6B, lanes 1 and 5) (data not shown). Thus, it appears that isoginkgetin blocks the A to B transition, resulting in sequestration of the pre-mRNA in a prespliceosome.
DISCUSSION
In comparison with other steps in gene expression, there is currently a paucity of small molecule modulators of pre-mRNA processing. Such modulators, particularly if their action is reversible, can be incredibly useful tools for both cell biological and biochemical analyses. Here we describe the development of a set of stable mammalian cell lines that we successfully employed in a high throughput screen to identify a new general inhibitor of pre-mRNA splicing. Our design principle was that inhibition of splicing should lead to a readily detectable positive readout, in our case an increase in luciferase activity. This design is similar to one previously employed in budding yeast, where a rise in copper resistance due to expression of Cup1p signaled increased use of one 5′ splice site over another (25). This CUP1 system has proven highly adaptable for studying other aspects of pre-mRNA processing as well (26, 27).
In addition to ours, a number of other reporter systems for monitoring either general or alternative splicing in mammalian cells via a protein activity readout have been described (9, 12, 28, 29). For example, Nasim et al. (12) characterized a dual reporter system in which a transiently transfected plasmid encodes β-galactosidase in the first exon and firefly luciferase in the second. The two exons are separated by an intron containing multiple in-frame stop codons. Thus, whereas β-galactosidase is produced from both pre-mRNA and mRNA, luciferase is only expressed from the latter. In this system, specific inhibition of splicing is signaled by a decrease in luciferase activity without a concomitant decrease in β-galactosidase activity. An alternate dual reporter scheme was developed by Lynch and co-workers (28) to monitor exon exclusion of CD45 exon 4. In their system, skipping-dependent expression of the transcriptional activator Gal4-VP16 drives expression of green fluorescent protein. Augmentation of the initial readout (i.e. an increase in Gal4-VP16 expression) by transcriptional synergy enabled amplification of a 3–5 fold change in splicing pattern into a 30–50 fold change in green fluorescent protein expression. This system was used successfully to identify two small molecules that promote exon 4 exclusion (28). More recently Stoilov et al. employed an alternative splicing reporter expressing either green fluorescent protein or red fluorescent protein in an alternative exon-dependent manner in a small scale screen (9). Of particular note was the 140-fold dynamic range of this cell-based assay obtained by monitoring the ratio of two alternative splicing reporters. From their screen, Stoilov et al. (9) identified a number of well known cardiotonic steroids, such as digoxin, as altering the splicing pattern of MAPT exon 10, their initial reporter construct. Although some of these same compounds were tested in our screen, they did not have any significant effects in our system. This is consistent with the idea that they are specific, not general, splicing effectors.
In contrast to the aforementioned systems, our cell-based assay was specifically designed to identify a general inhibitor of pre-mRNA splicing. It is based entirely on luciferase as a readout and employs three nearly identical reporters. By using the FLP/FRT system to generate cell lines stably transfected at identical genetic loci, we eliminated possible effects at the level of transcription due to different genomic contexts. By comparing the expression of two reporters that differed by only one nucleotide at the RNA level but were identical at the protein level, we eliminated possible differential effects of compounds on RNA or protein stability, RNA translatability, or protein activity. Here we used reporter cell lines II and III to identify a pre-mRNA splicing inhibitor. However, other combinations of these cell lines could be used to screen for compounds affecting a different step in mRNA biogenesis. For example, a compound promoting release of unspliced RNA from the nucleus without directly affecting splicing might be predicted to increase the luciferase signal from construct III while only minimally affecting those from constructs I and II. We note that several compounds having this particular set of differential effects were identified in our screen (Table 1, rows 6–11) and may be worthy of follow up.
Despite screening a relatively small number of compounds (<8000), we were able to identify a bona fide splicing inhibitor, the natural product isoginkgetin, which was by far our most positive hit. Isoginkgetin has previously been described as one of several biflavonoids isolated from G. biloba, many of which have been characterized as having antiproliferative, antioxidative, anti-inflammatory, and/or neuroprotective activities (30–32) (reviewed in Ref. 33). Perhaps the best characterized role of isoginkgetin is as an inhibitor of tumor cell invasion. At isoginkgetin concentrations similar to those we found inhibitory of splicing both in vivo and in vitro, Yoon et al. (15) showed that tumor cell invasion was inhibited. Supporting this, they documented changes in the levels of mRNAs encoding proteins involved in tumor cell invasion. These gene expression changes were suggested to result from inhibition of the kinase-dependent phosphatidylinositol 3-kinase/ATK/NF-κB signaling pathway, although the exact target of isoginkgetin was not identified in that study.
Recently, two other classes of cell-permeable compounds have emerged as general pre-mRNA splicing inhibitors: pladienolide-B and spliceostatin A, a derivative of FR901464 (13, 14, 34). Although these natural products were isolated from different organisms and have different core structures, they both target the SF3b complex. SF3b, which consists of the three proteins SAP130, SAP145, and SAP155, is a component of U2 and U12 snRNPs important for early spliceosome assembly and branch point adenosine definition. Extracts lacking SF3b fail to assemble A complex (35). Also known as the prespliceosome, A complex represents the first ATP-dependent stage of spliceosome assembly, and contains U2 snRNP stably associated with the branch site. Consistent with SF3b being the target of spliceostatin A, we recently showed that meayamycin, another FR901464 analog, which is 2 orders of magnitude more potent than FR901464 (36), completely blocks A complex formation in vitro.3 In contrast, isoginkgetin appears to act by a different mechanism, since its presence promotes the accumulation of A complex in in vitro splicing reactions. Although we have not yet identified the target of isoginkgetin, likely candidates would be components of both the major and minor spliceosomes required for the A to B complex transition. An example of such a component is the SRPK2 kinase, which phosphorylates PRP28, allowing stable association with the U4/U5/U6 tri-snRNP and progression to B complex (37). Regardless of its target, isoginkgetin should prove a useful reagent for the accumulation of prespliceosomes for mechanistic and structural studies.
Like isoginkgetin, pladienolide-B and FR901464 (and its derivatives) have been reported to have antiproliferative activity. Indeed, it was their promise as anti-tumor agents that prompted the studies leading to the identification of SF3b as the cellular target of both. That isoginkgetin, pladienolide-B, and spliceostatin A all have both anti-tumor and anti-splicing activity but inhibit splicing by two different mechanisms (inhibition of A complex formation versus the A to B complex transition) suggests that their shared anti-tumor activity is a consequence of failure to express some limiting component needed for tumor cell growth due to general splicing inhibition. In Schizosaccharomyces pombe, for example, mutation of the general splicing factor CDC5 leads to a cell cycle defect due to decreased splicing efficiency of α-tubulin pre-mRNA. The simple requirement for large quantities of α-tubulin at the G2/M transition leaves the cell vulnerable to anything limiting α-tubulin production (38). A similar mechanism may be at work with small molecule inhibitors of splicing. The antiproliferative effect may be the result of an increased sensitivity to decreases in certain limiting proteins in proliferating cells. Thus, small molecule inhibitors of general pre-mRNA splicing potentially represent an exciting new avenue for the development of novel anti-inflammatory and/or anti-cancer agents.
Supplementary Material
Acknowledgments
We thank Jing Yan for technical assistance and Amrit Singh and Danny Crawford for critical reading of the manuscript. We thank Kazunori Koide (University of Pittsburgh) for helpful discussions and sharing results prior to publication.
Author's Choice—Final version full access.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM35007 (to M. J. M.) and the National Institutes of Health, NCI, Initiative for Chemical Genetics, under Contract N01-CO-12400. This work was performed with the assistance of the Chemical Biology Platform of the Broad Institute of Harvard and MIT. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
Footnotes
The abbreviations used are: snRNA, small nuclear RNA; snRNP, small nuclear ribonucleoprotein; FRT, flp recombinase target; RT, reverse transcription; AdML, adenovirus major late; TPI, triose-phosphate isomerase.
M. J. Moore, K. O'Brien, and K. Koide, unpublished data.
References
- 1.Matlin, A. J., and Moore, M. J. (2007) Adv. Exp. Med. Biol. 623 14–35 [DOI] [PubMed] [Google Scholar]
- 2.Brody, E., and Abelson, J. (1985) Science 228 963–967 [DOI] [PubMed] [Google Scholar]
- 3.Grabowski, P. J., Seiler, S. R., and Sharp, P. A. (1985) Cell 42 345–353 [DOI] [PubMed] [Google Scholar]
- 4.Jurica, M. S. (2008) Nat. Chem. Biol. 4 3–6 [DOI] [PubMed] [Google Scholar]
- 5.Steitz, T. A. (2008) Nat. Rev. Mol. Cell. Biol. 9 242–253 [DOI] [PubMed] [Google Scholar]
- 6.Bakkour, N., Lin, Y. L., Maire, S., Ayadi, L., Mahuteau-Betzer, F., Nguyen, C. H., Mettling, C., Portales, P., Grierson, D., Chabot, B., Jeanteur, P., Branlant, C., Corbeau, P., and Tazi, J. (2007) PLoS Pathog. 3 1530–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nottrott, S., Hartmuth, K., Fabrizio, P., Urlaub, H., Vidovic, I., Ficner, R., and Luhrmann, R. (1999) EMBO J. 18 6119–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soret, J., Bakkour, N., Maire, S., Durand, S., Zekri, L., Gabut, M., Fic, W., Divita, G., Rivalle, C., Dauzonne, D., Nguyen, C. H., Jeanteur, P., and Tazi, J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8764–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoilov, P., Lin, C. H., Damoiseaux, R., Nikolic, J., and Black, D. L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumanasekera, C., Watt, D. S., and Stamm, S. (2008) Biochem. Soc. Trans. 36 483–490 [DOI] [PubMed] [Google Scholar]
- 11.Legrain, P., and Rosbash, M. (1989) Cell 57 573–583 [DOI] [PubMed] [Google Scholar]
- 12.Nasim, M. T., Chowdhury, H. M., and Eperon, I. C. (2002) Nucleic Acids Res. 30 e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaida, D., Motoyoshi, H., Tashiro, E., Nojima, T., Hagiwara, M., Ishigami, K., Watanabe, H., Kitahara, T., Yoshida, T., Nakajima, H., Tani, T., Horinouchi, S., and Yoshida, M. (2007) Nat. Chem. Biol. 3 576–583 [DOI] [PubMed] [Google Scholar]
- 14.Kotake, Y., Sagane, K., Owa, T., Mimori-Kiyosue, Y., Shimizu, H., Uesugi, M., Ishihama, Y., Iwata, M., and Mizui, Y. (2007) Nat. Chem. Biol. 3 570–575 [DOI] [PubMed] [Google Scholar]
- 15.Yoon, S. O., Shin, S., Lee, H. J., Chun, H. K., and Chung, A. S. (2006) Mol. Cancer Ther. 5 2666–2675 [DOI] [PubMed] [Google Scholar]
- 16.Nott, A., Meislin, S. H., and Moore, M. J. (2003) RNA 9 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seiler, K. P., George, G. A., Happ, M. P., Bodycombe, N. E., Carrinski, H. A., Norton, S., Brudz, S., Sullivan, J. P., Muhlich, J., Serrano, M., Ferraiolo, P., Tolliday, N. J., Schreiber, S. L., and Clemons, P. A. (2008) Nucleic Acids Res. 36 D351–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Hir, H., Izaurralde, E., Maquat, L. E., and Moore, M. J. (2000) EMBO J. 19 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abmayr, S. M., Workman, J. L., and Roeder, R. G. (1988) Genes Dev. 2 542–553 [DOI] [PubMed] [Google Scholar]
- 20.Dignam, J. D., Lebovitz, R. M., and Roeder, R. G. (1983) Nucleic Acids Res. 11 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reichert, V., and Moore, M. J. (2000) Nucleic Acids Res. 28 416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konarska, M. M., and Sharp, P. A. (1987) Cell 49 763–774 [DOI] [PubMed] [Google Scholar]
- 23.Tarn, W.-Y., and Steitz, J. A. (1996) Cell 84 801–811 [DOI] [PubMed] [Google Scholar]
- 24.Das, R., and Reed, R. (1999) RNA 5 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesser, C. F., and Guthrie, C. (1993) Genetics 133 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Query, C. C., and Konarska, M. M. (2004) Mol. Cell. 14 343–354 [DOI] [PubMed] [Google Scholar]
- 27.Stutz, F., and Rosbash, M. (1994) EMBO J. 13 4096–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levinson, N., Hinman, R., Patil, A., Stephenson, C. R., Werner, S., Woo, G. H., Xiao, J., Wipf, P., and Lynch, K. W. (2006) RNA 12 925–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts, G. C., Gooding, C., and Smith, C. W. (1996) EMBO J. 15 6301–6310 [PMC free article] [PubMed] [Google Scholar]
- 30.Kang, S. S., Lee, J. Y., Choi, Y. K., Song, S. S., Kim, J. S., Jeon, S. J., Han, Y. N., Son, K. H., and Han, B. H. (2005) Bioorg. Med. Chem. Lett. 15 3588–3591 [DOI] [PubMed] [Google Scholar]
- 31.Kim, S. J., Lim, M. H., Chun, I. K., and Won, Y. H. (1997) Skin Pharmacol. 10 200–205 [DOI] [PubMed] [Google Scholar]
- 32.Lee, S. J., Choi, J. H., Son, K. H., Chang, H. W., Kang, S. S., and Kim, H. P. (1995) Life Sci. 57 551–558 [DOI] [PubMed] [Google Scholar]
- 33.Kim, H. P., Park, H., Son, K. H., Chang, H. W., and Kang, S. S. (2008) Arch. Pharm. Res. 31 265–273 [DOI] [PubMed] [Google Scholar]
- 34.Nakajima, H., Hori, Y., Terano, H., Okuhara, M., Manda, T., Matsumoto, S., and Shimomura, K. (1996) J. Antibiot. (Tokyo) 49 1204–1211 [DOI] [PubMed] [Google Scholar]
- 35.Wang, Q., He, J., Lynn, B., and Rymond, B. C. (2005) Mol. Cell. Biol. 25 10745–10754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albert, B. J., Sivaramakrishnan, A., Naka, T., Czaicki, N. L., and Koide, K. (2007) J. Am. Chem. Soc. 129 2648–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathew, R., Hartmuth, K., Mohlmann, S., Urlaub, H., Ficner, R., and Luhrmann, R. (2008) Nat. Struct. Mol. Biol. 15 435–443 [DOI] [PubMed] [Google Scholar]
- 38.Burns, C. G., Ohi, R., Mehta, S., O'Toole, E. T., Winey, M., Clark, T. A., Sugnet, C. W., Ares, M., Jr., and Gould, K. L. (2002) Mol. Cell. Biol. 22 801–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.