Abstract
The B chain of ricin was expressed and delivered to the endoplasmic reticulum of tobacco protoplasts where it disappeared with time in a manner consistent with degradation. This turnover did not occur in the vacuoles or upon secretion. Indeed, several lines of evidence indicate that, in contrast to the turnover of endoplasmic reticulum-targeted ricin A chain in the cytosol, the bulk of expressed ricin B chain was degraded in the secretory pathway.
Ricin is a heterodimeric plant protein consisting of a catalytic ribosome-inactivating polypeptide (the A chain, or RTA)5 disulfide-bonded to a galactose-specific lectin (the B chain, or RTB) (1). In this form, it is able to enter mammalian cells to reach the endoplasmic reticulum (ER) where, following toxin reduction, the RTA subunit is exported to the cytosol in a process that probably exploits some or all phases of the quality control pathway known as ER-associated protein degradation (ERAD) (2, 3). Although a significant proportion of RTA is eventually degraded by proteasomes, a fraction appears to uncouple from this pathway to refold and inactivate substrate ribosomes (4). This inactivation results from a specific depurination of 28 S rRNA at a site essential for the binding of elongation factors during protein synthesis (5). In mammalian cells, the fate of endocytosed RTB is not known. During the biosynthesis of ricin in the producing castor oil plant, the protein initially folds within the ER lumen. However, retro-translocation of RTA is avoided by the translation and ER segregation of an A-B precursor (proricin) that is incompetent for such a step (6). Instead, the ER-sequestered precursor is transported to vacuoles by virtue of a targeting signal that lies in a propeptide linking the two polypeptides. This internal sequence is removed by proteolysis only when proricin reaches the safe haven of storage vacuoles (7). In this way, sensitive plant ribosomes remain undamaged in the wake of large scale synthesis of a highly toxic protein.
We have shown previously that in plant cells, in contrast to the fate of proricin, ER-sequestered RTA (rather like RTA reduced from ricin in the mammalian ER) was susceptible to proteasomal degradation following its retro-translocation and deglycosylation in the cytosol (8). As in mammalian cells, however, a fraction of dislocated RTA was able to refold to inhibit protein synthesis. This was the first demonstration of an operational retro-translocation pathway in plant cells (9, 10), and it highlighted the danger to the plant cell of expressing damaged transcripts or prematurely processed proricin. In contrast, when RTA was co-expressed with RTB, where both nascent proteins contained an ER signal peptide, a disulfide-bonded holotoxin was generated and subsequently secreted from the cell (8). The presence, on one or other of the subunits, of the previously characterized vacuolar targeting sequence, directed this holotoxin to vacuoles in a route akin to that of the proricin precursor (7, 11). These findings clearly showed that co-expression of RTB with RTA allowed entry of both subunits into the secretory pathway and strongly mitigated the toxic effects observed with RTA alone. Indeed, the rescue effect of RTB suggested that this polypeptide lacked the propensity to retrotranslocate across the ER membrane. To test how ER quality control deals with an excess of RTB (made in the absence of its normal partner RTA), we have examined the fate of this solo subunit when made in tobacco cells. Surprisingly, we found that much of RTB, like ER-localized RTA, was degraded intracellularly. Unlike RTA, however, RTB did not emerge into the cytosol for degradation but disappeared within the early secretory pathway. The data presented suggest that the plant cell secretory pathway may contain an alternative proteolytic system for the disposal of misfolded/orphan proteins, as may also be the case in mammalian cells.
EXPERIMENTAL PROCEDURES
Plasmid Constructs—All DNA constructs were generated in the CaMV 35S promoter-driven expression vectors pDHA (for toxin subunits, phaseolin, and Sec12 (12)), pamPAT-MCS (GenBank™ accession number AY436765 (13) for CDC48-based constructs), or pGreenII-0029 (14). Expression constructs encoding phaseolin (pDHE-T343F), RTA, RTB, γ-heavy chain of Guy's 13 IgG, Sec12, and CDC48 have been described previously (8, 13, 15, 16). RTB targeted to the ER using the signal peptide of phaseolin (8) is referred to as RTB throughout. To prepare the construct encoding RTB with its native signal peptide and propeptide, the 35-amino acid ricin presequence (containing the ricin ER signal peptide (17) and N-terminal propeptide (18)) was fused to the 5′ end of the mature RTB coding sequence via overlapping mutagenic PCR, using the primers 5′-TCTAGAATGAAACCGGGAGG-3′ and 5′-ACAAACATCAGCGTTGTTATCCTC-3′ to amplify the ricin presequence, 5′-GAGGATAACAACGCTGATGTTTGTATG-3′ and 5′-CTGCAGTCAAAATAATGGTAACCA-3′ to amplify RTB, and the first and fourth of these primers to fuse the two overlapping segments together. Restriction enzyme sites are underlined. The fusion construct was then inserted into the XbaI-PstI sites of pDHA. To generate pre-RTB (with native ricin signal sequence), DNA encoding the 9-residue N-terminal propeptide (18) was deleted from prepro-RTB using the QuickChange™ in vitro mutagenesis system (Stratagene, La Jolla, CA) and the mutagenic oligonucleotides 5′-GGATCCACCTCAGGGGCTGATGTTTGTATGG-3′ and 5′-CCATACAAACATCAGCCCCTGAGGTGGATCC-3′. Construction of the RTB open reading frame preceded by an uncleavable saporin signal peptide was achieved by again using overlapping mutagenic PCR, using the primers 5′-CGTACGTATCTAGAATGAAGATATATGTTG-3′ and 5′-GATCCATACAAACATCAGCCACATCATTTGTTG-3′ to amplify the uncleavable saporin signal peptide 5′-CAACAAATGATGTGATATTCCCCAAACAATACC-3′ and the same fourth primer from above to amplify RTB, and the first and fourth of these primers to fuse the two overlapping segments together. Restriction enzyme sites are again underlined, and the fusion construct was again inserted into the XbaI-PstI sites of pDHA. Finally, RTB was cloned into the XbaI-SacI sites of the CaMV 35S cassette using the primers 5′-GCGCGCGTCTAGAGCTGATGTTTGTATG-3′ and 5′-TAATGATGGAGCTCTCAAAATAATGGTAACCA-3′ for subsequent insertion into EcoRV-cut pGreenII-0029, used for generation of transgenic tobacco plants. Restriction enzyme sites are underlined.
Transient Transfection of Tobacco Leaf Protoplasts and Production of Transgenic Tobacco Plants—Protoplasts were prepared from axenic leaves (4–7 cm long) of Nicotiana tabacum cv. Petit Havana SR1 (19), or from transgenic plants where indicated, and were subjected to polyethylene glycol-mediated transfection with one or more plasmids as described previously (20). Agrobacterium tumefaciens transformed with pGreenII-0029 encoding RTB under the control of the CaMV 35S promoter and terminator was used to produce transgenic plants as described previously (20).
Pulse-Chase Analysis—Cells were radiolabeled with Pro-Mix (a mixture of [35S]cysteine and [35S]methionine (GE Healthcare)) and chased for the times indicated in the figures, as described previously (8). In some experiments, before radioactive labeling, protoplasts were incubated for 1 h at 25 °C in K3 medium (3.78 g/liter Gamborg's B5 basal medium with minimal organics, 750 mg/liter CaCl2·2H2O, 250 mg/liter NH4NO3, 136.2 g/liter sucrose, 250 mg/liter xylose, 1 mg/liter 6-benzylaminopurine (6-BAP), 1 mg/liter α-naphthaleneacetic acid) supplemented with either 36 μm brefeldin A (Sigma; 7 mm stock in 100% ethanol), 1 μm bafilomycin A (Sigma; 50 μm stock in dimethyl sulfoxide), or 5 mm 1-deoxymannojirimycin (Sigma; 0.2 m stock in sterile H2O). When indicated, clasto-lactacystin β-lactone (Calbiochem; 20 mm stock in dimethyl sulfoxide) or epoxomycin (Calbiochem; 20 mm stock in dimethyl sulfoxide) was added to a concentration of 80 μm at the beginning of the labeling period. At the desired time points, 3 volumes of cold W5 medium (9 g/liter NaCl, 0.37 g/liter KCl, 18.37 g/liter CaCl2·2H2O, 0.9 g/liter glucose) were added, and protoplasts were pelleted by centrifugation at 60 × g for 10 min at 4 °C. Separated or combined cells and media samples were frozen on dry ice and stored at -80 °C, unless further manipulations were to be performed as below.
Protoplast Fractionation—Protoplast pellets (from 500,000 cells) were resuspended in 140 μl of 12% sucrose buffer (100 mm Tris-HCl, pH 7.6, 10 mm KCl, 1 mm EDTA, 12% (w/w) sucrose, supplemented immediately before use with Complete™ protease inhibitor mixture (Roche Applied Science)) and homogenized by pipetting 50 times with a Gilson-type micropipette through a 200-μl tip. Intact cells and debris were removed by centrifugation at 500 × g for 5 min at 4 °C. 130 μl was loaded onto a 17% (w/w) sucrose pad and centrifuged at 100,000 × g for 30 min at 4 °C. Pellets (microsomes) and supernatants (cytosolic proteins) were frozen on dry ice and stored at -80 °C.
Preparation of Protein Extracts and Immunoprecipitation— Frozen samples were homogenized by adding 2 volumes of cold protoplast homogenization buffer (150 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1.5 mm EDTA, 1.5% (w/v) Triton X-100, supplemented immediately before use with Complete™ protease inhibitor mixture). Homogenates were used for immunoprecipitation with polyclonal sheep anti-RTB, or polyclonal rabbit anti-RTA, anti-phaseolin (20), anti-BiP (20) or anti-mouse IgG antisera (Sigma). To reduce any nonspecific immunoselection, anti-RTB and anti-phaseolin antisera were preincubated on ice for 2 h with unlabeled protoplast homogenate, before adding the radiolabeled samples. Immunoselected polypeptides were analyzed by 15% SDS-PAGE. Gels were fixed, treated with Amplify™ (GE Healthcare), and radioactive polypeptides revealed by fluorography. Band intensity was determined using TotalLab 2003 software (Nonlinear Dynamics, Newcastle-upon-Tyne, UK).
Assays for Protein Aggregation and Extracellular Degradation—Where indicated, the incubation medium of protoplasts transfected with pDHA alone was spiked with commercially available RTB (Vector Laboratories, Burlingame, CA; 50-ng per time point). Medium homogenates were trichloroacetic acid-precipitated, resolved by 15% SDS-PAGE, and immunoblotted with anti-RTB antiserum. Alternatively, protoplasts expressing RTB were subjected to immunoprecipitation in the presence or absence of anti-RTB antiserum, and unbound, nonimmunoprecipitable proteins trichloroacetic acid-precipitated and immunoblotted as above.
ATP-release Assay—Protein A- or Protein G-Sepharose beads carrying immunoprecipitated protein were washed once in ATP release buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% (w/v) Triton X-100, 6 mm MgCl2), resuspended in 500 μl of ATP release buffer containing 8 mm ATP pH 7.5, and incubated at 4 °C for 1 h.
Lactose Precipitation of Functional RTB—Cell and medium homogenates were vortexed with a 10% (bead w/v) suspension of α-lactose immobilized onto agarose in NET buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.1% (w/v) Nonidet P-40). After tumbling, the samples were washed three times with 1 ml of PBS-N buffer (phosphate-buffered saline, 0.1% (w/v) Nonidet P-40).
Endoglycosidase H Treatment—Protein G-Sepharose beads carrying immunoprecipitated protein were resuspended in 20 μl of sodium citrate buffer (0.25 m sodium citrate pH 5.5, 0.2% (w/v) SDS) and boiled for 5 min. Supernatants were treated with 10 milliunits of endoglycosidase H (Roche Applied Science; 5 milliunits/μl stock) at 37 °C for 16 h.
Peptide:N-Glycanase F Treatment—Protein G-Sepharose beads carrying immunoprecipitated protein were washed twice in sterile H2O, denatured by adding 28 μl of DNB buffer (0.5% (w/v) SDS, 1% (w/v) β-mercaptoethanol), and boiled for 5 min. Supernatants were treated with 1000 units of peptide:N-glycanase (New England Biolabs, Ipswich, MA) in 50 mm sodium phosphate buffer, pH 7.5, 1% (w/v) Nonidet P-40, at 37 °C for 1 h.
RESULTS
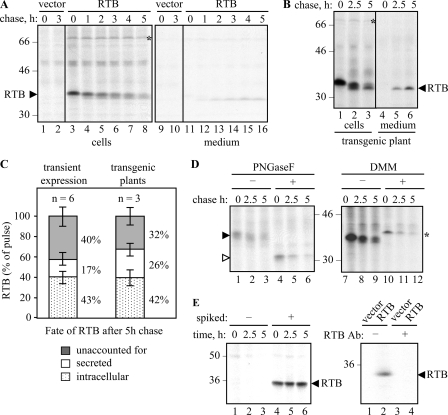
A Substantial Proportion of Ricin B Chain Is Degraded within Tobacco Protoplasts—Tobacco protoplasts transfected with RTB-expressing plasmids were pulse-labeled for 1 h, and the newly synthesized proteins were subsequently chased with non-radioactive amino acids. The RTB present at each time point was recovered by immunoprecipitation (IP). As shown in Fig. 1A, a significant amount of cellular RTB was lost during the chase. Sequential IP of the initial supernatants revealed quantitative recovery in the first IP (data not shown), suggesting that disappearance was not related to the efficiency of IP. Although a proportion of RTB could be accounted for by secretion, the bulk of newly made RTB disappeared, presumably a result of proteolysis or aggregation. Because, in some instances, the fate of a protein may differ when expressed transiently or permanently in tobacco (21), we also monitored the fate of RTB constitutively expressed in transgenic plants. It is clear that in tobacco plants adapted to the stable expression of RTB, its rate of disappearance was very similar to that observed following transient transfection of protoplasts (compare Fig. 1, A and B) Indeed, from Fig. 1C it can be seen that up to 40% of the newly made RTB was unaccounted for by the end of a 5-h chase upon both transient expression in transfected protoplasts and upon stable expression in transgenic plants.
FIGURE 1.
Ricin B chain disappears in transfected and transgenic tobacco protoplasts. A, protoplasts transfected with pDHA vector alone (vector) or plasmid encoding ER-targeted ricin B chain (RTB) were radiolabeled with 35S-amino-acids for 1 h and chased with unlabeled amino acids for the times indicated. RTB was immunoprecipitated from separated cell and medium homogenates and analyzed by SDS-PAGE and fluorography. B, protoplasts were treated as in A, but using leaf protoplasts prepared from transgenic tobacco plants constitutively expressing RTB. C, quantitation of the proportion of RTB remaining within cells, secreted into the medium, or unaccounted for after a 5-h chase in transiently expressing or transgenic protoplasts. Mean values are from six or three independent experiments, respectively. Error bars indicate standard deviation. D, protoplasts were transfected with plasmids encoding RTB and pulse-chased as in A. Where indicated, RTB immunoprecipitates were treated for 1 h in the absence or presence of PNGaseF, or protoplasts were preincubated for 1 h with 5 mm DMM before radiolabeling. E, left panel, immunoblot following incubation of medium from vector-transfected protoplasts with buffer (-) or with 150 ng of castor bean purified RTB (+). Right panel, protoplasts expressing vector or RTB were homogenized and incubated with (+) or without (-) anti-RTB antiserum and protein A-Sepharose before the unbound, nonimmunoreactive proteins were resolved by SDS-PAGE and immunoblotted for any nonimmunoprecipitable B chain. In all panels, numbers on the margins of gels indicate molecular mass markers in kilodaltons.
During the chase, it was noticeable that RTB was converted to slightly smaller, faster migrating bands in both transiently transfected protoplasts (Fig. 1A) and in transgenic plants (Fig. 1B). RTB is glycosylated in plant cells, as shown by the in vitro treatment of samples with peptide:N-glycanase F (PNGaseF) following IP (Fig. 1D). Such treatment converted this protein (Fig. 1D, black arrowhead) to a single, faster migrating form with the molecular size of nonglycosylated RTB (white arrowhead). Conversely, treating cells with 1-deoxymannojirimycin (DMM), to inhibit the action of ER mannosidase I (22), caused the appearance of a slightly larger, sharper RTB band (Fig. 1D, asterisk). By comparison, the smaller bands observed in the absence of DMM suggested that the mannose-rich core glycans of RTB were being rapidly processed, even during the pulse. Together, these analyses showed that the reduction in RTB size seen during the chase was a result of glycan modification. Importantly, the RTB species with unprocessed glycans disappeared at approximately the same rate as glycosylated RTB, where loss in each case is measured relative to the amount present at the start of the chase. Inhibition of ER mannosidase is known to prolong the retention of misfolded glycoproteins within the calnexin cycle, thereby reducing their delivery to the proteasome (23). However, the unaltered kinetics seen here ±DMM strongly suggests that the calnexin cycle is not normally involved in the loss of RTB (23, 24).
To rule out that the proportion of unaccounted lectin (Fig. 1C) was because of extracellular proteolysis, controls revealed that enzymes in the medium do not degrade RTB (Fig. 1E, left panel). Furthermore aggregation in vivo, to a form not recoverable by immunoprecipitation, was also excluded by immunoblotting denatured protein samples with or without prior removal of RTB by IP (Fig. 1E, right panel).
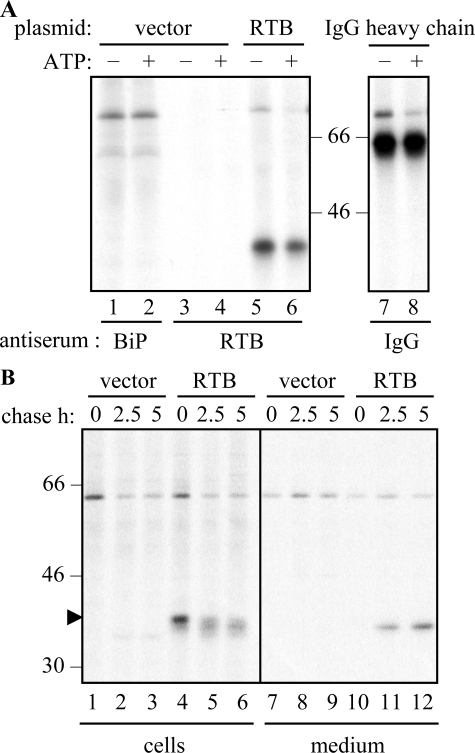
Ricin B Chain Interacts with BiP in the ER Lumen—In Fig. 1, A and B, it was noticed that in cells expressing RTB, a band of ∼75 kDa was co-precipitated (shown by the asterisks). Because a protein could be selected from control cells using anti-BiP antibodies (Fig. 2A, lanes 1 and 2) that was identical in size to that co-precipitated with anti-RTB antiserum (Fig. 2A, lane 5), this protein was tentatively identified as the ER chaperone BiP (the immunoglobulin heavy chain binding protein). The hallmark of a genuine BiP interaction is sensitivity to ATP, and this is indicated by the release of BiP from ATP-washed IPs (Fig. 2A, lane 6). Controls of the characterized chaperone-ligand interaction between BiP and immunoglobulin heavy chains in tobacco protoplasts (25) are shown for comparison (Fig. 2A, lanes 7 and 8).
FIGURE 2.
Fractions of ricin B chain can interact with BiP and lactose. A, protoplasts transfected with vector alone (vector), the RTB plasmid, or the γ-heavy chain of Guy's 13 immunoglobulin G plasmid (25) were radiolabeled with 35S-amino-acids for 1 h before homogenization and IP with anti-RTB, anti-BiP, or anti-IgG antisera. IPs were washed in the absence (-) or presence (+) of ATP and analyzed by SDS-PAGE and fluorography. B, transfected cells were subject to pulse-chase before separated cell and medium homogenates were incubated with α-lactose immobilized onto agarose to select RTB with native conformation. Lactose-agarose pulldowns were analyzed by SDS-PAGE and fluorography. Numbers on the left indicate molecular mass markers in kilodaltons.
BiP acts by binding newly made proteins to stabilize intermediates during protein folding and to prevent the formation of aggregates. This interaction, which can be transient or prolonged (26), suggests that RTB may require assistance to reach its biologically active conformation. An excellent assessment of the conformation of the RTB lectin is its ability to interact with galactose (27). As shown in Fig. 2B (lanes 4–6 and 11 and 12), a fraction of the expressed RTB could bind to immobilized lactose. Pulse-chase analysis confirmed that although the correctly folded RTB disappeared with time from the intracellular fractions, there was a concomitant appearance of the fully processed form in the medium (Fig. 2B). Interestingly, no BiP was detectable in association with the intracellular RTB population able to bind to lactose (note: the band at ∼60 kDa represents an unknown lactose-binding protein that is present in all lanes, including the controls). This finding supports the assertion that the fraction of RTB normally interacting with BiP (Fig. 2A) was in the process of folding but had not yet assumed its native, sugar-binding conformation. The secreted RTB is unlikely to be re-internalized (or to interact with cell surfaces) because plant cells lack β1,4-galactosyltransferase (28). Consequently, the characteristic galactose-containing complex N-glycans on glycoproteins that act as RTB receptors at the surface of mammalian cells are missing in plants.
Vacuolar Degradation Is Not Responsible for the Loss of Ricin B Chain—The vacuole is a major site for the degradation of proteins in the secretory pathway of yeast (29). Indeed, in plant cells, the central lytic vacuole is a major repository of proteolytic enzymes (30) and is responsible for protein degradation during seed germination and autophagic processes (31). ER residents themselves may be constitutively transported to vacuoles, particularly when interactions with ligands are prolonged (32, 33). We therefore investigated whether vacuolar degradation was responsible for the loss of RTB.
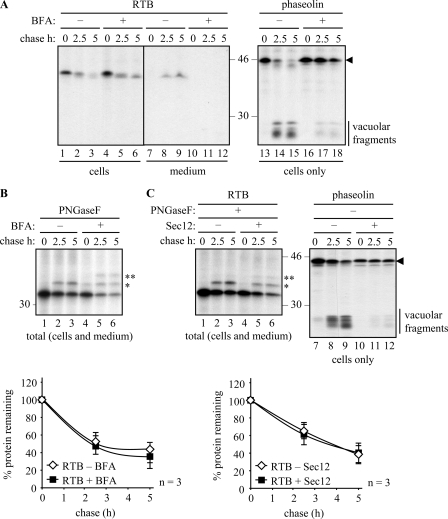
The classical pathway to vacuoles involves transport from the ER via the Golgi complex, a process that can be disrupted in the presence of brefeldin A (BFA) (8), a reagent that has the same molecular target in plant cells as in other eukaryotes (34). We can reveal this pathway by biochemically visualizing changes that occur to the newly synthesized vacuolar protein phaseolin. This protein is known to be proteolytically processed into a characteristic set of low molecular weight fragments (20–25 kDa) upon arrival in the vacuoles of tobacco leaf cells (Fig. 3A) (21). The trafficking (and therefore processing) of phaseolin is blocked when cells are treated with BFA (Fig. 3A, compare lanes 14 and 15 with lanes 17 and 18), confirming previous observations (35) that effective BFA treatment efficiently prevents the transport of this protein to the vacuole. BFA did not, however, have any stabilizing effect on RTB (Fig. 3A, lanes 1–6), although it did effectively block its secretion (Fig. 3A, lanes 11 and 12). The appearance of a slightly faster form of RTB in the presence of BFA by the end of the pulse (Fig. 3A, lane 4), and its processing during the chase (lanes 5 and 6) to a size that is comparable with the secreted RTB fraction in the absence of BFA (lanes 8 and 9), most likely arises from the exposure of ER-localized RTB to the full range of Golgi oligosaccharide-modifying enzymes that occurs when the Golgi stack collapses into the ER (20).
FIGURE 3.
Ricin B chain is not stabilized when secretion is blocked. A, protoplasts were transfected with plasmids encoding ER-targeted RTB or phaseolin and subjected to pulse-chase as described under “Experimental Procedures.” Where indicated, protoplasts were preincubated for 1 h with 36 μm BFA before radiolabeling. IPs were analyzed by SDS-PAGE and fluorography. The black arrowhead shows the position of full-length phaseolin, and the vertical bar indicates the vacuolar-generated fragments of phaseolin. B, protoplasts were treated as in A but IPs, from combined cell and medium samples, were treated for 1 h in the presence of PNGaseF prior to SDS-PAGE and fluorography. The single and double asterisks indicate the position of PNGase-resistant singly and doubly glycosylated RTB, respectively. The graph shows densitometric quantifications of RTB made in the presence or absence of BFA, expressed as percentages of the total RTB present at the end of the pulse. Average values from three independent experiments are shown; Error bars indicate standard deviation. C, protoplasts were transfected with constructs encoding RTB or phaseolin and, where indicated, co-transfected with a Sec12 expressing plasmid. Protoplasts were treated as in B. The black arrowhead and vertical bar are as described in A. The asterisks are the same as in B. The graph shows quantification calculated as in B. In all panels, numbers at the margins of gels indicate molecular mass markers in kilodaltons.
To simplify quantitation of fuzzy glycosylated bands to assess the kinetics of degradation, we treated total RTB samples (i.e. combined cell and media fractions) with PNGaseF to remove PNGaseF-sensitive glycans from RTB (Fig. 3B). The higher molecular weight band seen in the chase samples in the absence of BFA, and denoted by a single asterisk, appears to represent RTB with a PNGaseF-resistant glycan (Fig. 3B, lanes 2 and 3) (note that RTB has two glycans, but upon secretion from tobacco cells only one of these is PNGaseF-resistant). The only documented way in which a glycan gains PNGaseF resistance in plants is by fucosylation. Plant α1,3-fucosyltransferase is located in the late Golgi (36), suggesting that the species of RTB acquiring PNGase-F resistance in the absence of BFA represents the fraction being secreted. The singly and doubly glycosylated PNGaseF-resistant forms present during the pulse and the chase in the presence of BFA, denoted by single and double asterisks (Fig. 3B, lanes 4–6), are likely to have been generated by Golgi-modifying enzymes present in the ER-Golgi hybrid compartment. By quantifying the combined bands in each lane, it is clear from the graph in Fig. 3B that RTB is not stabilized in the presence of BFA.
Next, to show that RTB was not exiting the ER within COPII vesicles, we exploited knowledge that an overproduction of Sec12 can reduce ER export (16). Anterograde transport classically occurs in a COPII-dependent manner requiring the GTPase Sar1, the Sar1-specific guanosine nucleotide exchange factor Sec12, COPII coat components, and GTP (37–40). Overexpression of Sec12 is therefore an alternative and more specific approach to disrupt secretion. Under these conditions, the abundance of Sec12 is believed to titrate the limiting pool of Sar1 to cause a backlog of secretory proteins within the ER. This was confirmed upon expression of phaseolin where the precursor is maintained throughout the chase (Fig. 3C, compare band with arrowhead in lanes 8 and 9 with 11 and 12). Overexpression of Sec 12 did not, however, affect the disappearance of RTB, as measured by quantifying PNGaseF-treated IPs (Fig. 3C, lanes 1–6, and its graphical representation).
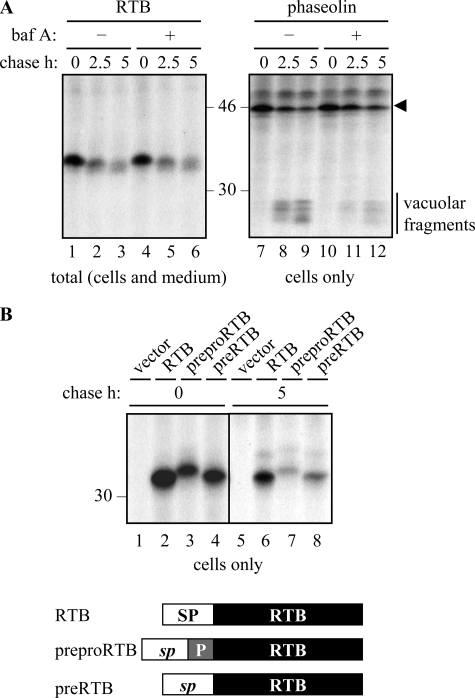
Because there is experimental evidence for an atypical trafficking route to vacuoles, albeit minor, that allows some proteins to bypass the Golgi apparatus altogether (32, 35), we felt it was necessary to rule out vacuolar involvement in RTB degradation in additional ways. Lytic vacuoles and prevacuolar compartments have low pH (41). Increasing the pH in such acidic organelles using bafilomycin A (Fig. 4A) (42) or ammonium chloride (data not shown), which would predictably reduce the catalytic activity of the resident proteases, was without effect on RTB degradation (Fig. 4A, left panel), as was treatment with E64d, an inhibitor of vacuolar cysteine proteinases (32) (data not shown). That bafilomycin A was having an effect at the concentration used is supported by the reduced level of vacuolar fragmentation of the phaseolin control (Fig. 4A, right panel).
FIGURE 4.
Degradation of ricin B chain does not occur in vacuoles. A, protoplasts expressing RTB or phaseolin were subject to pulse-chase as described under “Experimental Procedures.” Where indicated, protoplasts were preincubated for 1 h with 1 μm bafilomycin A (baf A) before radiolabeling. Subsequent IPs were analyzed by SDS-PAGE and fluorography. The arrowhead indicates the size of full-length phaseolin, and the vertical bar indicates vacuolar-generated fragments of phaseolin. B, protoplasts were transfected with vector alone (vector), or plasmids encoding RTB, prepro-RTB, or pre-RTB, where RTB is targeted to the ER via the phaseolin signal peptide (RTB), or via the native ricin signal peptide followed by a 9-residue propeptide (prepro-RTB) that is removed in vacuoles, or via the native ricin signal peptide alone (pre-RTB). Following pulse-chase, RTB was immunoprecipitated from separated cell homogenates and analyzed by SDS-PAGE and fluorography. In the schematic, SP represents the phaseolin signal peptide; sp represents the ricin signal peptide, and P represents the N-terminal propeptide of the ricin precursor. Numbers at the margins of gels indicate molecular mass markers in kilodaltons.
Finally, we investigated the vacuolar-dependent processing of a normally cleavable propeptide (9). During ricin biosynthesis, proricin reaches the protein storage vacuoles of the Ricinus communis endosperm with a 9-residue propeptide at the N terminus of RTA and a 12-residue linker peptide joining RTA to RTB (7, 18). In the vacuole, both of these are cleaved to generate mature ricin holotoxin (43). We therefore added the native signal peptide and a 9-residue propeptide to the N terminus of RTB (prepro-RTB; Fig. 4B), rationalizing that if RTB was being turned over within vacuoles, it was likely that the normally dispensable propeptide would be rapidly removed. After the pulse, ER-sequestered (signal peptide-cleaved) RTB containing the N-terminal propeptide was visible as a slightly larger band than mature RTB alone (Fig. 4B, compare lane 3 with lanes 2 and 4). Even upon the chase, when substantial degradation had occurred, the N-terminal propeptide remained attached (Fig. 4B, compare lane 7 with lanes 6 and 8). The failure to observe vacuolar processing of this naturally occurring and topologically correct propeptide further supports the assertion that RTB is not degraded within these organelles.
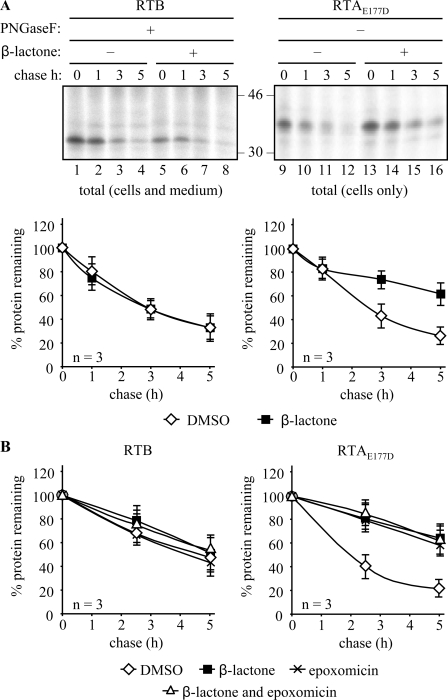
RTB Does Not Appear to Reach the Cytosol—In the absence of vacuolar degradation, it was plausible that loss of nonsecreted RTB, like that of RTA, could involve the ERAD pathway, especially as RTB was being made as an orphan polypeptide and was thus presumably exposing an interface to solvent that is normally obscured by its partner polypeptide. A significant fraction of retrotranslocated RTA is targeted for destruction by proteasomes (10), a process that can be significantly blocked when cells are treated with the specific proteasome inhibitor, clasto-lactacystin β-lactone (β-lactone) (44). As expected, both RTB and RTA were degraded with time in the absence of proteasome inhibitor (Fig. 5A, lanes 1–4 and 9–12). Repeating in the presence of β-lactone significantly reduced the rate of RTA degradation (Fig. 5A, lanes 13–16) as reported earlier (10) but, surprisingly, was without effect on RTB (Fig. 5A, lanes 5–8). To circumvent difficulties in quantitating RTB bands that appear fuzzy because of heterogeneous glycans, immunoprecipitates of RTB were treated with PNGaseF as before. Treatment was almost completely effective (data not shown) leaving just a minor fraction of PNGaseF-resistant RTB that has been observed previously (Fig. 3). Quantitation of these data is shown graphically in Fig. 5A. Whereas this conventional inhibitor of mammalian proteasomes reduces the degradation rate of RTA, it has no detectable effect on the kinetics of RTB degradation. Similarly, the proteasome inhibitor epoxomycin (45), which is known to inhibit chymotrypsin- and trypsin-like activities of mammalian and plant proteasomes (46–48), had no impact on the loss of RTB when added alone or with β-lactone, whereas both these reagents impeded the degradation of RTA (Fig. 5B).
FIGURE 5.
Inhibition of the proteasome does not stabilize ricin B chain. A, protoplasts were transfected with plasmids encoding RTB or RTA and subjected to pulse-chase analysis as under “Experimental Procedures” but with immunoprecipitation from combined cell and medium samples. Where indicated, protoplasts were incubated with 80 μm clasto-lactacystin β-lactone during the pulse. Immunoprecipitates were subjected to analysis by SDS-PAGE and fluorography. Graphs show densitometric quantification of the RTB or RTA IPs made in the presence or absence of clasto-lactacystin β-lactone, expressed as percentages of the total RTB or RTA present at the end of the pulse, and are the average values from three independent experiments. Error bars indicate standard deviation. B, densitometric quantification of immunoprecipitates from cells treated with DMSO, 80 μm clasto-lactacystin β-lactone, 80 μm epoxomycin, or a combination of these. Data are expressed as percentages of the total RTB or RTA present at the end of the pulse. These show the average values from three independent experiments. Error bars indicate standard deviation.
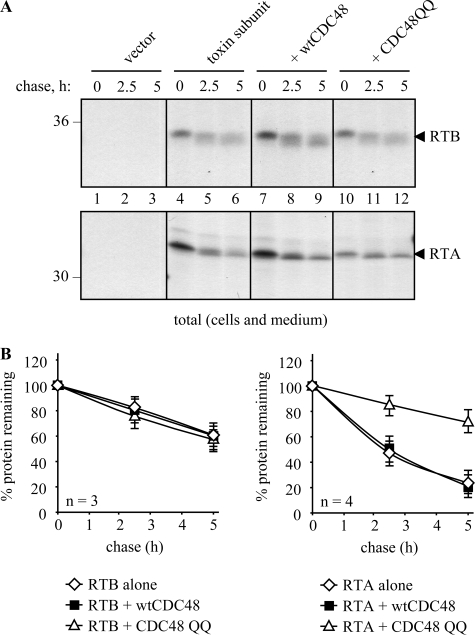
In a different approach, we attempted to perturb the actual arrival of RTB into the cytosol. The extraction of classical ERAD substrates from the ER membrane usually requires the involvement of an AAA ATPase called CDC48 in yeast or p97 in mammals (49–51). Following recruitment to the ER membrane, this ATPase works in association with its partners Ufd1 and Npl4 (49) to provide the driving force for the extraction process. Three homologues of this ATPase are present in the Arabidopsis genome, and one has been shown to functionally complement a yeast CDC48 mutant (52, 53). CDC48 has also been implicated in plant cell ERAD for both a mutant membrane protein (13) and for RTA (54). A trans-dominant CDC48 mutant would be expected to stabilize ERAD substrates, as it did for RTA (54). We therefore co-expressed ER-targeted RTB with a mutant CDC48 in which the conserved glutamate residues of the Walker B motifs (Glu-308 and Glu-581) of the two ATPase domains had been replaced by glutamine (denoted CDC48QQ) (13). We have noted previously that, even though the expression of CDC48QQ is quite toxic to cells, its presence promotes the stabilization of proteins that are normally extracted from the ER membrane by preventing their retrotranslocation to the cytosol (this is measured by relating the amount of test protein at the end of a chase period with the amount that is present at the end of the pulse) (54). However, and unlike RTA (Fig. 6) (54), when RTB is co-expressed with CDC48QQ, there is no observable stabilization (Fig. 6) suggesting that RTB is not progressing to the cytosol in a CDC48-dependent manner.
FIGURE 6.
Mutant CDC48 does not increase the stability of ER-sequestered ricin B chain. Protoplasts were transfected with vector alone (vector) or plasmids encoding RTB or RTA and, where indicated, co-transfected with plasmids encoding wtCDC48 or CDC48QQ. Following pulse-chase analysis as under “Experimental Procedures,” RTB and RTA were immunoprecipitated from combined cell and medium samples. Numbers at the margins of gels indicate molecular mass markers in kilodaltons. Graphs show densitometric quantification of the RTB or RTA IPs when expressed alone or co-expressed with wtCDC48 or CDC48QQ, expressed as percentages of the total RTB or RTA present at the end of the pulse, and are the average values from three independent experiments. Error bars indicate standard deviation.
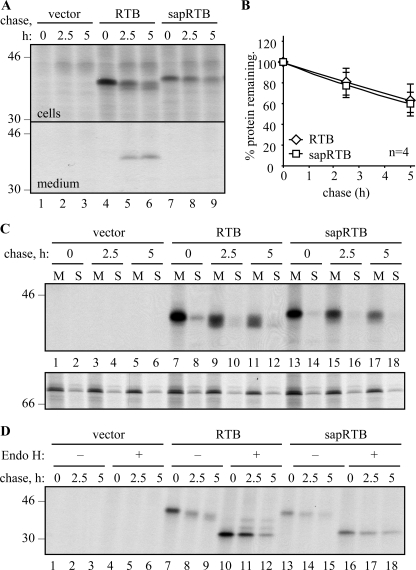
Finally, we prepared a construct expressing RTB with an ER signal peptide that was rendered uncleavable by mutation. In this way, we could ensure that the hydrophobic signal peptide would anchor the protein into the membrane, allowing its retention within the early secretory pathway. The selected signal peptide was that of the type I ribosome-inactivating protein saporin (55), containing T22N and A24V to prevent cleavage by signal peptidase without affecting ER import.6 When we followed the fate of this glycosylated protein (sapRTB) by pulse-chase, it was noticeable that, in contrast to native RTB (Fig. 7A, lanes 5 and 6), none of it was secreted (Fig. 7A, lanes 8 and 9), and yet the kinetics of disappearance of both proteins was almost identical (Fig. 7B). Membrane fractionation revealed that the bulk of RTB expressed with either its usual cleavable signal peptide or the uncleavable signal peptide was found within the membrane fraction of cells (Fig. 7C). The small proportion of RTB found with increasingly processed glycans in the soluble fractions (Fig. 7C, lanes 8, 10, and 12) was fucosylated (data not shown), and probably represents material from broken secretory vesicles. Note that the downsizing of the sapRTB was less pronounced than that of native RTB, suggesting it was not encountering Golgi glycan modification enzymes. Furthermore, although a fraction of native RTB acquired resistance to endoglycosidase H (Endo H) during the chase (Fig. 7D), presumably reflecting the fraction that is secreted and that reaches the late Golgi where glycans become Endo H-resistant, sapRTB did not acquire Endo H resistance. Nevertheless, this protein, presumably anchored in a membrane of the early secretory pathway, was clearly unstable.
FIGURE 7.
Ricin B chain disappears from the membrane fraction. A, protoplasts were transfected with vector alone (vector), or with plasmids encoding RTB carrying a cleavable phaseolin signal peptide (RTB), or with the saporin signal peptide rendered uncleavable by mutation (sapRTB). Protoplasts were subjected to pulse-chase, and RTB IPs from separated cell homogenates and medium were analyzed by SDS-PAGE and fluorography. B, quantification of RTB was carried out as in Fig. 3. The average of four independent experiments is shown. Error bars indicate standard deviation. C, protoplasts were transfected as in A and subjected to pulse-chase before being homogenized in the absence of detergent and fractionated to yield microsomal membranes (M) and soluble fractions (S). Proteins were sequentially immunoprecipitated using anti-RTB and anti-BiP antisera and analyzed by SDS-PAGE and fluorography. D, protoplasts were transfected as in A and subjected to pulse-chase analysis as before. RTB immunoprecipitates were treated for 16 h in the presence or absence of Endo H before analysis by reducing SDS-PAGE and fluorography. The single and double asterisks indicate the position of Endo H-resistant singly and doubly glycosylated RTB, respectively. In all gel panels, numbers down the margin indicate molecular mass markers in kilodaltons.
DISCUSSION
Errors in transcription/translation, inefficient folding, unbalanced synthesis of the individual subunits, and protein aging are among the many factors that can contribute to the accumulation of misfolded or unassembled polypeptides within the plant secretory pathway. As in other eukaryotes, accumulation of solo polypeptides and incomplete proteins is potentially very damaging to the plant cell and so must be eliminated. Lytic vacuoles are a repository of proteolytic enzymes and are responsible for protein degradation during seed germination and autophagic processes (30, 31). However, following the ER targeting of RTA in the absence of its partner B chain, we have previously demonstrated that in this particular case, degradation did not occur in vacuoles but in the cytosol (9). This location for turnover required the ER-to-cytosol delivery of RTA, a process that was precluded by co-expression of RTB (8). The ameliorating effect of RTB suggested this protein may not be retro-translocated in a similar way.
As part of a continuing study into protein quality control in the plant secretory pathway, we therefore studied the fate of RTB in plant cells, principally by pulse-chase analysis. This demonstrated a significant loss of newly made protein. Because it has been reported that, in some instances, the fate of a protein may differ when expressed transiently versus stably (21), we confirmed the loss of RTB in both transient expression experiments and in transgenic plants adapted to the synthesis of this protein (Fig. 1). Although a small fraction of ER-targeted RTB clearly folded into a sugar-binding conformation that became PNGaseF-resistant and secreted (Figs. 2B and 3), there remained a marked loss of intracellular material when both transiently and permanently expressed (Fig. 1C). Controls established that the unaccounted fraction was not a result of degradation in the medium, nor was it due to aggregation to a form unrecoverable by immunoprecipitation (Fig. 1E).
Given that acidic plant vacuoles are known as a site for protein digestion (30, 31), we disrupted both the trafficking to these organelles and the likely processing events within them. However, BFA (Fig. 3, A and B), Sec12 (Fig. 3C), and bafilomycin A (Fig. 4A) were unable to stabilize RTB, and the N-terminal propeptide, normally processed in vacuoles, was not removed from the N terminus of the B chain (Fig. 4B). These data clearly support the view that RTB was not reaching the degradative vacuole by either a classical COPII- and Golgi-dependent pathway or by any alternative route (32, 56).
Because the RTB fraction that was pulled down with lactose-agarose was never observed to interact with BiP (Fig. 2), it was reasonable to assume that the retained BiP-binding fraction represented a population of B chain that was either misfolded or that was in the act of folding. We noticed that even when cells were treated with cycloheximide after the 1-h pulse (to prevent further protein synthesis), the interaction of BiP with a fraction of already synthesized RTB persisted throughout the chase (data not shown). This led us to consider the possibility that this fraction of orphan B chain might be failing to acquire its correct conformation and be deemed terminally misfolded. Proteins delivered to the ER that fail to fold and or assemble often become iteratively associated with chaperones or, if glycosylated, become engaged with calnexin cycle components (57). Terminally misfolded proteins are eventually targeted for degradation. In mammalian and yeast cells, this process involves their Sec61- or Derlin/VIMP-mediated retrotranslocation across the ER membrane, polyubiquitination of (most) substrates on lysyl residues, CDC48/p97 complex-mediated extraction from the ER membrane, and targeting to proteasomes for proteolytic digestion. If the unfolded substrate is glycosylated, its glycans will be removed by a cytosolic peptide:N-glycanase prior to degradation (58). This pathway is known as ERAD (for reviews see Refs. 2, 3, 59).
It is now clear that a protein disposal pathway similar to the mammalian and yeast ERAD pathway also occurs in plant cells (9, 10, 13, 60). The first study in plants described the fate of an orphan RTA delivered into the ER of tobacco protoplasts in the absence of its partner B chain (9). RTA has been shown to be largely degraded in the cytosol following its CDC48-dependent retro-translocation from the ER lumen (54), but it can be partially stabilized there if cells are pretreated with the proteasome inhibitor, clasto-lactacystin β-lactone (9, 10, 54). Here we tested the fate of newly made RTB with a range of proteasome inhibitors that have been shown to be effective in plant cells (clasto-lactacystin β-lactone (61, 62), epoxomycin (47, 48) or a combination of the two), to achieve a more effective inhibition of proteasomes. However, although both inhibitors partially stabilized RTA, none of them had any observable impact on the degradation kinetics of RTB (Fig. 5). Precedents do exist for a proteasome-independent ERAD pathway (63), and loss of proteasome function can lead to induction of alternative proteolytic systems (64). Indeed, a study using ER-localized amyloid β peptide has shown this protein to be degraded in the cytosol by both proteasome-dependent and proteasome-independent pathways (65). Alternatively, it is possible that the level of proteasome inhibition achieved in vivo, although sufficient to slow down RTA degradation, might be inadequate to stabilize other substrates (46), including RTB.
Before invoking a novel site for protein turnover in plant cells, we needed to test for the cytosolic delivery of RTB. This was done by examining the fate of RTB in the presence of a mutant CDC48 complex. The function of this complex in facilitating the dislocation of ERAD substrates can be disrupted by replacing two conserved glutamic acid residues in the ATPase domains (Glu-308 and Glu-581) with glutamine residues (E308Q and E581Q, to generate CDC48QQ). Overexpression of this mutant inhibits ER-to-cytosol retro-translocation and consequently ERAD-mediated degradation (66). An Arabidopsis orthologue can functionally complement Saccharomyces cerevisiae mutants (52, 53) and has been shown to be critical for the ER-to-cytosol extraction (and subsequent degradation) of a mutant barley powdery mildew resistance protein (MLO) and the orphan RTA. When mutant AtCDC48QQ was expressed in plant cells, both these proteins failed to reach the cytosol and instead were significantly stabilized (13, 54). However, expression of CDC48QQ did not observably affect the degradation of RTB (Fig. 6). Although this does not completely rule out the cytosol as its site of degradation (for there are examples of p97-independent ERAD substrates (67, 68)), it would suggest that, at the very least, RTB is atypical, because its cytosolic disappearance would be independent of both CDC48 and the proteasome.
In a more recent study, a misfolded form of the ER-resident chaperone BiP was actively degraded when proteasome-dependent ERAD was blocked by either proteasome inhibitors or by ATP depletion (69). The data suggested that BiP degradation actually occurred within the ER lumen. However, direct evidence for the complete degradation of proteins within the ER itself is lacking. Rather, specific cleavages or trimming reactions have more typically been observed. For example, signal peptidase can sometimes act post-translationally as an endopeptidase, cleaving proteins at specific exposed sites (70–72), whereas signal peptide peptidase has been shown to have an unexpected role in the disposal of particular substrates by ERAD (73). One very well studied protease within the mammalian ER lumen is the ER aminopeptidase associated with antigen processing (ERAAP/ERAP1), which is responsible for generating trimmed peptides for loading onto major histocompatibility complex class I proteins (74). Although such peptidases are likely to be absent in the plant cell ER, other classes of secretory pathway proteases cannot be entirely excluded at this stage. Perhaps the most intriguing evidence for the novel secretory pathway turnover of RTB comes from its continued disappearance when anchored as a glycoprotein via an uncleaved signal peptide (Fig. 7). At this juncture, however, we have been unable to stabilize RTB with pepstatin, E64d, calpeptin, or 4-(2-amino-ethyl)benzenesulfonyl fluoride (data not shown). Although further work will be needed to identify the protease(s) responsible for RTB degradation, the evidence to date points to proteolysis within the early secretory pathway itself. This would represent a novel location for protein degradation in plant cells that adds to classical ERAD pathway exceptions already reported in other systems (63).
Acknowledgments
We thank Prof. Ralph Panstruga (Max-Planck Institute for Plant Breeding Research, Köln, Germany) for provision of the plasmids encoding wild-type and mutant Arabidopsis CDC48 and Prof. Jurgen Denecke (Centre for Plant Sciences, University of Leeds, UK) for provision of the plasmid encoding Sec12.
This work was supported in part by Wellcome Trust Programme Grant 080566/Z/06/Z (to L. M. R. and J. M. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: RTA, ricin A chain; RTB, ricin B chain; ER, endoplasmic reticulum; ERAD, ER-associated protein degradation; DMM, 1-deoxymannojirimycin; PNGaseF, peptide:N-glycanase F; CaMV, cauliflower mosaic virus; BFA, brefeldin A; IP, immunoprecipitation; Endo H, endoglycosidase H.
A. Ceriotti, unpublished results.
References
- 1.Roberts, L. M., and Lord, J. M. (2004) Mini Rev. Med. Chem. 4 505-512 [DOI] [PubMed] [Google Scholar]
- 2.Romisch, K. (2005) Annu. Rev. Cell Dev. Biol. 21 435-456 [DOI] [PubMed] [Google Scholar]
- 3.Meusser, B., Hirsch, C., Jarosch, E., and Sommer, T. (2005) Nat. Cell Biol. 7 766-772 [DOI] [PubMed] [Google Scholar]
- 4.Lord, M. J., Jolliffe, N. A., Marsden, C. J., Pateman, C. S., Smith, D. C., Spooner, R. A., Watson, P. D., and Roberts, L. M. (2003) Toxicol. Rev. 22 53-64 [DOI] [PubMed] [Google Scholar]
- 5.Endo, Y., and Tsurugi, K. (1987) J. Biol. Chem. 262 8128-8130 [PubMed] [Google Scholar]
- 6.Frigerio, L., and Roberts, L. (1998) J. Exp. Bot. 49 1473-1480 [Google Scholar]
- 7.Frigerio, L., Jolliffe, N. A., Di Cola, A., Felipe, D. H., Paris, N., Neuhaus, J. M., Lord, J. M., Ceriotti, A., and Roberts, L. M. (2001) Plant Physiol. 126 167-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frigerio, L., Vitale, A., Lord, J. M., Ceriotti, A., and Roberts, L. M. (1998) J. Biol. Chem. 273 14194-14199 [DOI] [PubMed] [Google Scholar]
- 9.Di Cola, A., Frigerio, L., Lord, J. M., Ceriotti, A., and Roberts, L. M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14726-14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Cola, A., Frigerio, L., Lord, J. M., Roberts, L. M., and Ceriotti, A. (2005) Plant Physiol. 137 287-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jolliffe, N. A., Ceriotti, A., Frigerio, L., and Roberts, L. M. (2003) Plant Mol. Biol. 51 631-641 [DOI] [PubMed] [Google Scholar]
- 12.Tabe, L. M., Wardley-Richardson, T., Ceriotti, A., Aryan, A., McNabb, W., Moore, A., and Higgins, T. J. (1995) J. Anim. Sci. 73 2752-2759 [DOI] [PubMed] [Google Scholar]
- 13.Muller, J., Piffanelli, P., Devoto, A., Miklis, M., Elliott, C., Ortmann, B., Schulze-Lefert, P., and Panstruga, R. (2005) Plant Cell 17 149-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S., and Mullineaux, P. M. (2000) Plant Mol. Biol. 42 819-832 [DOI] [PubMed] [Google Scholar]
- 15.Ma, J. K., Lehner, T., Stabila, P., Fux, C. I., and Hiatt, A. (1994) Eur. J. Immunol. 24 131-138 [DOI] [PubMed] [Google Scholar]
- 16.Phillipson, B. A., Pimpl, P., daSilva, L. L., Crofts, A. J., Taylor, J. P., Movafeghi, A., Robinson, D. G., and Denecke, J. (2001) Plant Cell 13 2005-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrini, J. B., Martin, M., Taupiac, M. P., and Beaumelle, B. (1995) Eur. J. Biochem. 233 772-777 [DOI] [PubMed] [Google Scholar]
- 18.Jolliffe, N. A., Di Cola, A., Marsden, C. J., Lord, J. M., Ceriotti, A., Frigerio, L., and Roberts, L. M. (2006) J. Biol. Chem. 281 23377-23385 [DOI] [PubMed] [Google Scholar]
- 19.Maliga, P., Sz-Breznovits, A., and Marton, L. (1973) Nat. New Biol. 244 29-30 [DOI] [PubMed] [Google Scholar]
- 20.Pedrazzini, E., Giovinazzo, G., Bielli, A., de Virgilio, M., Frigerio, L., Pesca, M., Faoro, F., Bollini, R., Ceriotti, A., and Vitale, A. (1997) Plant Cell 9 1869-1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frigerio, L., de Virgilio, M., Prada, A., Faoro, F., and Vitale, A. (1998) Plant Cell 10 1031-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vitale, A., Zoppe, M., and Bollini, R. (1989) Plant Physiol. 89 1079-1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Virgilio, M., Kitzmuller, C., Schwaiger, E., Klein, M., Kreibich, G., and Ivessa, N. E. (1999) Mol. Biol. Cell 10 4059-4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellgaard, L., and Helenius, A. (2003) Nat. Rev. Mol. Cell Biol. 4 181-191 [DOI] [PubMed] [Google Scholar]
- 25.Nuttall, J., Vine, N., Hadlington, J. L., Drake, P., Frigerio, L., and Ma, J. K. (2002) Eur. J. Biochem. 269 6042-6051 [DOI] [PubMed] [Google Scholar]
- 26.Nuttall, J., Vitale, A., and Frigerio, L. (2003) Plant Mol. Biol. 51 885-894 [DOI] [PubMed] [Google Scholar]
- 27.Wales, R., Richardson, P. T., Roberts, L. M., Woodland, H. R., and Lord, J. M. (1991) J. Biol. Chem. 266 19172-19179 [PubMed] [Google Scholar]
- 28.Bakker, H., Bardor, M., Molthoff, J. W., Gomord, V., Elbers, I., Stevens, L. H., Jordi, W., Lommen, A., Faye, L., Lerouge, P., and Bosch, D. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2899-2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong, E., Davidson, A., and Kaiser, C. (1996) J. Cell Biol. 135 623-633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter, C., Pan, S., Zouhar, J., Avila, E. L., Girke, T., and Raikhel, N. V. (2004) Plant Cell 16 3285-3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Contento, A. L., Kim, S. J., and Bassham, D. C. (2004) Plant Physiol. 135 2330-2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura, K., Yamada, K., Shimada, T., and Hara-Nishimura, I. (2004) Plant J. 39 393-402 [DOI] [PubMed] [Google Scholar]
- 33.Pimpl, P., Taylor, J. P., Snowden, C., Hillmer, S., Robinson, D. G., and Denecke, J. (2006) Plant Cell 18 198-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nebenfuhr, A., Ritzenthaler, C., and Robinson, D. G. (2002) Plant Physiol. 130 1102-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frigerio, L., Pastres, A., Prada, A., and Vitale, A. (2001) Plant Cell 13 1109-1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saint-Jore-Dupas, C., Faye, L., and Gomord, V. (2007) Trends Biotechnol. 25 317-323 [DOI] [PubMed] [Google Scholar]
- 37.d'Enfert, C., Wuestehube, L. J., Lila, T., and Schekman, R. (1991) J. Cell Biol. 114 663-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barlowe, C., d'Enfert, C., and Schekman, R. (1993) J. Biol. Chem. 268 873-879 [PubMed] [Google Scholar]
- 39.Barlowe, C., and Schekman, R. (1993) Nature 365 347-349 [DOI] [PubMed] [Google Scholar]
- 40.d'Enfert, C., Barlowe, C., Nishikawa, S., Nakano, A., and Schekman, R. (1991) Mol. Cell. Biol. 11 5727-5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taiz, L. (1992) J. Exp. Biol. 172 113-122 [DOI] [PubMed] [Google Scholar]
- 42.Brauer, D., Uknalis, J., Triana, R., Shachar-Hill, Y., and Tu, S. I. (1997) Plant Physiol. 113 809-816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lord, J. M. (1985) Eur. J. Biochem. 146 411-416 [DOI] [PubMed] [Google Scholar]
- 44.Fenteany, G., Standaert, R. F., Lane, W. S., Choi, S., Corey, E. J., and Schreiber, S. L. (1995) Science 268 726-731 [DOI] [PubMed] [Google Scholar]
- 45.Meng, L., Mohan, R., Kwok, B. H., Elofsson, M., Sin, N., and Crews, C. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10403-10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kisselev, A. F., Callard, A., and Goldberg, A. L. (2006) J. Biol. Chem. 281 8582-8590 [DOI] [PubMed] [Google Scholar]
- 47.Speranza, A., Scoccianti, V., Crinelli, R., Calzoni, G. L., and Magnani, M. (2001) Plant Physiol. 126 1150-1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng, X., Hu, Z., Lu, H., Wang, X., Baluska, F., Samaj, J., and Lin, J. (2006) Plant Physiol. 141 1578-1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye, Y., Meyer, H. H., and Rapoport, T. A. (2001) Nature 414 652-656 [DOI] [PubMed] [Google Scholar]
- 50.Braun, S., Matuschewski, K., Rape, M., Thoms, S., and Jentsch, S. (2002) EMBO J. 21 615-621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D. H., and Sommer, T. (2002) Nat. Cell Biol. 4 134-139 [DOI] [PubMed] [Google Scholar]
- 52.Feiler, H. S., Desprez, T., Santoni, V., Kronenberger, J., Caboche, M., and Traas, J. (1995) EMBO J. 14 5626-5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rancour, D. M., Dickey, C. E., Park, S., and Bednarek, S. Y. (2002) Plant Physiol. 130 1241-1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marshall, R. S., Jolliffe, N. A., Ceriotti, A., Snowden, C. J., Lord, J. M., Frigerio, L., and Roberts, L. M. (2008) J. Biol. Chem. 283 15869-15877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benatti, L., Saccardo, M. B., Dani, M., Nitti, G., Sassano, M., Lorenzetti, R., Lappi, D. A., and Soria, M. (1989) Eur. J. Biochem. 183 465-470 [DOI] [PubMed] [Google Scholar]
- 56.Tormakangas, K., Hadlington, J. L., Pimpl, P., Hillmer, S., Brandizzi, F., Teeri, T. H., and Denecke, J. (2001) Plant Cell 13 2021-2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parodi, A. J. (2000) Annu. Rev. Biochem. 69 69-93 [DOI] [PubMed] [Google Scholar]
- 58.Katiyar, S., Li, G., and Lennarz, W. J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13774-13779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai, B., Ye, Y., and Rapoport, T. A. (2002) Nat. Rev. Mol. Cell Biol. 3 246-255 [DOI] [PubMed] [Google Scholar]
- 60.Brandizzi, F., Hanton, S., DaSilva, L. L., Boevink, P., Evans, D., Oparka, K., Denecke, J., and Hawes, C. (2003) Plant J. 34 269-281 [DOI] [PubMed] [Google Scholar]
- 61.Reichel, C., and Beachy, R. N. (2000) J. Virol. 74 3330-3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gillespie, T., Boevink, P., Haupt, S., Roberts, A. G., Toth, R., Valentine, T., Chapman, S., and Oparka, K. J. (2002) Plant Cell 14 1207-1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitz, A., and Herzog, V. (2004) Eur. J. Cell Biol. 83 501-509 [DOI] [PubMed] [Google Scholar]
- 64.Glas, R., Bogyo, M., McMaster, J. S., Gaczynska, M., and Ploegh, H. L. (1998) Nature 392 618-622 [DOI] [PubMed] [Google Scholar]
- 65.Schmitz, A., Schneider, A., Kummer, M. P., and Herzog, V. (2004) Traffic 5 89-101 [DOI] [PubMed] [Google Scholar]
- 66.Ye, Y., Meyer, H. H., and Rapoport, T. A. (2003) J. Cell Biol. 162 71-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong, X., Lin, Z., Liang, D., Fath, D., Sang, N., and Caro, J. (2006) Mol. Cell. Biol. 26 2019-2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Asher, G., Tsvetkov, P., Kahana, C., and Shaul, Y. (2005) Genes Dev. 19 316-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Donoso, G., Herzog, V., and Schmitz, A. (2005) Biochem. J. 387 897-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith, D. C., Gallimore, A., Jones, E., Roberts, B., Lord, J. M., Deeks, E., Cerundolo, V., and Roberts, L. M. (2002) J. Immunol. 169 99-107 [DOI] [PubMed] [Google Scholar]
- 71.Tolchinsky, S., Yuk, M. H., Ayalon, M., Lodish, H. F., and Lederkremer, G. Z. (1996) J. Biol. Chem. 271 14496-14503 [DOI] [PubMed] [Google Scholar]
- 72.Yuk, M. H., and Lodish, H. F. (1993) J. Cell Biol. 123 1735-1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loureiro, J., Lilley, B. N., Spooner, E., Noriega, V., Tortorella, D., and Ploegh, H. L. (2006) Nature 441 894-897 [DOI] [PubMed] [Google Scholar]
- 74.Blanchard, N., and Shastri, N. (2008) Curr. Opin. Immunol. 20 82-88 [DOI] [PMC free article] [PubMed] [Google Scholar]