Abstract
To facilitate qualitative and quantitative analysis of glycosaminoglycans, we tagged the reducing end of lyase-generated disaccharides with aniline-containing stable isotopes (12C6 and 13C6). Because different isotope tags have no effect on chromatographic retention times but can be discriminated by a mass detector, differentially isotope-tagged samples can be compared simultaneously by liquid chromatography/mass spectrometry and quantified by admixture with known amounts of standards. The technique is adaptable to all types of glycosaminoglycans, and its sensitivity is only limited by the type of mass spectrometer available. We validated the method using commercial heparin and keratan sulfate as well as heparan sulfate isolated from mutant and wild-type Chinese hamster ovary cells, and select tissues from mutant and wild-type mice. This new method provides more robust, reliable, and sensitive means of quantitative evaluation of glycosaminoglycan disaccharide compositions than existing techniques allowing us to compare the chondroitin and heparan sulfate compositions of Hydra vulgaris, Drosophila melanogaster, Caenorhabditis elegans, and mammalian cells. Our results demonstrate significant differences in glycosaminoglycan structure among these organisms that might represent evolutionarily distinct functional motifs.
Metazoans make several types of sulfated glycosaminoglycans (GAGs),2 including keratan sulfate (KS), chondroitin sulfate/dermatan sulfate (CS/DS), and heparan sulfate/heparin (HS). Each type of chain consists of unique disaccharide units. KS consists of galactose (Gal) and GlcNAc ([Galβ1, 4GlcNAcβ1,3]n) with variable sulfation at C6 of either sugar. CS/DS assembles as a copolymer of GlcAβ1,3GalNAcβ1,4 and then undergoes various processing reactions, including C5 epimerization of a portion of GlcA units to iduronic acid in DS, O-sulfation at C2 and more rarely at C3 of the uronic acids, and O-sulfation at C4 and C6 of the GalNAc residues (1). HS is the most highly modified GAG, consisting initially of GlcAβ1,4GlcNAcα1,4 units, which then undergo variable processing by GlcNAc N-deacetylation and N-sulfation, C5 epimerization of some GlcA units to iduronic acid, and O-sulfate addition to C2 of the uronic acids and C6 and more rarely at C3 of the glucosamine units (2). The arrangement of the modified residues along the chain creates binding sites for numerous growth factors, enzymes, and extracellular matrix proteins. The structural variation that can occur makes sulfated GAG chains one of the most complex classes of macromolecules found in nature.
GAG fine structure is typically assessed by analyzing the disaccharide composition of an isolated mixture of chains. A number of techniques have been developed to accomplish this task that rely on chemical or enzymatic depolymerization of the chains into their constituent disaccharides, followed by separation via anion exchange chromatography, reversed-phase chromatography with ion pairing agents, or capillary electrophoresis. These techniques separate disaccharides based on charge, position of sulfate groups, and uronic acid composition. The individual disaccharide residues are detected using different methods, such as metabolic radiolabeling of chains prior to isolation, radiochemical labeling of liberated disaccharides by borotritide reduction, UV absorbance of enzymatically generated products, or fluorescence of fluorophore-tagged derivatives. These techniques have a limit of sensitivity of ∼10-11 mol (∼10 ng). The actual identification of the disaccharides depends on determining the retention time relative to authentic standards, which are sometimes difficult to procure. At times the profile can be confusing due to spurious peaks or contaminants (3-5).
The adaptation of high performance liquid chromatographymass spectrometry (LC/MS) to analyze disaccharides circumvents some of these problems (6-16). The technology affords simultaneous measurement of chromatographic retention time, absolute mass, ion adduction characteristics, and fragment ion analysis of each disaccharide, eliminating the need for comparison to disaccharide standards. In addition to revealing more structural and chemical information for each disaccharide, the use of LC/MS eliminates the need for radiolabeling or the requirement for UV or fluorescent derivatization. Furthermore, depending on mass detector design, LC/MS can be significantly more sensitive (∼10-15 mol, or ∼1 pg).
Although the use of LC/MS has greatly enhanced disaccharide analysis, the technology suffers in one important aspect: the technique is semiquantitative due to variation in the ionization efficiency of different disaccharides, solvent effects on ionization, and suppression by contaminants in the preparation. To circumvent these problems, LC/MS procedures have been developed that utilize linear equations to correlate the concentration of underivatized disaccharides with the detection of molecular standards (10, 17-20). Another approach is to use mass-tagging techniques based on differentially isotopically labeled tags introduced into oligosaccharides derived from intact chains (16, 21-24). Here we describe a mass-tagging technique that allows quantitative analysis of disaccharide composition and ratiometric comparisons between samples. This method employs glycan reductive isotope labeling (GRIL) with [12C6]- and [13C6]aniline3, modification of N-unsubstituted glucosamine residues by propionylation and resolution of the derivatives by LC/MS (GRIL-LC/MS). In this study, we demonstrate the reliability and sensitivity of this new method compared with the most commonly used means of GAG compositional analysis based on post column fluorescence derivatization. Improving LC/MS analysis in this way revealed that heparan sulfates from the invertebrates Drosophila melanogaster and Hydra vulgaris have a high content of free-amine-containing disaccharides compared with samples obtained from Chinese hamster ovary cells, mice, and Caenorhabditis elegans.
EXPERIMENTAL PROCEDURES
Culture Conditions—Wild-type and mutant pgsF17 cells (26) were grown in Ham's F-12 medium containing 50 units/ml penicillin, 50 μg/ml streptomycin, 2 mm glutamine, and 10% fetal bovine serum. Mice were housed in Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-approved vivaria in the School of Medicine, University of California San Diego, following standards and procedures approved by the local Institutional Animal Care and Use Committee for the ethical use of animals in experiments. A line bearing a conditional loxP-flanked allele of uronyl-2-O-sulfotransferase (Hs2st) was bred to ZP3Cre to generate heterozygous mice.4 These were then in-bred to produce homozygous null, heterozygous, and wild-type littermate embryos. Another set of mice was crossbred with AlbCre mice to generate a hepatocyte-specific mutant. Embryos and tissues were genotyped by PCR. H. vulgaris (Zurich strain), D. melanogaster, and C. elegans were grown according to established procedures.
Purification and Enzymatic Depolymerization of HS and CS Chains—GAG chains were extracted from cell monolayers, liver, or homogenized whole mouse embryos by exhaustive digestion with Pronase (Sigma-Aldrich) in phosphate-buffered saline at 37 °C for 24 h followed by anion-exchange chromatography (DEAE-Sephacel, GE Healthcare). After loading the columns, they were washed with 0.25 m NaCl, and GAGs were eluted with 1 m NaCl (27). GAGs from H. vulgaris, D. melanogaster, and C. elegans were prepared in a similar way except that the samples were first delipidated with acetone (3) before Pronase digestion and subsequent anion-exchange chromatography. All preparations were desalted by gel filtration (PD-10, GE Healthcare).
GAGs were digested with 1 milliunit each of heparin lyases I, II, and III (Seikagaku, Tokyo) for 16 h at 37 °C in 50 μl of buffer containing 40 mm ammonium acetate and 3.3 mm calcium acetate, pH 7, or for 3 h at 37 °C with 25 milliunits of chondroitinase ABC (Seikagaku, Tokyo) in 100 μl of buffer containing 50 mm Tris and 50 mm sodium acetate, pH 8.0 (28). Bovine Corneal KS (Sigma-Aldrich) was digested with either 100 milliunits of keratanase (Seikagaku, Tokyo) or 5 milliunits of keratanase II (Seikagaku, Tokyo) per 20 μg of GAG in 50 μl of buffer containing 7.5 mm sodium acetate, pH 6, for 16 h at 37 °C. The extent of conversion was complete based on analysis of standard GAGs incubated under the same conditions.
Disaccharide Standards and Purification of 3-O-Sulfated Disaccharides—HS disaccharides D0A6, D2A0, and D2A65 were purchased from Calbiochem, whereas all other HS disaccharide standards were purchased from Sigma-Aldrich. CS disaccharides were obtained from Oxford Glycosystems. Stock solutions (1 mm) were prepared by dissolution of each disaccharide in water. The final concentration was verified by quantitating the amount of uronic acid present using high performance anion-exchange chromatography with pulsed amperometric detection (29). The 3-O-sulfated disaccharides were isolated from a stable CHO cell line CHO[ECO] containing a retroviral expression construct for the human 3-OST-3A cDNA (12). HS isolated from these transductants was subjected to exhaustive digestion with heparan lyases as described above, and the disaccharides corresponding to D2S3 and D2S9 were collected (12). Purity and identity was checked by LC/MS.
Reductive Amination and Acylation of Disaccharides—1 pmol to 10 nmol of HS and CS disaccharides was transferred to 1.5-ml microcentrifuge tubes and dried down in a centrifugal evaporator. [12C6]Aniline or [13C6]aniline (15 μl, 165 μmol) and 15 μl of 1 m NaCNBH3 (Sigma-Aldrich) freshly prepared in dimethyl sulfoxide:acetic acid (7:3, v/v) were added to each sample. Reactions were carried out at 65 °C for 4 h or alternatively at 37 °C for 16 h and then dried in a centrifugal evaporator. In most cases subsequent purification steps are not necessary. However, if samples contain excessive contaminants, non-glycan components can be removed using paper chromatography cartridges (GlycoClean S cartridges, Prozyme) according to the manufacturer's instructions without loss of aniline-tagged disaccharides (as judged by LC/MS analysis of flow through and wash fractions). Unsubstituted amines were reacted with propionic anhydride (Sigma-Aldrich). Dried samples were reconstituted in 20 μl of 50% methanol and 3 μl of propionic anhydride (Sigma-Aldrich, 23.3 μmol) was added. Reactions were carried out at room temperature for 2 h. Acylated disaccharides were subsequently aniline-tagged as described above.
LC/MS Analysis of HS and CS Disaccharides and KS Digestion Products—An LCQ classic quadrupole ion trap mass spectrometer equipped with an electrospray ionization source, and a quaternary high-performance liquid chromatography pump (Thermo-Finnigan, San Jose, CA) was used for disaccharide analyses. Derivatized and non-derivatized disaccharide residues were separated on a C18 reversed-phase column (0.46 × 25 cm, Vydac) with the ion pairing agent dibutylamine (DBA, Sigma-Aldrich) (9, 12, 30). The isocratic steps were: 100% buffer A (8 mm acetic acid, 5 mm DBA) for 10 min, 17% buffer B (70% methanol, 8 mm acetic acid, 5 mm DBA) for 15 min; 32% buffer B for 15 min, 40% buffer B for 15 min, 60% buffer B for 15 min; 100% buffer B for 10 min; and 100% buffer A for 10 min. The most highly substituted disaccharide, D2S9 (see Table 1), eluted at 60% buffer B (42% methanol). Ions of interest were monitored in negative ion mode, and signal intensity was optimized for a representative species (D2S0, see Table 1). To minimize in-source fragmentation of sulfated disaccharides, the capillary temperature and spray voltage were kept at 140 °C and 4.75 kV, respectively. The accumulative extracted ion current (XIC) was computed, and further data analysis was carried out as described in the documentation for the Qual Browser software provided by Thermo-Finnigan. For CID analysis of CS-monosulfated disaccharides, ions with m/z 535 were selected using a 2-atomic mass unit window and activated with 30% normalized collision energy.
TABLE 1.
Molecular masses for component disaccharides from HS, CS/DS, and KS used in generating disaccharide XIC traces for both quantitative and qualitative compositional analysis
The disaccharide structure code (DSC) (25), the abbreviated chemical structure (ΔU = 4,5-unsaturated uronic acid), and the observed m/z values in negative ion mode for each enzymatically derived disaccharide are given. The values for both underivatized and [12C6]aniline-derivatized forms are provided as well as the m/z values for unsubstituted amine-containing HS disaccharides modified with both propionic anhydride and [12C6]aniline (PA/aniline). For aniline-labeled disaccharides, both the free molecular ion ([M-H]−1), the adduction ions formed by many of the more highly sulfated disaccharides with the ion pairing reagent ([M-2H+DBA]−1), and any in-source desulfation products (which correspond in mass to the masses of disaccharides that naturally have one less sulfate) were used in determining the XIC traces. The HS and CS/DS disaccharides listed in bold font are commercially available, and the [13C6]aniline-derivatized forms of these disaccharides were used for quantitative analysis by GRIL-LC/MS. The m/z values of the [13C6]aniline-derivatized standards differ from the [12C6]aniline derivatives listed in this table by +6 atomic mass units.
|
DSC
|
Structure
|
Observedm/z
|
|||
|---|---|---|---|---|---|
| Non-derivatized | [12C6]Aniline-derivatized | [12C6]Aniline-derivatized | PA/aniline | ||
| [M-H]−1 | [M-H]−1 | [M-2H+DBA]−1 | [M-H]−1 | ||
| HS disaccharides | |||||
| D0H0 | ΔUA-GlcNH2 | 336 | 413 | 469 | |
| D0H6 | ΔUA-GlcNH2-6S | 416 | 493 | 549 | |
| D2H0 | ΔUA2S-GlcNH2 | 416 | 493 | 549 | |
| D2H6 | ΔUA2S-GlcNH26S | 496 | 573 | 629 | |
| D0A0 | ΔUA-GlcNAc | 378 | 455 | ||
| D0A6 | ΔUA-GlcNAc6S | 458 | 535 | ||
| D2A0 | ΔUA2S-GlcNAc | 458 | 535 | ||
| D2A6 | ΔUA2S-GlcNAc6S | 538 | 615 | 744 | |
| D0S0 | ΔUA-GlcNS | 416 | 493 | ||
| D0S6 | ΔUA-GlcNS6S | 496 | 573 | 702 | |
| D0S3 | ΔUA-GlcNS3S | 496 | 573 | 702 | |
| D2S0 | ΔUA2S-GlcNS | 496 | 573 | 702 | |
| D2S6 | ΔUA2S-GlcNS6S | 576 | 653 | 782 | |
| D2S3 | ΔUA2S-GlcNS3S | 576 | 653 | 782 | |
| D0S9 | ΔUA-GlcNS3S6S | 576 | 653 | 782 | |
| D2S9 | ΔUA2S-GlcNS3S6S | 656 | 733 | 862 | |
| CS disaccharides | |||||
| D0a0 | ΔUA-GalNAc | 378 | 455 | ||
| D0a4 | ΔUA-GalNAc4S | 458 | 535 | ||
| D2a0 | ΔUA2S-GalNAc | 458 | 535 | ||
| D0a6 | ΔUA-GalNAc6S | 458 | 535 | ||
| D2a4 | ΔUA2S-GalNAc4S | 538 | 615 | 744 | |
| D2a6 | ΔUA2S-GalNAc6S | 538 | 615 | 744 | |
| D0a10 | ΔUA-GalNAc4S6S | 538 | 615 | 744 | |
| D2a10 | ΔUA2S-GalNAc4S6S | 618 | 695 | 824 | |
| KS disaccharides | |||||
| g0A6 | Gal-GlcNAc6S | 462 | 539 | ||
| g6A6 | Gal6S-GlcNAc6S | 542 | 619 | 748 | |
| A6g0 | GlcNAc6S-Gal | 462 | 539 | ||
Heparin (Sigma) disaccharides were also separated by high-performance liquid chromatography on a ProPac PA1 anion-exchange column (Dionex) with a linear gradient of sodium chloride (50 mm to 1 m, pH 3.5, 60 min). Post column derivatization with 2-cyanoacetamide was performed as described (31). Fluorescent products were detected with a flow-through fluorescence detector (Jasco) set at excitation 346 nm and emission 410 nm.
RESULTS
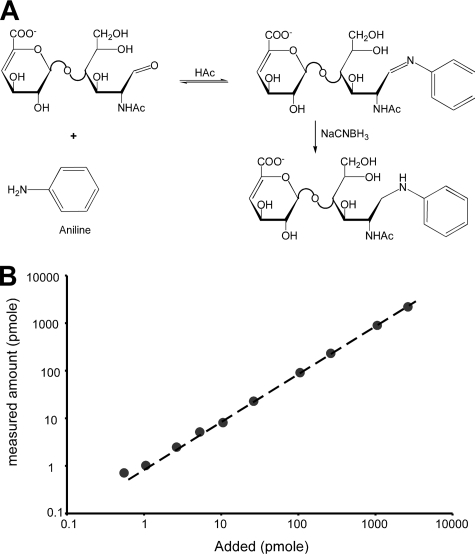
Stable Isotope Labeling and LC/MS Analysis of Glycosaminoglycan Disaccharides—Of the different technologies available to analyze the disaccharide composition of GAG chains, LC/MS is most robust because of its higher sensitivity and its capacity for multiparametric analysis. However, due to intrinsic molecular differences in structure and extrinsic effects of solvent and contaminants on ionization, traditional LC/MS is not quantitative. To overcome the non-quantitative aspects of LC/MS, we adapted the GRIL technique.3 As applied to glycosaminoglycans, the technique first involves enzymatic depolymerization of partially purified chains using bacterial hydrolases or eliminases, followed by reductive amination of the reducing ends of the disaccharides with aniline to form a secondary amine derivative (Fig. 1A). The conjugation reaction is rapid and quantitative. Incubation of standard disaccharides derived from heparan sulfate and chondroitin sulfate with a 100-fold molar excess of [12C6]aniline at 65 °C yielded >98% derivatization in 4 h, which was determined by measuring unmodified disaccharide present in the reaction mixture. No detectable O-desulfation byproducts and only a trace amount of N-deacetylated or N-desulfated byproducts were detected under these conditions. Less than 2% of these byproducts were observed after incubation at 65 °C for 4 h (supplemental Fig. S1). Reducing the temperature to 37 °C (for 16 h) almost completely eliminated these minor byproducts, without sacrificing the efficiency of conjugation (>98%).
FIGURE 1.
Aniline-tagging of GAG disaccharides. A, the reducing form of each GAG disaccharide can form a Schiff base with the amine group of aniline. The resulting imine is then reduced with sodium cyanoborohydride resulting in a stable derivative. B, to determine the sensitivity and linear range of GRIL disaccharide analysis, D2S0 was labeled with either [12C6]aniline or [13C6]aniline. Differing amounts of [12C6]aniline-labeled D2S0 (between 0.5 pmol and 1 nmol) were mixed in triplicate with 25 pmol of [13C6]aniline-labeled D2S0 prior to LC/MS analysis. The measured amount (y-axis) is plotted against the amount of [12C6]aniline-labeled disaccharide added (x-axis). The standard error was less than the diameter of the symbols and <5%.
We utilized a Finnigan LCQ Classic quadrupole mass spectrometer during the development phase of this method, which is capable of moderate levels of sensitivity. Based on titration studies, the limit of sensitivity was 1 pmol/disaccharide. Because of multiparametric analysis, detection was reliable for disaccharides present at low levels in samples containing significant background contaminants that tend to obscure the disaccharide profile obtained by UV or fluorescence detection. Furthermore, all species present in the sample, including underivatized disaccharides, can be detected, so that the extent of derivatization can be internally monitored.
The linear range of GRIL-LC/MS was evaluated by measuring the XIC ratio between increasing amounts of the disulfated disaccharide standard D2S0 (Table 1) tagged with [12C6]aniline and a set amount (25 pmol) of D2S0 tagged with [13C6]aniline. Three replicate measurements were carried out with each concentration of [12C6]aniline-tagged D2S0 and showed the linear range to be from 1 pmol to >2500 pmol (Fig. 1B). Similar results were observed in titration assays for other purified disaccharide standards (data not shown). The standard curve deviated somewhat from linearity below 1 pmol, but this lower limit reflects the mass spectrometer configuration. At least 100-fold greater sensitivity should be achievable on a more modern instrument (e.g. a linear ion trap or quadrupole time-of-flight mass spectrometer).
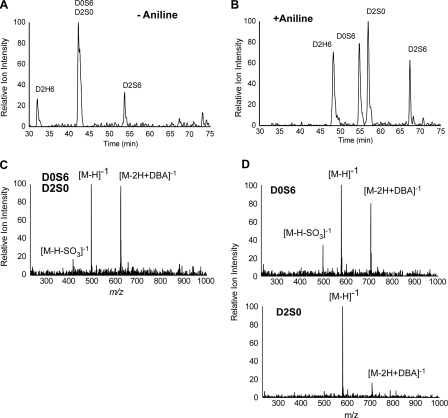
Aniline Derivatization Enhances Detection and Resolution of HS and CS Disaccharides—Derivatization of disaccharides with aniline led to enhanced resolution by liquid chromatography and greater sensitivity of detection by mass spectrometry. For example, the isobaric disaccharides designated D0S6 and D2S0 in their underivatized states did not separate by reversed-phase chromatography on C18 resin (Fig. 2A), whereas the aniline derivatives completely resolved from each other (Fig. 2B). Higher sensitivity was also observed for the aniline derivatives presumably due to greater desolvation at the ion source of conjugated disaccharides, which eluted at a higher concentration of methanol (notice the increase in peak height of D2H6 and D2S6 in Fig. 2B compared with Fig. 2A). The extracted ion current (XIC) for m/z corresponding to the underivatized forms of D0S6 and D2S0 showed the presence of the free molecular ions (m/z = 496.1 [M-H]-1), adduction ions formed with the ion pairing reagent (DBA) (m/z = 625.1 [M-2H+DBA]-1), and a small amount of desulfated species (m/z = 416.1 [M-H-SO3]-1) caused by in-source fragmentation (Fig. 2C), but the relative contribution of each disaccharide to the XIC could not be determined. However, the separated aniline derivatives had mass spectra that differed significantly (Fig. 2D). The aniline derivative of D0S6 formed DBA adducts more readily than the derivative of D2S0 and underwent partial in-source desulfation to a greater extent, which presumably reflects structural differences between these isobaric disaccharides.
FIGURE 2.
Enhanced detection and resolution of disaccharides. A, the accumulated XIC for disulfated disaccharide ions present in equimolar mixtures of untagged D2H6, D0S6, D2S0, and D2S6 are shown. The accumulative XIC is the summation of individual ionic currents for the free molecular ion, [M-H]-1 (m/z = 496), the adduction ion formed with the ion-pairing reagent, [M-2H+DBA]-1 (m/z = 625), and in-source desulfation of D2S6 [M-H-SO3]-1 (m/z = 496). The two isobaric isomers D0S6 and D2S0 did not resolve. B, the accumulated XIC for D2H6, D0S6, D2S0, and D2S6 tagged with [13C6]aniline is shown. The accumulative XIC is the summation of individual ionic currents for the free molecular ion, [M-H]-1 (m/z = 573), the adduction ion formed with the ion-pairing reagent, [M-2H+DBA]-1 (m/z = 702), and the in-source desulfation of D2S6 [M-H-SO3]-1 (m/z = 573). D0S6 and D2S0, which were not resolved in their underivatized state, are resolved as aniline derivatives. C, corresponding mass spectrum for underivatized D0S6 and D2S0 showing the free molecular ions, [M-H]-1, adduction ions formed with the ion-pairing reagent, [M-2H+DBA]-1, and desulfated in-source fragment ions [M-H-SO3]-1. D, the corresponding mass spectra for D0S6 and D2S0 tagged with [13C6]aniline.
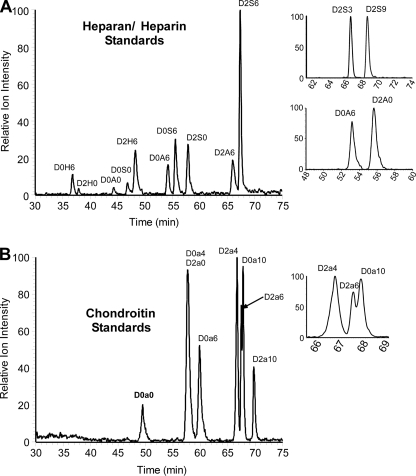
To determine if all of the disaccharides typically found in naturally occurring HS chains can be separated and quantitated by GRIL-LC/MS, mixtures of equimolar amounts of disaccharide standards were analyzed (Fig. 3A). Peak assignments were made by comparing the retention times and mass spectra to individual aniline-tagged standards (Table 1). The results showed that all of the HS disaccharide standards resolved with the exception of the trisulfated disaccharides D2S3 and D2S6, which co-elute. In all tissues analyzed to date, D2S3 is either absent or exists in very low amounts compared with D2S6 (12). Thus, its contribution to the combined peak can usually be considered negligible. If necessary, the presence of D2S3 can be determined by analyzing the disaccharides in their underivatized states in which they are fully resolved by conventional LC/MS (12). GRIL-LC/MS can detect the 3-O-sulfated disaccharides D2S3 and D2S9, which were prepared from CHO cells transduced with human Hs3st-3a (12). Two other disaccharide containing 3-O-sulfated glucosamine units (D0S3, D0S9) are not yet available for analysis. Other relatively rare disaccharides containing unsubstituted amino groups (D0H6, D2H0, and D2H6) were also resolved.
FIGURE 3.
GRIL-LC/MS analysis of aniline-tagged HS and CS disaccharides. A, XIC chromatographs for aniline-tagged HS disaccharide standards resolved by LC/MS. The panels on the right show that the two 3-O-sulfated HS disaccharides D2S3 and D2S9 can also be resolved after aniline tagging as can the two N-acetylated isobaric isomers D0A6 and D2A0. The only standard not resolved was D0H0, which co-elutes with the salt peak, thus suppressing its ionic signature. B, XIC chromatographs for CS disaccharide standards after aniline tagging. The panel on the right expands the region where disulfated CS disaccharide standards elute showing partial separation of D2a6 and D0a10. The two isobaric isomers D0a4 and D2a0 did not separate under these conditions but can be easily distinguished by CID (see Fig. 7). All XIC traces were generated with the appropriate m/z values for the HS and CS disaccharides listed in Table 1.
All disaccharides derived from CS by chondroitinase ABC digestion also resolved, with the exception of the two monosulfated isomers D0a4 and D2a0 (Fig. 3B). The latter is a component of dermatan sulfate. As discussed below, collision-induced dissociation (CID) can be used to distinguish these isobaric species. This approach is not useful for discriminating the HS disaccharides D2S3 and D2S6, because desulfation predominates during ionic fragmentation of these more highly sulfated species.
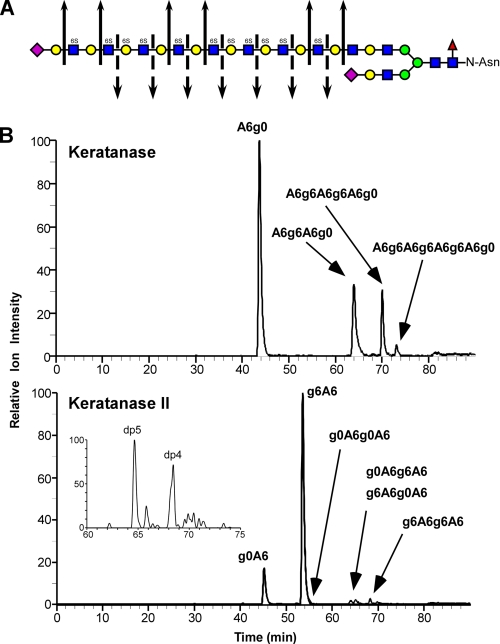
Qualitative Analysis of GAG Chain Structure Using GRIL-LC/MS—Corneal KS consists of variably sulfated poly-N-acetyllactosamine units, bearing sulfate groups at C6 of N-acetylglucosamine and galactose residues. The non-reducing end termini can also include sialic acid linked α2-3 to a penultimate galactose residue (Fig. 4A). To demonstrate the suitability of GRIL-LC/MS for analyzing KS oligosaccharides, a sample of bovine corneal KS was subjected to exhaustive digestion with either keratanase or keratanase II, which depolymerize the chains dependent on the sulfation of the N-acetylglucosamine and galactose subunits (Fig. 4A). The digestion products were then labeled with [12C6]aniline and analyzed by LC/MS. Keratanase cleaves between unsulfated galactose and 6-O-sulfated or unsulfated N-acetylglucosamine (Fig. 4A) resulting in the release of sequential A6g0 disaccharides and blocks of A6g6 oligosaccharides (Fig. 4B, upper panel). The results show that these KS I chains contain significant amounts of fully sulfated blocks up to the size of octamers, which are separated by one or more A6g0 disaccharides. Keratanase II cleaves between 6-O-sulfated N-acetylglucosamine and galactose with or without sulfation (Fig. 4A) resulting in the release of both g0A6 and g6A6 disaccharides (Fig. 4B, lower panel). In addition, small amounts of a number of tetrasaccharides and a single pentasaccharide were detected, consistent with previous studies (32). The pentasaccharide has a m/z value consistent with the sialylated species Neu5Ac-g0A6g0A6 (Fig. 4B, lower panel inset), which has previously been shown to be the predominant non-reducing end terminal structure in corneal KS(32). Thus, GRIL-LC/MS can be used to evaluate block structure and disaccharide composition of KS GAG chains. More importantly, using differential isotope labeling of two samples, GRIL-LC/MS can be a very convenient and effective means of showing ratiometric differences in KS fine structure between two samples differentially labeled with aniline containing different stable isotopes (see Fig. 6C for an example using HS isolated from two genetically different mice).
FIGURE 4.
Qualitative structural analysis of KS GAG chains using GRIL-LC/MS. A, generalized structure of corneal KS I showing the cleavage sites for keratanase (solid arrows) and keratanase II (broken arrows). The symbols used for monosaccharides subunits: sialic acid (purple diamond), galactose (yellow circle), N-acetylglucosamine (blue squares), mannose (green circles), and fucose (red triangle) are as previously described (grtc.ucsd.edu/symbol.html). The presence of 6-O-sulfate is also indicated (6S) above each sulfated monosaccharide. B, bovine corneal KS was digested with either keratanase (“Experimental Procedures”), aniline-tagged and subjected to LC/MS for analysis of digestion products. The chromatographs show the relative ion intensity (XIC) of digestion products eluting from the C18 column. The indicated peak assignments were based on retention time, mass, and the known activity of the keratanases used. The inset in the lower panel shows the elution of two species released by keratanase II digestion with degree of polymerization of 4 and 5 (dp4 and dp5) based on mass. The XIC traces were generated with the appropriate m/z values for KS disaccharides listed in Table 1 and for m/z values for the major expected ions of oligosaccharides released after keratanase digestion. For keratanase, these included: dp4 ([M-3H+DBA]-2, m/z = 596), dp6 ([M-5H+3DBA]-2, m/z = 987), and dp8 ([M-5H+3DBA]-2, m/z = 1250). For keratanase II these included dp4 species g0A6g0A6 ([M-2H]-2, m/z = 491.5), g0A6g6A6 and g6A6g0A6 ([M-3H+DBA]-2, m/z = 596), and g6A6g6A6 ([M-4H+2DBA]-2, m/z = 700.5). The inset XIC trace in the Keratanase II panel was generated for the putative dp5 species Neu5Ac-g0A6g0A6 ([M-2H]-2, m/z = 637), which also reveals a putative minor adduction ion for g6A6g6A6 ([M-3H+DBA]-2, m/z = 636). The mass spectra of the small peak eluting just after 80 min in the upper chromatogram and the small peak eluting after the putative g6A6g6A6 peak in the lower chromatogram do not correspond to any known keratan sulfate-derived oligosaccharides and are most likely contaminants.
FIGURE 6.
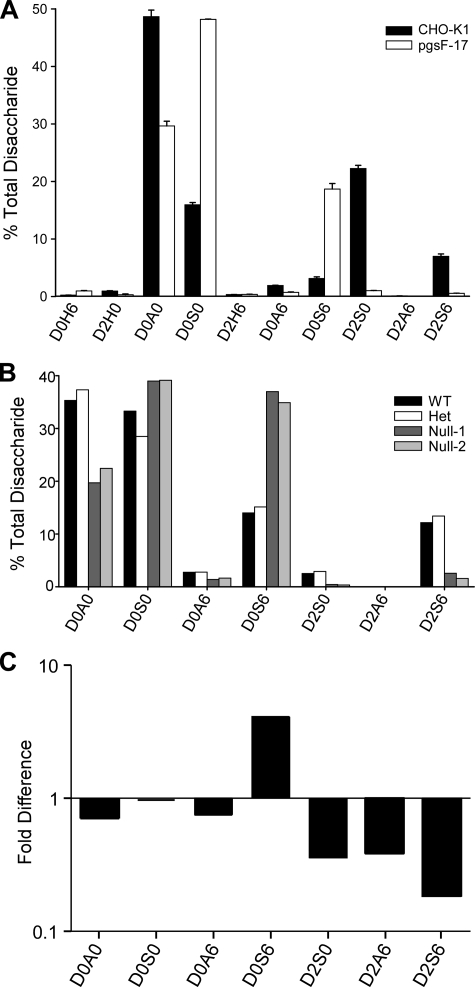
GRIL-LC/MS analysis of disaccharides in wild-type and Hs2st-deficient mutant cells and mice. A, the disaccharide profile for HS isolated from wild-type CHO-K1 cells and from pgsF-17 mutant cells (Hs2st-deficient), error bars represent the standard error of the means (n = 3). The results are reported as percentage of total disaccharide. B, the disaccharide profile for HS isolated from Hs2st-/-, Hs2st+/-, or wild-type mouse embryos. The results are reported as percentage of total disaccharide recovered from each sample. C, the disaccharides derived from HS isolated from the liver of Hs2stf/fAlbCre+ and Hs2stf/fAlbCre- mice were tagged with [13C6]aniline or [12C6]aniline, respectively. The samples were mixed and then analyzed by LC/MS. The ratio of 13C and 12C recovered for each disaccharide is shown.
Quantitative Analysis of Heparin and HS Disaccharides from Cells and Tissues—The advantage of differential mass tagging using aniline containing different stable isotopes (12C6 or 13C6). is that samples tagged with different isotopically labeled anilines can be mixed and simultaneously analyzed to yield quantitative comparisons. The addition of the [12C6]aniline group adds 77 atomic mass units to the mass of the underivatized free molecular ion ([M-H]-1) as well as to any adducts or decomposition products, whereas [13C6]aniline group adds 83 atomic mass units (for example, compare Fig. 2C and 2D). The difference of 6 atomic mass units between [12C6]aniline- and [13C6]aniline-tagged disaccharides is readily detected by mass spectrometry.
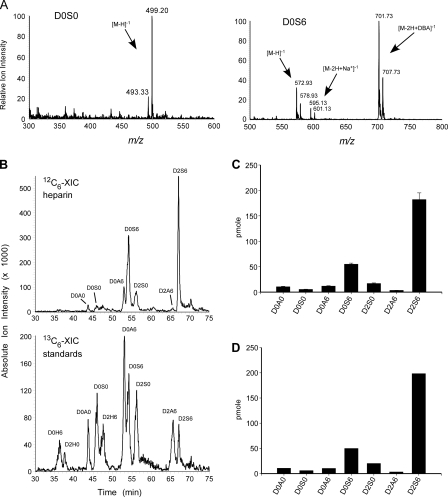
To use GRIL-LC/MS for quantitative measurements, all available disaccharide standards were prepared with [13C6]aniline. A sample of heparin was then digested with heparin lyases, and the liberated disaccharides were derivatized with [12C6]aniline. 25 pmol of each [13C6]aniline-tagged disaccharide standard was mixed with the [12C6]aniline-tagged heparin disaccharides and separated by LC/MS. As expected, LC/MS showed that the mass difference between each pair of [12C6]aniline-and [13C6]aniline-tagged disaccharides was 6 atomic mass units, which was reflected in the free molecular ion and all adduction ions (e.g. D0S6 in Fig. 5A), which are produced independently of heavy and light isotopes. The accumulative XIC for each isotope was then determined, by summing all of the [12C6]- or [13C6]aniline-tagged XIC values for the standard disaccharides listed in Table 1 (and any in-source desulfation products). Because the [13C6]aniline-tagged standards are added at known amounts and behave in the same way as the corresponding [12C6]aniline-tagged disaccharides, the actual amount of these residues can be calculated from the ratio of the XIC profiles of each isotope (Fig. 5B). For example, the peak area for the D0S6 standard shown in the 12C6-XIC trace is 2.38-fold that of [13C6]aniline-tagged D0S6 shown in the 13C6-XIC trace (Fig. 5B), which is reflected by the relative ion intensities of both molecular and adduction ions seen in the corresponding mass spectrum (Fig. 5A). Thus, the amount of D0S6 in the sample can be calculated as 2.38*25 pmol = 59.5 pmol. The results from three independent analyses of commercially obtained porcine intestinal mucosal heparin are shown in Fig. 5C, with standard errors for GRIL-LC/MS of 2-8% (n = 3). The composition was nearly identical to that obtained by anion-exchange chromatography and post-column derivatization with 2-cyanoacetamide (post column fluorescence derivatization, see “Experimental Procedures”) (Fig. 5D). The background in the XIC profile was due to unidentified peaks, a few of which had masses similar to those predicted for known tetrasaccharides, but further analysis will be necessary to confirm their identity.
FIGURE 5.
GRIL-LC/MS quantitation of porcine intestinal mucosal heparin. Disaccharides derived from heparin were tagged with [12C6]aniline and mixed with [13C6]aniline disaccharide standards (25 pmol each). A, the mass spectra for a representative low abundance disaccharide, D0S0, and a higher abundance disaccharide, D0S6, are shown. Note the 6 atomic mass units difference between the [12C6]aniline-tagged residue and the corresponding [13C6]aniline-tagged standard for the free molecular ions ([M-H]-1), the sodium adducts ([M-2H+Na+]-1), and the ion pairing reagent adducts ([M-2H+DBA]-1). Adduction ions are typically detected for disaccharides with two or more sulfates (compare the mass spectra for D0S0 and D0S6, see Table 1). B, the individual XIC profiles for the [12C6]aniline-tagged sample and [13C6]aniline-tagged standards for one of three separate analyses. The XIC traces were generated using the m/z values for HS disaccharide standards listed in Table 1. C, GRIL-LC/MS determined disaccharide composition of porcine heparin (n = 3), error bars represent the standard error. D, analysis of an identical sample using the anion-exchange high performance liquid chromatography with post column derivatization and fluorescence detection. The preparation of heparin used in this study did not contain measurable amounts of D0H0, D0H6, D2H0, or D2H6 detected by either LC/MS-GRIL or post column derivatization with 2-cyanoacetamide. The rare disaccharide D2A0, which is also detectable by GRIL-LC/MS (Fig. 3A), was not analyzed in this experiment.
To establish the accuracy and the reliability of the GRIL-LC/MS analysis method, we evaluated the disaccharide profiles of HS derived from wild-type CHO cells and the mutant CHO line pgsF-17, deficient in uronyl 2-O-sulfotransferase activity (Hs2st). The results along with the standard errors for three independent HS extractions from wild-type and mutant cells are presented in Fig. 6A. As expected, the mutant exhibited very little of the 2-O-sulfated disaccharides D2S0 and D2S6, and their decrease correlated with a concomitant increase in N-sulfated and 6-O-sulfated disaccharides D0S0 and D0S6, confirming previously published results (26). We also analyzed the disaccharide composition of HS extracted from wild-type mouse embryos and embryos bearing null alleles of Hs2st (Fig. 6B). Although the differences in disaccharide compositions of wild-type and heterozygous embryos were negligible, the HS composition from the homozygous null animals showed a loss of 2-O-sulfated disaccharides and an associated rise in D0S0 and D0S6 residues (33). Thus similar changes in the disaccharide profiles occurred in both CHO and mouse Hs2st mutants. The low, but statistically relevant, amount of D2S6 detected in both homozygous null embryos was verified by post column fluorescence derivatization (data not shown). The source of this material is unknown.
The reliability of GRIL-LC/MS quantitation of disaccharides was further evaluated by measuring the recovery of individual disaccharides as a function of sample concentration using liver HS. GRIL-LC/MS analysis was performed at four different dilutions of the same sample. The results showed a strong linear correlation (R2 values from 0.9954 to 1.000) between disaccharide concentration and the disaccharide/standard XIC ratio for all of the disaccharides detected in the sample (supplemental Fig. S2). As a further test of analytical recovery, a bolus of heparin byproduct (70 ng) was added to a heart tissue homogenate (containing ∼250 ng of HS) and to an equal volume of phosphate-buffered saline. The two samples and an equal aliquot of heart homogenate were subjected to GAG purification, heparan lyase digestion, and analysis by GRIL-LC/MS. The results showed that the byproduct was recovered from the tissue sample and from phosphate-buffered saline in the same proportion (supplemental Fig. S3). The average difference in recovery of the individual disaccharides in the combined sample and the control was <5%. Overall, these results demonstrate the high degree of reliability provided by this technique to carry out comparative analysis.
GRIL-LC/MS also permits comparative analyses of samples without the introduction of standards thus showing relative differences for all disaccharides and not just those for which standards are available. In this format, a sample of disaccharides derived from one experimental condition was tagged with [12C6]aniline and a sample from a different set of conditions was tagged with [13C6]aniline. The samples were then mixed and analyzed by LC/MS, and the ratio of XICs for each isotope was determined. Fig. 6C shows the analysis of liver HS from wild-type mice (Hs2stf/fAlbCre-) tagged with [12C6]aniline and liver HS from mice bearing a hepatocyte-specific deletion of the Hs2st gene (Hs2stf/fAlbCre+) tagged with [13C6]aniline. The analysis showed diminished amounts of all disaccharides containing 2-O-sulfated uronic acids and an increase in D0S6. Complete loss of 2-O-sulfated disaccharides did not occur as in the CHO cell mutant due to incomplete deletion of Hs2st in the hepatocytes and the presence of wild-type HS from ∼10% untargeted endothelial cells in the liver preparations. No significant increase in D0S0 occurred as in CHO and whole mouse embryos, suggesting liver-specific changes in composition (Fig. 6, compare C to A and B).
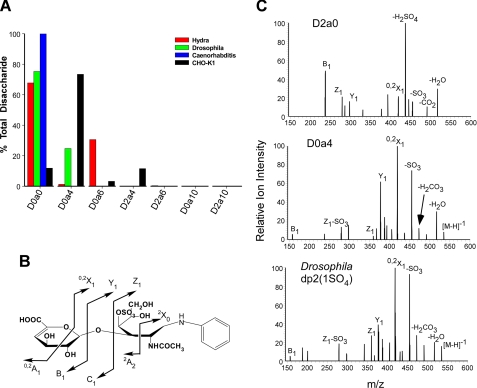
Survey of GAGs from Model Organisms—To evaluate the differences in GAG chain fine structure present in evolutionarily distant organisms, we analyzed GAGs isolated from H. vulgaris (cnidarian), C. elegans (nematode), D. melanogaster (arthropod), and CHO cells (vertebrate). Chondroitin chains were depolymerized with chondroitinase ABC, tagged with [12C6]aniline, mixed with [13C6]aniline-tagged standards (Table 1), and the mixture was analyzed by GRIL-LC/MS. Each invertebrate organism showed a distinct disaccharide composition. In all three invertebrates, D0a0 dominated the disaccharide composition, indicating a low overall degree of sulfation compared with CHO cells (Fig. 7A). H. vulgaris chondroitin contained ∼30% D0a6 and <2% D0a4, in contrast to a recent report on chondroitin derived from Hydra magnipapillata showing equal amounts of both 4- and 6-O-sulfated disaccharides, which may reflect species-specific differences (5). D. melanogaster chondroitin contained 25% monosulfated disaccharide resembling D0a4 or D2a0, two isobaric disaccharides that co-elute. To determine the dominant species that elutes at this position, both D0a4 and D2a0 standards were subjected to CID tandem MS/MS. From the mass of the inter-ring cleavage products, these two isobaric species can be easily distinguished because the position of the sulfate differs (Fig. 7B). For example, the B1 ion for D2a0 has an m/z = 237 and the B1 ion for D0a4 has an m/z = 157. When compared, the CID spectrum of the unknown monosulfated species in the D. melanogaster most closely resembled that of D0a4 (Fig. 7C). The presence in the CID spectrum of Y1 with m/z = 377, Z1 with m/z = 359, and B1 with m/z = 157, and lack of the diagnostic product ion for D2a0 (18, 34) show that the predominant and perhaps only species eluting at this position has sulfate on the hexosamine side of the glycosidic bond. C. elegans chondroitin consists entirely of D0a0, as described previously (3, 35, 36). CHO cell chondroitin consists primarily of D0a4, but interestingly ∼3% D0a6, ∼10% D0a0, and ∼10% D2a4 were detected. The presence of D2a4 suggests the presence of either 2-O-sulfated iduronic or glucuronic acids, which have not been previously reported in CHO cells.
FIGURE 7.
GRIL-LC/MS disaccharide analysis of CS isolated from Hydra, nematodes, and arthropods. A, chondroitins from H. vulgaris, D. melanogaster, C. elegans, and CHO-K1 cells was enzymatically depolymerized and tagged with [12C6]aniline. An equimolar mixture (25 pmol) of [13C6]aniline-tagged CS/DS disaccharide standards was added prior to LC/MS analysis. Results are shown as the percentage of total disaccharides recovered. B, schematic showing possible tandem mass spectra intra-and inter-ring cleavages of aniline-conjugated D0a4, using the nomenclature proposed by Domon and Costello (49). C, tandem mass spectra of D2a0, D0a4, and the uncharacterized monosulfated disaccharide (Drosophila dp2(1SO4)) present in the D. melanogaster chondroitin sample.
Previous studies of HS from C. elegans and D. melanogaster and a recent report on H. magnipapillata suggested that these invertebrates produce an array of disaccharides similar to those found in vertebrates (3-5, 35, 37). However, inspection of the published data suggested that other disaccharides might be present, because several unidentified peaks were present in the chromatograms. Application of GRIL-LC/MS revealed the presence of three N-unsubstituted disaccharides (D0H6, D2H6, and D2H0), which fully resolve (Fig. 3A). Due to the lack of any sulfates on D0H0, this species does not absorb to the C18 matrix and is thus not significantly retained by the reversed-phase column. Its co-elution with salts causes suppression of the ion current, and attempts to quantify D0H0 failed. To circumvent this problem, disaccharides bearing free amino groups were amidated (supplemental Fig. S4A) with propionic anhydride (20), which results in stronger interaction with the C18 matrix and separation from salts in the sample. Propionylation generates a product (D0R0) with a mass 56 atomic mass units larger than the corresponding free amino disaccharide (supplemental Fig. S4B). Furthermore, aniline-tagged D0R0 separates completely from aniline-tagged D0A0 (supplemental Fig. S4C). These two unsulfated disaccharides are structurally similar but vary in the length of the amide substituent.
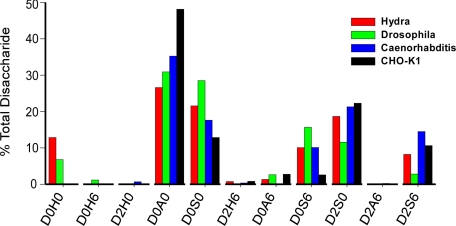
Derivatization of disaccharides from D. melanogaster and H. vulgaris HS with both propionic anhydride and aniline demonstrated surprisingly high levels of D0H0 (6.8% and 12.8%, respectively) (Fig. 8), which were absent from C. elegans and CHO cell HS. Minor amounts of D0H6 were present in D. melanogaster (1.2%) but absent in other samples. D2H6 was present in trace amounts in all samples except HS from D. melanogaster.
FIGURE 8.
GRIL-LC/MS disaccharide analysis of HS from Hydra, nematodes, and arthropods. HS samples were depolymerized by exhaustive digestion with a mixture of heparan lyases, reacted with propionic anhydride, tagged with [12C6]aniline, and mixed with [13C6]aniline-tagged HS disaccharide standards. The proportion of each disaccharide in the samples was then determined by comparison to the standards, including [13C6]aniline-tagged standards for propionylated N-unsubstituted disaccharides. Significant levels of D0H0 were detected in H. vulgaris and D. melanogaster HS but not in C. elegans or CHO cells.
DISCUSSION
In this report we demonstrate the use of differential isotope tagging and LC/MS (GRIL-LC/MS) to facilitate quantitative compositional analysis of disaccharides derived from depolymerized GAG chains. Reductive amination with isotopically labeled anilines is rapid, quantitative, and sensitive, and when coupled to reversed-phase ion-pairing liquid chromatography permits separation of nearly all known disaccharides, including difficult to separate species bearing unsubstituted glucosamine residues. Based on titration studies, the limit of sensitivity was 1 pmol/disaccharide (see Fig. 1B), a value 5-10 times more sensitive than the post column fluorescence detection system using 2-cyanoacetamide. Our results demonstrate the high sensitivity and wide linear range (between 1 pmol and 3 nmol) of this new method, which makes it an attractive alternative to other techniques for conducting compositional GAG analysis especially on small and difficult to obtain samples. We validated the method by analysis of enzymatically depolymerized KS and heparin chains, HS from mutant and wild-type CHO cells, HS from whole mouse liver, HS from mouse embryos bearing null alleles of Hs2st, and HS from a tissue-specific mutation of Hs2st in mouse hepatocytes. Although the compositions that were obtained resembled that obtained by traditional methods, GRIL-LC/MS revealed the presence of previously undescribed disaccharides in CHO cells (D2a4) and invertebrates (D0H0, D0H6, and D2H6). Furthermore, because a second dimension of analysis by MS/MS can be imposed, GRIL-LC/MS provides structural information that methods based solely on co-chromatography with standards do not. Another advantage is that proportional comparisons between samples can be made by mixing samples tagged with different isotopically labeled anilines, which eliminates differential effects due to varying levels of contaminants.
When comparing the capabilities of GRIL-LC/MS with those of other detection methods such as post column fluorescence derivatization, we found GRIL-LC/MS to be more reliable for analyzing disaccharides present in low abundance, easier to set up and maintain (assuming the availability of a mass spectrometer) and easier to troubleshoot if problems arise with the derivatization reaction or sample preparation (because the presence of underivatized species or problematic contaminants can be detected). The sensitivity of this method is only limited by the mass spectrometer used for the analysis. We expect a further dramatic increase in sensitivity when a more advanced mass spectrometer is used (femtomoles). Furthermore, because of the multiparametric data generated by LC/MS, this new technique is significantly more reliable for detecting low abundance species whose signal can be obscured by co-eluting contaminates.
GRIL-LC/MS is also a significant enhancement over LC/MS procedures that rely on linear equations to correlate the concentration of underivatized disaccharides with the detection of molecular standards (10, 17, 18, 20). The presence of non-GAG contaminants, which is unavoidable in many samples, can affect analyte ionization and the calculated results. GRIL-LC/MS doesn't suffer from this limitation, because standards for each disaccharide are included in each sample and the sample disaccharide and its corresponding standard co-elute. Thus, buffer conditions and contaminants that affect disaccharide ionization will affect both the sample and the standard to the same extent and the ratio of disaccharide to standard remains the same regardless of changing LC/MS conditions.
GRIL-LC/MS also can be used for other types of GAG analyses. For example, the method easily resolves disaccharides derived from hyaluronan using either lyases or hydrolases.6 Furthermore, the non-reducing end of the chain resolves from the internal disaccharides, making measurement of chain length possible. Thus, the technique should also allow partial sequencing of short oligosaccharides, e.g. by mass tagging the reducing terminus with [13C6]aniline followed by lyase digestion and mass tagging the liberated disaccharides with [12C6]aniline. The suitability of GRIL-LC/MS for nitrous acid digests of GAGs such as HS, which preserves the stereochemistry of the uronic acid residues but converts the amino sugars to anhydromannose, has yet to be determined. Because standards for nitrous acid-liberated disaccharides are not readily available, the potential of GRIL-LC/MS for analyzing nitrous acid-liberated disaccharides samples will require further work.
Most organisms express one or more members of the HS 3-O-sulfotransferasese family, which generates 3-O-sulfated glucosamine residues, and some CS contains 3-O-sulfated glucuronic acids (1, 38). In theory, GRIL-LC/MS should be able to separate and quantify these disaccharides, but standards are not readily available to optimize chromatography conditions or to use as mass standards. Thus GRIL-LC/MS, in the absence of suitable 3-O-sulfated standards, cannot be used to quantitate these particular species. It is important to note that all other analysis schemes that rely on comparison to standards, such as post column fluorescence derivatization, also suffer from this limitation. However, expressing 3-OST-3A in CHO cells yield two disaccharides, D2S3 and D2S9, which can be easily separated by GRIL-LC/MS analysis (Fig. 3A). Although other 3-O-sulfated disaccharides might be obtained in this way, chemoenzymatic methods may be needed to generate sufficient quantities of rare disaccharides for further analysis and for preparation as heavy isotope-tagged standards.
Using GRIL-LC/MS, we found significant differences in chondroitins from H. vulgaris, C. elegans, D. melanogaster, and CHO cells. The major sulfated disaccharides present in D. melanogaster CS were D0a4, whereas H. vulgaris CS contained D0a6 as its major sulfated disaccharide. In both cases these sulfated disaccharides represented <25-30% of the total, and the remainder was unmodified D0a0. CHO cell chondroitin is much more highly sulfated with D0a4 being the principle species. CHO cells also make a significant amount of D2a4, which has not been previously described in this cell line. Chondroitin chains from C. elegans are unsulfated and therefore consists of 100% D0a0, as previously observed (3, 35, 36).
GRIL-LC/MS analysis of HS from D. melanogaster and H. vulgaris revealed significant structural differences highlighted by residues containing N-unsubstituted amines. Disaccharides containing N-unsubstituted glucosamine residues are relatively rare in vertebrate HS (≤5%), and their detection depends on the source of material and the method of analysis (sensitivity to high pH nitrous acid, reactivity to NHS-biotin, binding of specific antibodies) (20, 39-46). As shown here, D. melanogaster and H. vulgaris HS possess substantial amounts of free amino-containing disaccharides, consisting almost entirely of D0H0. Presumably, D0H0 arises from incomplete GlcNAc N-deacetylation/N-sulfation by GlcNAc N-deacetylase/N-sulfotransferases encoded in these organisms, although we cannot exclude the possibility of a specific N-deacetylase or N-sulfatase. The significance of high levels of disaccharides with free amino groups in HS from these species is not known, but may signal the existence of an evolutionarily ancient class of biologically important functional motifs. Earlier studies have shown that GAGs are expressed in several other invertebrates (37, 47, 48). While the earliest diverging animal phylum, Porifera (sponges) appears to express only CS, the expression of both CS and HS is manifest throughout the rest of the animal kingdom (37). The advantage of GRIL-LC/MS is that even minor amounts of free amine-containing disaccharides can be detected, e.g. D0H6 in D. melanogaster. Evaluation of the type and degree of GAG chain modification across multiple species will help determine if a correlation exists between GAG chain modifications and biological function during evolution.
Supplementary Material
Acknowledgments
We thank Kristin Stanford and Rusty Bishop for providing heparan sulfate samples from liver. Analysis of heparin by the post-column derivatization method was done by Dr. Sulabha Argade, and all mass spectrometry experiments were preformed in the Glycotechnology Core Resource in the Glycobiology Research and Training Center at the University of California at San Diego.
This work was supported, in whole or in part, by National Institutes of Health Grant GM33063 and HL57345 (to J. D. E.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental additional text and Figs. S1-S4.
Footnotes
The abbreviations used are: GAG, glycosaminoglycan; GRIL, glycan reductive isotope labeling; LC/MS, liquid chromatography/mass spectrometry; CID, collision-induced dissociation; HS, heparan sulfate; CS, chondroitin sulfate; DS, dermatan sulfate; DSC, disaccharide structure code; CHO, Chinese hamster ovary; Hs2st, uronyl 2-O-sulfotransferase; GlcA, d-glucuronic acid; GlcNS, N-sulfo-d-glucosamine; GlcNH2, N-unsubstituted d-glucosamine; XIC, extracted ion current; DBA, dibutylamine; dp, degree of polymerization.
B. Xia, C. L. Feasley, G. P. Sachdev, D. F. Smith, and R. D. Cummings, submitted for publication.
O. Garner, B. E. Crawford, J. Castagnola, D. Song, R. Lawrence, J. R. Brown, J. R. Bishop, D. Y. Zhang, L. Wang, and J. D. Esko, unpublished results.
Throughout this report we use a shorthand nomenclature called “disaccharide structure code (DSC)” to simplify the description of glycosaminoglycan disaccharides. See Lawrence et al. (25) for details and Table 1.
R. Lawrence, R. Gallo, and J. D. Esko, unpublished results.
References
- 1.Kinoshita-Toyoda, A., Yamada, S., Haslam, S. M., Khoo, K. H., Sugiura, M., Morris, H. R., Dell, A., and Sugahara, K. (2004) Biochemistry 43 11063-11074 [DOI] [PubMed] [Google Scholar]
- 2.Esko, J. D., and Selleck, S. B. (2002) Annu. Rev. Biochem. 71 435-471 [DOI] [PubMed] [Google Scholar]
- 3.Toyoda, H., Kinoshita-Toyoda, A., and Selleck, S. B. (2000) J. Biol. Chem. 275 2269-2275 [DOI] [PubMed] [Google Scholar]
- 4.Toyoda, H., Kinoshita-Toyoda, A., Fox, B., and Selleck, S. B. (2000) J. Biol. Chem. 275 21856-21861 [DOI] [PubMed] [Google Scholar]
- 5.Yamada, S., Morimoto, H., Fujisawa, T., and Sugahara, K. (2007) Glycobiology 17 886-894 [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita, A., Yamada, S., Haslam, S. M., Morris, H. R., Dell, A., and Sugahara, K. (1997) J. Biol. Chem. 272 19656-19665 [DOI] [PubMed] [Google Scholar]
- 7.Kim, Y. S., Ahn, M. Y., Wu, S. J., Kim, D. H., Toida, T., Teesch, L. M., Park, Y. M., Yu, G. Y., Lin, J. H., and Linhardt, R. J. (1998) Glycobiology 8 869-877 [DOI] [PubMed] [Google Scholar]
- 8.Yang, H. O., Gunay, N. S., Toida, T., Kuberan, B., Yu, G. L., Kim, Y. S., and Linhardt, R. J. (2000) Glycobiology 10 1033-1040 [DOI] [PubMed] [Google Scholar]
- 9.Kuberan, B., Lech, M., Zhang, L. J., Wu, Z. L. L., Beeler, D. L., and Rosenberg, R. D. (2002) J. Am. Chem. Soc. 124 8707-8718 [DOI] [PubMed] [Google Scholar]
- 10.Saad, O. M., and Leary, J. A. (2003) Anal. Chem. 75 2985-2995 [DOI] [PubMed] [Google Scholar]
- 11.Saad, O. M., and Leary, J. A. (2004) J. Am. Soc. Mass Spectrom. 15 1274-1286 [DOI] [PubMed] [Google Scholar]
- 12.Lawrence, R., Kuberan, B., Lech, M., Beeler, D. L., and Rosenberg, R. D. (2004) Glycobiology 14 467-479 [DOI] [PubMed] [Google Scholar]
- 13.Saad, O. M., and Leary, J. A. (2005) Anal. Chem. 77 5902-5911 [DOI] [PubMed] [Google Scholar]
- 14.Barroso, B., Didraga, M., and Bischoff, R. (2005) J. Chromatogr. A 1080 43-48 [DOI] [PubMed] [Google Scholar]
- 15.Karlsson, N. G., Schulz, B. L., Packer, N. H., and Whitelock, J. M. (2005) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 824 139-147 [DOI] [PubMed] [Google Scholar]
- 16.Hitchcock, A. M., Costello, C. E., and Zaia, J. (2006) Biochemistry 45 2350-2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korir, A. K., Limtiaco, J. F., Gutierrez, S. M., and Larive, C. K. (2008) Anal. Chem. 80 1297-1306 [DOI] [PubMed] [Google Scholar]
- 18.Desaire, H., and Leary, J. A. (2000) J. Am. Soc. Mass Spectrom. 11 916-920 [DOI] [PubMed] [Google Scholar]
- 19.Saad, O. M., Ebel, H., Uchimura, K., Rosen, S. D., Bertozzi, C. R., and Leary, J. A. (2005) Glycobiology 15 818-826 [DOI] [PubMed] [Google Scholar]
- 20.Behr, J. R., Matsumoto, Y., White, F. M., and Sasisekharan, R. (2005) Rapid Commun. Mass Spectrom. 19 2553-2562 [DOI] [PubMed] [Google Scholar]
- 21.Yuan, J., Hashii, N., Kawasaki, N., Itoh, S., Kawanishi, T., and Hayakawa, T. (2005) J. Chromatogr. A 1067 145-152 [DOI] [PubMed] [Google Scholar]
- 22.Hitchcock, A. M., Yates, K. E., Shortkroff, S., Costello, C. E., and Zaia, J. (2007) Glycobiology 17 25-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowman, M. J., and Zaia, J. (2007) Anal. Chem. 79 5777-5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hitchcock, A. M., Yates, K. E., Costello, C. E., and Zaia, J. (2008) Proteomics 8 1384-1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence, R., Lu, H., Rosenberg, R. D., Esko, J. D., and Zhang, L. (2008) Nat. Methods 5 291-292 [DOI] [PubMed] [Google Scholar]
- 26.Bai, X. M., and Esko, J. D. (1996) J. Biol. Chem. 271 17711-17717 [DOI] [PubMed] [Google Scholar]
- 27.Bame, K. J., and Esko, J. D. (1989) J. Biol. Chem. 264 8059-8065 [PubMed] [Google Scholar]
- 28.Linhardt, R. J., Galliher, P. M., and Cooney, C. L. (1986) Appl. Biochem. Biotechnol. 12 135-176 [DOI] [PubMed] [Google Scholar]
- 29.Manzi, A. E., Norgard-Sumnicht, K., Argade, S., Marth, J. D., Van Halbeek, H., and Varki, A. (2000) Glycobiology 10 669-689 [DOI] [PubMed] [Google Scholar]
- 30.Guo, Y. C., and Conrad, H. E. (1988) Anal. Biochem. 168 54-62 [DOI] [PubMed] [Google Scholar]
- 31.Toyoda, H., Nagashima, T., Hirata, R., Toida, T., and Imanari, T. (1997) J. Chromatogr. B Biomed. Sci. Appl 704 19-24 [DOI] [PubMed] [Google Scholar]
- 32.Tai, G. H., Huckerby, T. N., and Nieduszynski, I. A. (1996) J. Biol. Chem. 271 23535-23546 [DOI] [PubMed] [Google Scholar]
- 33.Wilson, V. A., Gallagher, J. T., and Merry, C. L. (2002) Glycoconj. J. 19 347-354 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., Conrad, A. H., Tasheva, E. S., An, K., Corpuz, L. M., Kariya, Y., Suzuki, K., and Conrad, G. W. (2005) Invest. Ophthalmol. Vis. Sci. 46 1604-1614 [DOI] [PubMed] [Google Scholar]
- 35.Yamada, S., Van Die, I., Van den Eijnden, D. H., Yokota, A., Kitagawa, H., and Sugahara, K. (1999) FEBS Lett. 459 327-331 [DOI] [PubMed] [Google Scholar]
- 36.Hwang, H. Y., Olson, S. K., Esko, J. D., and Horvitz, H. R. (2003) Nature 423 439-443 [DOI] [PubMed] [Google Scholar]
- 37.Medeiros, G. F., Mendes, A., Castro, R. A. B., Baú, E. C., Nader, H. B., and Dietrich, C. P. (2000) Biochim. Biophys. Acta Gen. Subj. 1475 287-294 [DOI] [PubMed] [Google Scholar]
- 38.Sugahara, K., Tanaka, Y., Yamada, S., Seno, N., Kitagawa, H., Haslam, S. M., Morris, H. R., and Dell, A. (1996) J. Biol. Chem. 271 26745-26754 [DOI] [PubMed] [Google Scholar]
- 39.Norgard-Sumnicht, K. E., and Varki, A. (1995) J. Biol. Chem. 270 12012-12024 [DOI] [PubMed] [Google Scholar]
- 40.Van Den Born, J., Gunnarsson, K., Bakker, M. A. H., Kjellén, L., Kusche-Gullberg, M., Maccarana, M., Berden, J. H. M., and Lindahl, U. (1996) J. Biol. Chem. 270 31303-31309 [DOI] [PubMed] [Google Scholar]
- 41.Toida, T., Yoshida, H., Toyoda, H., Koshishi, I., Imanari, T., Hileman, R. E., Fromm, J. R., and Linhardt, R. J. (1997) Biochem. J. 322 499-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding, K., Jönsson, M., Mani, K., Sandgren, S., Belting, M., and Fransson, L. Å. (2001) J. Biol. Chem. 276 3885-3894 [DOI] [PubMed] [Google Scholar]
- 43.Westling, C., and Lindahl, U. (2002) J. Biol. Chem. 277 49247-49255 [DOI] [PubMed] [Google Scholar]
- 44.Holmborn, K., Ledin, J., Smeds, E., Eriksson, I., Kusche-Gullberg, M., and Kjellen, L. (2004) J. Biol. Chem. 279 42355-42358 [DOI] [PubMed] [Google Scholar]
- 45.van den Born, J., Salmivirta, K., Henttinen, T., Ostman, N., Ishimaru, T., Miyaura, S., Yoshida, K., and Salmivirta, M. (2005) J. Biol. Chem. 280 20516-20523 [DOI] [PubMed] [Google Scholar]
- 46.Wei, Z., Lyon, M., and Gallagher, J. T. (2005) J. Biol. Chem. 280 15742-15748 [DOI] [PubMed] [Google Scholar]
- 47.Nader, H. B., Ferreira, T. M., Paiva, J. F., Medeiros, M. G., Jeronimo, S. M., Paiva, V. M., and Dietrich, C. P. (1984) J. Biol. Chem. 259 1431-1435 [PubMed] [Google Scholar]
- 48.Cesaretti, M., Luppi, E., Maccari, F., and Volpi, N. (2004) Glycobiology 14 1275-1284 [DOI] [PubMed] [Google Scholar]
- 49.Domon, B., and Costello, C. E. (1988) Glycoconj. J. 5 397-409 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.