Abstract
Signal peptide peptidase (SPP) and γ-secretase are intramembrane aspartyl proteases that bear similar active site motifs but with opposite membrane topologies. Both proteases are inhibited by the same aspartyl protease transition-state analogue inhibitors, further evidence that these two enzymes have the same basic cleavage mechanism. Here we report that helical peptide inhibitors designed to mimic SPP substrates and interact with the SPP initial substrate-binding site (the “docking site”) inhibit both SPP and γ-secretase, but with submicromolar potency for SPP. SPP was labeled by helical peptide and transition-state analogue affinity probes but at distinct sites. Nonsteroidal anti-inflammatory drugs, which shift the site of proteolysis by SPP and γ-secretase, did not affect the labeling of SPP or γ-secretase by the helical peptide or transition-state analogue probes. On the other hand, another class of previously reported γ-secretase modulators, naphthyl ketones, inhibited SPP activity as well as selective proteolysis by γ-secretase. These naphthyl ketones significantly disrupted labeling of SPP by the helical peptide probe but did not block labeling of SPP by the transition-state analogue probe. With respect to γ-secretase, the naphthyl ketone modulators allowed labeling by the transition-state analogue probe but not the helical peptide probe. Thus, the naphthyl ketones appear to alter the docking sites of both SPP and γ-secretase. These results indicate that pharmacological effects of the four different classes of inhibitors (transition-state analogues, helical peptides, nonsteroidal anti-inflammatory drugs, and naphthyl ketones) are distinct from each other, and they reveal similarities and differences with how they affect SPP and γ-secretase.
One of the top therapeutic strategies for the prevention and treatment of Alzheimer disease (AD)2 is suppression of the production of the amyloid β-protein (Aβ). Aβ is the primary protein component of the hallmark plaques in the AD brain, and aggregated Aβ is widely thought to cause the onset of AD (1). The 4-kDa Aβ is produced from the amyloid β-protein precursor (APP), a type I integral membrane protein, through sequential proteolysis by β-secretase and γ-secretase. γ-Secretase is composed of four essential membrane proteins, including presenilin (PS), Pen-2, nicastrin, and Aph-1 (2), with one of each component being sufficient for proteolytic activity (3). PS is the catalytic component of the enzyme (4), and missense mutations in PS cause early onset familial AD and alter the length of the products, Aβ (5) and the APP intracellular domain (AICD) (6). Although a minor species, the 42-residue Aβ42 is initially deposited in the AD brain instead of the more predominant 40-residue Aβ40 (7), and Aβ42 is especially implicated in the pathogenesis of AD. As the proportion of Aβ42 to Aβ40 is determined by γ-secretase, this membrane-embedded aspartyl protease is a major target for the development of AD drugs (8).
Signal peptide peptidase (SPP) is an intramembrane aspartyl protease with homology to PS (9). SPP cleaves membrane protein signal sequences (i.e. with type II orientation), including the major histocompatibility complex class I signal sequence for generating human leukocyte antigen E epitopes (10), and is also responsible for the maturation of the hepatitis C virus core protein (11), the latter suggesting that modulation of SPP activity may be suitable for antiviral therapy. Similar to PS, SPP has aspartate-containing YD and LGLGD motifs within adjacent transmembrane domains that include the active site and a PAL motif near the C terminus (12); however, each of these motifs is flipped in the membrane when comparing PS and SPP, correlating with the opposite orientation of their respective substrates.
Despite their opposite membrane orientations, the biochemical properties of these two proteases are similar, especially upon detergent solubilization from the asymmetric environment of the lipid bilayer. SPP is inhibited by transition-state analogue inhibitors for γ-secretase (13), and analogous to γ-secretase, which requires prior substrate cleavage by α- or β-secretase, SPP requires prior cleavage of the substrate by signal peptidase (14). Recently, we developed an in vitro cell-free SPP assay system, which uses n-dodecyl β-d-maltoside (DDM)-solubilized membrane fractions and a synthetic substrate based on an SPP-cleaved signal sequence, and we showed that a subset of nonsteroidal anti-inflammatory drugs (NSAIDs) shifted the cleavage site of SPP. This shift in the SPP cleavage site by certain NSAIDs was similar to that observed for γ-secretase cleavage sites (15), suggesting that the NSAID-binding site on the γ-secretase complex might reside on PS, at a site conserved in SPP.
On the other hand, there are also critical differences between SPP and γ-secretase. Unlike γ-secretase, SPP does not form a proteolytic complex with other components (9), and SPP expressed in and purified from Escherichia coli has the appropriate proteolytic activity without coexpression or copurification of any other proteins (16). PS undergoes endoproteolysis into an N-terminal fragment (NTF) and a C-terminal fragment (CTF) during maturation to an active protease, whereas SPP is active as its full-length protein (9). Moreover, γ-secretase cleaves the APP transmembrane domain at least twice, whereas SPP cleaves its substrate mainly at one site (15).
Investigation of similarities and differences between SPP and γ-secretase is important for developing specific inhibitors as AD drugs as well as for understanding common features shared by intramembrane aspartyl proteases. In this study, we examined the effect of inhibitors on SPP and γ-secretase activities using photoaffinity probes based on either a helical peptide inhibitor or a transition-state analogue inhibitor. Taking advantage of these two classes of probes, we carried out competition studies with other compounds, including two classes of γ-secretase modulators, NSAIDs and naphthyl ketones, and demonstrate that all four pharmacological classes (transition-state analogues, helical peptides, NSAIDs and naphthyl ketones) affect SPP and γ-secretase in distinct ways.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Characterization—Helical peptide inhibitors were synthesized by standard solution or solid-phase procedures using amino acids and α-amino-isobutyric acid as described previously (17, 18). Peptides were purified by reverse phase-high performance liquid chromatography (RP-HPLC) using a C18 column and subsequently characterized by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometric analysis on a Voyager-DE STR biospectrometry workstation (Applied Biosystems). CD spectra were acquired using an Aviv 62A DS spectropolarimeter. Photoaffinity probes were prepared by replacing each residue of a 10-residue peptide (compound 8, Fig. 1A) with 4-benzoyl-l-phenylalanine (Bpa; Sigma) to identify those that retained SPP inhibitory activity. For the two Bpa-containing compounds that showed the best SPP inhibitory activity (Fig. 4A), biotin (Sigma) was coupled to the N terminus. All peptides were dissolved in dimethyl sulfoxide (DMSO) to prepare stock solutions.
FIGURE 1.
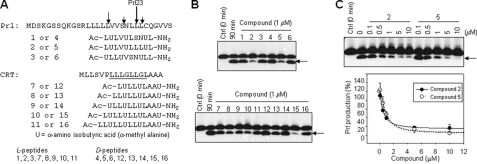
Inhibitory profiles of helical peptide inhibitors. A, amino acid sequences of helical peptide inhibitors. Arrows above the Prl signal sequence indicate the main cleavage site (Prl23) and some minor cleavage sites. Underline below CRT signal sequence is the putative cleavage region. B, inhibitory profiles of the helical peptides. Solubilized membrane fractions were incubated for 90 min in the presence of 1 μm of the helical peptides. N-terminal Prl cleavage product was detected by Western blot with anti-Myc antibody. Arrows indicate the product bands. C, inhibitory profiles of Cpds 2 and 5. Solubilized membrane fractions were incubated for 90 min in the presence of various concentrations of Cpds 2 and 5. Production of the Prl cleavage product, indicated with an arrow, was quantified and plotted.
FIGURE 4.
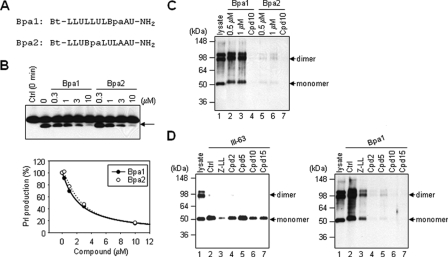
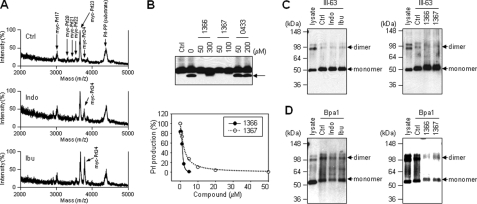
Photolabeling of SPP and competition with inhibitors. A, helical peptide photoaffinity probes. Bt and Bpa indicate biotin and 4-benzoyl-l-phenylalanine, respectively. B, inhibitory profiles of Bpa1 and Bpa2. Solubilized membrane fractions from SPP-transfected CHO cells were incubated for 90 min with Prl substrate in the presence of various concentrations of Bpa1 and Bpa2. SPP-mediated Prl cleavage product (arrow) was quantified and plotted. The results shown are the average values of two independent experiments. Ctrl, control. C, position of Bpa affects the photolabeling of SPP. SPP was photolabeled with 0.5 or 1 μm Bpa1 and Bpa2 in the absence or presence of Cpd 10 (10 μm), respectively. Photolabeled SPP was detected by streptavidin precipitation (to pull down biotinylated proteins) and Western blot with anti-V5 antibody (to detect SPP). D, SPP was photolabeled with 2 μm III-63 (left panel) or 0.5 μm Bpa1 (right panel) in the presence of 20 μm (Z-LL)2 ketone (Z-LL), Cpds 2, 5, 10, and 15. Photolabeled SPP was detected with anti-V5 antibody.
Cell-free SPP and γ-Secretase Assays—Membranes from Chinese hamster ovary (CHO) cells stably overexpressing V5- and His-tagged human SPP (15) or γ-secretase components (γ-30 cells) (19) were collected and homogenized in homogenization (H) buffer (50 mm Hepes, pH 7.0, 250 mm sucrose, 5 mm EDTA) by passing once through a French press at 1,000 p.s.i., and cell debris and nuclei were removed at 3,000 × g for 10 min. The supernatants were centrifuged at 100,000 × g for 1 h, and the resultant pellets were washed with 0.1 m sodium carbonate, pH 11.4, and then centrifuged again. The membrane pellets were solubilized with 2% DDM for SPP and 1% CHAPSO for γ-secretase in H buffer for 90 min on ice and then centrifuged at 100,000 × g for 1 h. The protein concentration of the supernatant was determined with BCA protein assay reagent (Pierce). DDM- or CHAPSO-solubilized membrane fractions were diluted to 0.25% and incubated with substrate Prl-PP for SPP (15) or C100FLAG for γ-secretase (20) at 37 °C for 90 min or 2 h. The reaction was stopped by adding 2× SDS sample buffer containing 5% 2-mercaptoethanol. The samples were subjected to SDS-PAGE and Western blot as described previously (3, 15). The N-terminal Prl cleavage products were detected with mouse monoclonal anti-Myc antibody (Invitrogen), and Aβ and AICD were detected with mouse monoclonal 6E10 (Covance Research Products) and anti-FLAG M2 (Sigma) antibodies, respectively.
Inhibitor Treatment—Reaction mixtures were incubated in the presence of inhibitors. The inhibition levels of the products were quantified by Western blot with program ImageJ 1.34s (National Institutes of Health), and IC50 values were estimated with SigmaPlot (Systat Software). An SPP inhibitor ((Z-LL)2 ketone (Calbiochem)), γ-secretase inhibitor (III-31-C (20)), kinase inhibitor type γ-secretase modulators (1366, 1367, and 0433 (21)), and the helical peptide inhibitors reported here were dissolved in DMSO. NSAIDs, indomethacin (Sigma), and (S)-ibuprofen (Biomol) were dissolved in ethanol. Aspirin (Sigma) was dissolved in water. All reaction mixtures were preincubated on ice for 15 min and then placed at 37 °C. DMSO or ethanol was used as a vehicle control. The final concentration of vehicle was 1%. To identify the length of N-terminal Prl cleavage products after incubation, those products were immunoprecipitated with anti-Myc antibody and then subjected to MALDI-TOF mass spectrometric analysis on the Voyager-DE STR Biospectrometry workstation (Applied Biosystems) as described previously (15).
Photolabeling of SPP and γ-Secretase—0.25% CHAPSO-solubilized membrane fractions of SPP-transfected cells or γ-30 cells were incubated with the helical photoaffinity probe Bpa1 (at 0.5 μm for SPP and 4 μm for γ-secretase) or the transition-state analogue photoaffinity probe III-63 (22) (at 2 μm for SPP and 0.5 μm for γ-secretase) for 30 min in the presence of helical peptide inhibitors, (Z-LL)2 ketone or III-31-C, NSAIDs, kinase inhibitor-type γ-secretase modulators, or control vehicle DMSO or ethanol and then irradiated for 30 min at 350 nm. To isolate and detect photolabeled SPP, the irradiated samples were mixed with an equal volume of RIPA buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) and rocked with immobilized streptavidin (Pierce) overnight. Biotinylated proteins were eluted with SDS sample buffer by incubating at 37 °C for 15 min. The samples were subjected to SDS-PAGE and Western blot, detecting with a mouse monoclonal anti-V5 antibody (Invitrogen) for SPP. To isolate and detect photolabeled γ-secretase complexes, the irradiated samples plus an equal volume of 1% digitonin in H buffer were incubated with a monoclonal anti-HA-agarose antibody (Sigma) or a monoclonal anti-His-agarose antibody (Sigma) overnight (the former recognizes HA-tagged Aph-1 expressed in γ-30 cells, and the latter is a control). The immunoprecipitated samples were eluted with SDS sample buffer by incubating at 37 °C for 15 min. The samples were subjected to SDS-PAGE and Western blot, detecting with a rabbit polyclonal anti-biotin antibody (Bethyl) for photolabeled proteins, rabbit polyclonal AB14 (23) for PS1-NTF, rabbit polyclonal 4627 (24) for PS1-CTF, a rabbit polyclonal ECS antibody (Bethyl) for FLAG-Pen-2, and a rat monoclonal anti-HA antibody (Roche Applied Science) for Aph-1aL-HA.
RESULTS
Design and Characterization of Helical Peptide Inhibitors—γ-Secretase apparently possesses an initial substrate-binding site (or “docking site”) on PS (25). We showed previously that certain biochemical characteristics of SPP were very similar to those of γ-secretase, suggesting that SPP is a simple model for the PS-containing γ-secretase complex (15). To further understand substrate- and inhibitor-binding sites on SPP, we designed helical peptide inhibitors based on two of its substrates, the signal sequences of prolactin (Prl) and calreticulin (CRT) (14). We created both helical l- and d-peptides, containing between 9 and 13 amino acids (Fig. 1A). These peptides contain α-amino-isobutyric acid (Aib), which is well known to favor helical conformations (26). These Aibs were placed every three or four residues so that the Aibs would be on one face of the helix and signal sequence residues would be along the other face for interaction with SPP. After purification by RP-HPLC, the identity and purity of the final compounds were verified by MALDI-TOF mass spectrometry (data not shown). The helical conformation of these peptides was demonstrated by CD spectra, and we confirmed that the helical character increased with the length of the peptides (data not shown), similar to what we have previously shown for Aib-containing helical peptide inhibitors of γ-secretase (17, 18).
We first characterized the inhibitory effect of these peptides using a proteolytic assay for SPP that we had previously developed, which utilizes a Prl signal sequence with an N-terminal Myc tag for detection and a double proline substitution to prevent confounding processing by signal peptidase (15). Thus, we measured the conversion of this “Prl-PP” substrate to Prl cleavage products in a cell-free SPP assay system. Solubilized membrane fractions from CHO cells stably transfected with V5- and His-tagged SPP were incubated with Prl-PP in the presence of 1 μm of the helical peptide inhibitors for 90 min, and the samples were subjected to Western blot (Fig. 1B). We found that several inhibitors could block SPP-mediated cleavage of the Prl-PP substrate at 1 μm. The dose dependence of these helical peptides was then examined by quantifying the intensity of immunoblot signals (Fig. 1C). The results are shown in Tables 1 and 2.
TABLE 1.
Structures of Aib-containing peptides and their inhibitory potencies IC50 values were determined by measuring the level of Prl cleavage products. The errors represent standard deviations (n = 3).
| Cell-free assay, IC50 (nm) | ||
|---|---|---|
| l-peptides | ||
| 1 | Ac-LULVULSNUL-NH2 | >50,000 |
| 2 | Ac-LULVULLLUL-NH2 | 618 ± 83 |
| 3 | Ac-ULLUVSNULL-NH2 | >50,000 |
| d-peptides | ||
| 4 | Ac-LULVULSNUL-NH2 | >50,000 |
| 5 | Ac-LULVULLLUL-NH2 | 884 ± 143 |
| 6 | Ac-ULLUVSNULL-NH2 | >50,000 |
TABLE 2.
Structures of Aib-containing peptides and their inhibitory potencies IC50 values were determined by measuring the level of Prl cleavage products. The errors represent standard deviations (n = 3).
| Cell-free assay, IC50 (nm) | ||
|---|---|---|
| l-peptides | ||
| 7 | Ac-LULLULAAU-NH2 | 1,583 ± 147 |
| 8 | Ac-LLULLULAAU-NH2 | 1,267 ± 92 |
| 9 | Ac-ULLULLULAAU-NH2 | 696 ± 72 |
| 10 | Ac-LULLULLULAAU-NH2 | 414 ± 155 |
| 11 | Ac-LLULLULLULAAU-NH2 | 1,546 ± 403 |
| d-peptides | ||
| 12 | Ac-LULLULAAU-NH2 | 7,744 ± 1,419 |
| 13 | Ac-LLULLULAAU-NH2 | 1,579 ± 201 |
| 14 | Ac-ULLULLULAAU-NH2 | 3,337 ± 1,311 |
| 15 | Ac-LULLULLULAAU-NH2 | 274 ± 108 |
| 16 | Ac-LLULLULLULAAU-NH2 | 1,702 ± 281 |
Compounds (Cpds) 1–6 are each 10-residue peptides designed from the Prl signal sequence, with different placement of the Aibs or replacement of an SN motif with LL (Fig. 1A). The SN motif has been suggested to be critical for substrate cleavage by virtue of helix-destabilizing effects (14). The LL replacement was designed to keep the Aib-containing peptides helical throughout. Cpds 7–16 are 9–13-residue peptides designed from the CRT signal sequence, the key difference being length. Even though Cpds 1, 3, 4, and 6 are more similar to the wild type Prl sequence than Cpds 2 and 5 (which have the SN to LL alteration), the enantiomeric Cpds 2 and 5 were very effective inhibitors. Similarly, results in Table 2 with the CRT-based peptides show that both l- and d-forms could be effective inhibitors with 12 residues being the optimal length in both cases (Cpds 10 and 15). This is similar to what we have reported previously for helical peptide inhibitors of γ-secretase, in which APP-based l- and d-peptides potently inhibited the protease with an optimal length of 12–13 residues (18). d-peptide 15 was particularly impressive, with an IC50 value of 274 nm. A key difference between these results and those seen with Aib-containing inhibitors of γ-secretase, although γ-secretase is much more effectively inhibited by d-peptides (17, 18), SPP was inhibited by both l- and d-peptides to similar degrees, with similar IC50 values for enantiomeric pairs.
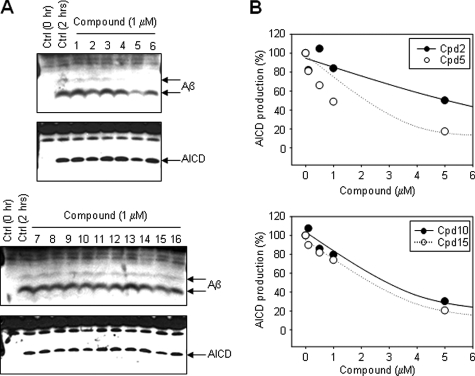
Inhibitory Effects on γ-Secretase and SPP Cleavage Site Specificity—A number of γ-secretase inhibitors can also inhibit SPP (13). Therefore, we analyzed whether these helical peptide SPP inhibitors affect γ-secretase activity and, if so, whether the inhibitory potencies versus the two enzymes correlate. CHAPSO-solubilized membrane fractions of γ-30 cells (19) were used to evaluate the effect of these helical peptide SPP inhibitors. After 2 h of incubation at 37 °C with the APP-based recombinant substrate C100FLAG (20) in the presence of various inhibitors, the production of Aβ and AICD-FLAG was analyzed by Western blot. Cpds 2, 5, 10, and 15 at 1 μm slightly inhibited Aβ and AICD production (Fig. 2A). These inhibitors, Cpds 2, 5, 10, and 15, also displayed stronger potencies against SPP activity, demonstrating that both SPP and γ-secretase prefer the same sequences and length of helical peptide inhibitors at the initial substrate-binding site (docking site), thereby suggesting that the structure of the docking site of γ-secretase may partially resemble that of SPP. We then examined these inhibitors for dose-dependent effects (Fig. 2B). Cpds 2 and 5 inhibited the production of AICD-FLAG with IC50 values of 5.3 and 1.0 μm, respectively, whereas Cpds 10 and 15 inhibited the production of AICD-FLAG with IC50 values of 2.4 and 2.1 μm, respectively. These results demonstrate that the potencies of the inhibitors for SPP are severalfold higher than for γ-secretase. These Prl and CRT signal sequence-based d-peptides inhibited the γ-secretase activity with slightly higher potencies than their l-peptide counterparts; however, the differences in potency between l- and d-peptides was not high, as observed previously with helical peptide inhibitors based on APP (17, 18).
FIGURE 2.
Inhibitory effects of the helical peptide inhibitors on γ-secretase. A, inhibitory profiles of helical peptides on γ-secretase cleavage products of APP-based recombinant substrate C100FLAG. Solubilized membrane fractions from γ-30 cells were incubated for 2 h in the presence of 1 μm of the helical peptide inhibitors. Aβ was detected with anti-Aβ antibody 6E10, and AICD-FLAG was detected with anti-FLAG antibody M2. Ctrl, control. B, inhibitory profiles of Cpds 2, 5, 10, and 15. Solubilized membrane fractions from γ-30 cells were incubated for 2 h in the presence of various concentrations of Cpds 2, 5, 10, and 15. The results shown are the average values of two independent experiments.
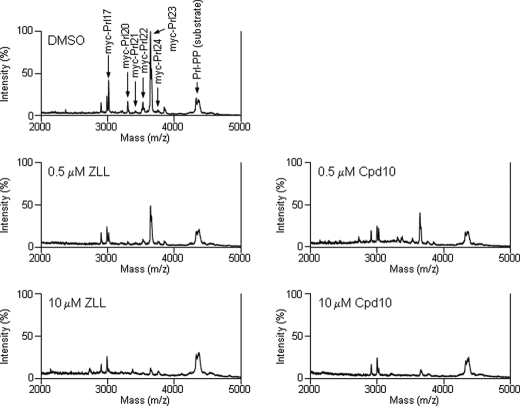
We next examined the effect of these inhibitors on the SPP cleavage sites. Several γ-secretase inhibitors increase Aβ42 production at low concentrations, even though the Aβ40 production is effectively decreased (27, 28). To determine whether SPP cleavage sites are similarly affected by SPP inhibitors at low concentrations, we performed MALDI-TOF mass spectrometric analysis. Solubilized membrane fractions with Prl-PP were incubated in the presence of the inhibitors for 90 min, and the samples were immunoprecipitated with anti-Myc antibody. The products in the resultant pellets were eluted and then subjected to the mass spectrometric analysis. This analysis revealed several cleaved products (Fig. 3, top panel), with the main cleavage site being between Leu-23 and Leu-24, as shown in our previous report (15). The SPP transition-state analogue inhibitor (Z-LL)2 ketone at 0.5 μm did not change the cleavage site, and the signal intensities of the products were reduced proportionately (Fig. 3, left panels). We then analyzed SPP cleavage products formed in the presence of a helical peptide inhibitor. In the presence of Cpd 10, the cleavage site of the Prl products was also not shifted at a low concentration (0.5 μm), and the various products were inhibited proportionately (Fig. 3, right panels), demonstrating that both transition-state analogue and helical peptide inhibitors globally inhibit SPP activity without specificity for particular cleavage sites.
FIGURE 3.
Effect of low concentrations of inhibitors on the cleavage sites by SPP. Solubilized membrane fractions from CHO cells stably transfected with V5- and His-tagged human SPP were incubated for 90 min with Myc-tagged Prl substrate in the presence of various concentrations of inhibitors. The products were immunoprecipitated with anti-Myc antibody and subjected to MALDI-TOF mass spectrometric analysis. Arrows indicate the cleavage products or substrate. DMSO and ZLL indicate control DMSO vehicle and (Z-LL)2 ketone, respectively.
Photoaffinity Labeling of SPP by Helical Peptide Inhibitors and Competition with Other Inhibitors—To obtain direct evidence that helical peptide inhibitors bind to SPP, we developed photoaffinity probes based on the 10-residue l-peptide Cpd 8 and tested labeling of the protease by this probe. We synthesized two different photoaffinity probes that included the photoactivable Bpa residue at different positions (Fig. 4A). Bpa has benzophenone as a side chain, a moiety commonly used to react covalently with C-H bonds upon irradiation at 350 nm (29). Bpa1 contains Bpa at the eighth residue, and Bpa2 contains it at the fourth residue. Biotin, which allows isolation of the labeled proteins with avidin beads, was installed at the N terminus as described previously (25). Both Bpa1 and Bpa2 inhibited SPP activity with closely similar potencies (IC50 values of 2.2 and 2.3 μm, respectively; see Fig. 4B).
Solubilized membrane fractions of cells expressing SPP fused with a V5 tag (15) were incubated with Bpa1 or Bpa2 in the absence or presence of unbiotinylated helical peptide inhibitor Cpd 10 and subjected to UV irradiation. The proteins labeled with the probe were pulled down with streptavidin beads and analyzed for the presence of SPP by Western blot with anti-V5 antibody. Both monomeric SPP and SDS-stable dimeric SPP were detected by labeling with Bpa1 (Fig. 4C), and the labeling was blocked with the helical peptide Cpd 10 (lane 4). The mobility of SPP was slightly slower than SPP in the original lysates (Fig. 4C, lanes 1 and 2), presumably because the photoaffinity probe, which is >1 kDa, binds covalently to SPP. Although the inhibitory potencies of Bpa1 and Bpa2 were virtually the same, SPP was barely labeled by Bpa2 (Fig. 4C, lanes 5 and 6). The only difference between these probes was the position of Bpa, which for Bpa2 may not be in close proximity or oriented properly with respect to SPP to allow covalent attachment.
We next analyzed the labeling of SPP with the transition-state analogue photoaffinity probe III-63 (22), which specifically binds to the active site of SPP (15, 30). Photoprobe III-63 labeled monomeric SPP in solubilized membrane fractions, and this labeling was blocked by (Z-LL)2 ketone (Fig. 4D, lane 3 in left panel). Among the helical peptide inhibitors, Cpds 2, 10, and 15 showed partial reduction of the labeling (Fig. 4D, lanes 4–7 in left panel), suggesting that the helical peptide inhibitors bind to a distinct albeit partially overlapping site on SPP from where the transition-state analogue inhibitor binds, as demonstrated for γ-secretase (25). This observation was consistent with the finding that (Z-LL)2 ketone partially blocked the labeling of Bpa1 (Fig. 4D, lane 3 in right panel). In contrast, all the helical peptide inhibitors tested (Cpds 2, 5, 10, and 15) very effectively blocked the labeling of SPP by Bpa1 (Fig. 4D, lanes 4–7 in right panel).
We previously showed that a subset of NSAIDs that affect γ-secretase-mediated cleavage of APP to specifically decrease Aβ42 production while simultaneously increasing Aβ38 production (31) (thus, their potential as AD therapeutics) could also shift the SPP-mediated cleavage site of the Prl products from Leu23–Leu24 to Leu24–Cys25 (15). Therefore, we investigated whether these compounds affect the labeling of SPP by the photoaffinity probes. Moreover, because certain naphthyl ketone kinase inhibitors specifically inhibit γ-secretase cleavage of APP without affecting that of another key substrate, the Notch receptor (21), we also investigated the effect of these selective inhibitors (naphthyl ketones) and one nonselective inhibitor (a sulfonamide) on the labeling of SPP by the photoaffinity probes.
As shown previously (15), when the SPP-containing membrane fractions were incubated with Prl-PP in the presence of indomethacin or ibuprofen, the cleavage site of the Prl products was shifted from Leu23–Leu24 to Leu24–Cys25 (Fig. 5A). Production formation was not suppressed at high concentrations of these compounds (data not shown). We then examined whether kinase inhibitors of γ-secretase also inhibit SPP activity. The solubilized membrane fractions with Prl-PP were incubated in the presence of the kinase inhibitors 1366, 1367 or 0433, which inhibit γ-secretase activity (21). We found that the APP-selective naphthyl ketones 1366 and 1367 effectively inhibited the SPP activity, whereas the nonselective sulfonamide 0433 did not (Fig. 5B, top panel). 1366 and 1367 inhibited SPP activity with IC50 values of 1.2 and 2.1 μm, respectively (Fig. 5B, bottom panel). These two compounds were also effective inhibitors for the γ-secretase-mediated cleavage of APP substrate (21), suggesting that the inhibitory mechanisms for SPP by the naphthyl ketones are similar to that for γ-secretase.
FIGURE 5.
Effects ofγ-secretase modulators on SPP. A, solubilized membrane fractions from SPP-transfected cells were incubated with Prl substrate for 90 min in the presence of 500 μm indomethacin (Indo) or 2 mm ibuprofen (Ibu). The N-terminal Prl cleavage products were immunoprecipitated with anti-Myc antibody and subjected to MALDI-TOF mass spectrometric analysis. Arrows indicate the cleavage products or substrate. Ctrl indicates control DMSO vehicle (no compound). B, SPP inhibitory profiles of APP-selective γ-secretase inhibitors. Solubilized membrane fractions from SPP-transfected cells were incubated for 90 min with Prl substrate in the presence of various concentrations of 1366, 1367, and 0433; cleavage products (arrow) were detected with anti-Myc antibody, quantified, and plotted. The results shown are the average values of two independent experiments. C and D, SPP was photolabeled with 2 μm III-63 (C) or 0.5 μm Bpa1 (D) in the presence of 500 μm indomethacin (Indo), 2 mm ibuprofen (Ibu) (left panels), 20 μm 1366, and 20 μm 1367 (right panels), respectively. Photolabeled SPP was detected with anti-V5 antibody.
We then investigated the labeling of SPP by photoaffinity probes in the presence of these compounds. Solubilized membrane fractions were incubated with the photoaffinity probes in the presence of NSAIDs or the naphthyl ketones and subjected to UV irradiation and isolation. When III-63, a transition-state analogue inhibitor, was used as a probe, 500 μm indomethacin and 2000 μm ibuprofen and 20 μm 1366 and 1367 (all concentrations well above that needed to affect SPP activity) did not block the labeling of SPP (Fig. 5C). When Bpa1, a helical peptide inhibitor, was used as a probe, NSAIDs did not block this labeling either (Fig. 5D, left panel); however, 20 μm 1366 and 1367 substantially reduced the labeling (Fig. 5D, right panel). These results demonstrate that NSAIDs do not affect either the substrate-binding site or the active site in SPP; however, the naphthyl ketone inhibitors may disrupt the interaction between substrates and SPP.
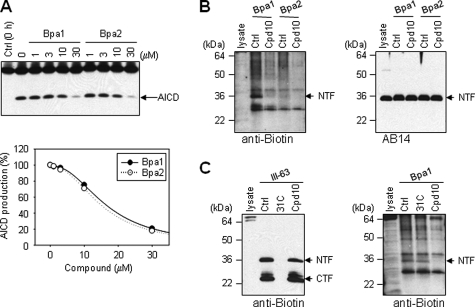
Effects of Compounds on Photoaffinity Labeling of γ-Secretase—We next investigated whether these SPP affinity labeling reagents could also inhibit and specifically label γ-secretase and whether NSAIDs or the naphthyl ketone inhibitors could affect this labeling. Solubilized membrane fractions of γ-30 cells, which overexpress γ-secretase components, were incubated with C100FLAG in the presence of various concentrations of Bpa1 or Bpa2. Both Bpa1 and Bpa2 inhibited the γ-secretase activity with closely similar IC50 values of 16.6 and 15.2 μm, respectively (Fig. 6A). To label γ-secretase with these photoaffinity probes, solubilized γ-30 membrane fractions were incubated with 4 μm Bpa1 or Bpa2 in the absence or presence of the helical peptide inhibitor (Cpd 10) and subjected to UV irradiation. The γ-secretase complexes were immunoprecipitated with anti-HA-agarose (Aph-1 with a C-terminal HA tag is overexpressed in γ-30 cells), and we confirmed that PS1 was coimmunoprecipitated with Aph-1 (Fig. 6B, right panel). The molecular size showed that Bpa1 labeled only PS1 NTF, not CTF or other components, and this labeling was specifically blocked by Cpd 10 (Fig. 6B, left panel). As observed with SPP (Fig. 4C), photoprobe Bpa2 essentially did not label any components of γ-secretase (Fig. 6B, left panel). The labeling of PS1 NTF and CTF with transition-state analogue affinity reagent III-63 was not blocked by helical peptide Cpd 10, and conversely, labeling of PS1 NTF with helical peptide affinity reagent Bpa1 was not blocked by transition-state analogue III-31-C, the parent compound of III-63 (Fig. 6C), indicating that these two classes of inhibitors bind to distinct sites on presenilin.
FIGURE 6.
Photolabeling of PS1 with helical peptide SPP inhibitors. A, γ-secretase inhibitory profiles of Bpa1 and Bpa2. Solubilized membrane fractions from γ-30 cells were incubated for 2 h with C100FLAG in the presence of various concentrations of Bpa1 and Bpa2. γ-Secretase-mediated production of AICD-FLAG, indicated with an arrow, was quantified and plotted. The results shown are the average values of two independent experiments. Ctrl, control. B, position of Bpa affects the photolabeling of PS1. PS1 was photolabeled with 5 μm Bpa1 and Bpa2 in the absence or presence of helical peptide inhibitor Cpd 10 (50 μm). Photolabeled PS1 in immunoprecipitated samples was detected with anti-biotin (left panel), and total PS1 NTF was detected with anti-PS1 AB14 (right panel) antibodies. C, PS1 was photolabeled with 0.5 μm III-63 (left panel) or 4 μm Bpa1 (right panel) in the presence of 5 μm III-31-C (31C), 40 μm Cpd10. Photolabeled and immunoprecipitated PS1 was detected with anti-biotin antibody.
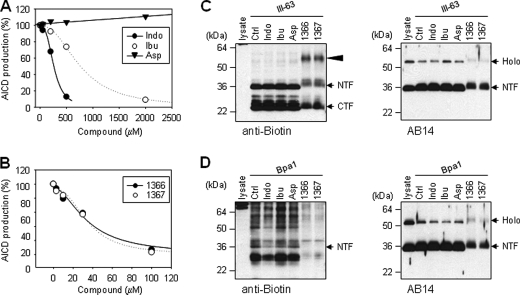
We then examined the effect of NSAIDs and the naphthyl ketone inhibitors on the photoaffinity labeling of γ-secretase. Unlike what was observed with SPP, Aβ42-lowering NSAIDs inhibited γ-secretase activity at the high concentrations (Fig. 7A), as described previously (32, 33). The IC50 values of indomethacin and ibuprofen were 263 and 770 μm, respectively. Aspirin, which does not reduce the Aβ42 production (31), did not inhibit the γ-secretase activity at all (Fig. 7A). The naphthyl ketone inhibitors 1366 and 1367 inhibited the γ-secretase activity with the IC50 values of 34.8 and 35.0 μm, respectively (Fig. 7B). After incubation and irradiation in the presence of these compounds, the γ-complexes were immunoprecipitated with anti-HA-agarose, and Western blot of the resultant pellets with anti-biotin antibody confirmed the labeling of PS1 (Fig. 7, C and D, right panels). When the solubilized membrane fractions were incubated with III-63, NSAIDs did not block the labeling of PS1 NTF and CTF (Fig. 7C, left panel). However, when the solubilized membrane fractions were incubated with III-63 in the presence of 1366 or 1367, a higher molecular weight band (∼55 kDa) labeled with III-63 appeared (Fig. 7C, left panel). Next, when the solubilized membrane fractions were incubated with Bpa1, NSAIDs again did not block the labeling of PS1 NTF (Fig. 7D, left panel). However, when the solubilized membrane fractions were incubated with Bpa1 in the presence of 1366 or 1367, these compounds completely blocked the labeling of PS1 NTF by Bpa1 (Fig. 7D, left panel). The gel mobility of PS1 NTF appeared to be slightly slower in the presence of 1366 or 1367 (Fig. 7, C and D, right panels), suggesting that these compounds directly bind to PS tightly and inhibit the activity.
FIGURE 7.
PS1 photolabeling and competition with γ-secretase modulators. A, γ-secretase inhibitory profiles of NSAIDs. Solubilized membrane fractions from γ-30 cells were incubated with C100FLAG for 2 h in the presence of various concentrations of indomethacin (Indo), ibuprofen (Ibu), and aspirin (Asp), respectively. B, inhibitory profiles of APP-selective γ-secretase inhibitors. Solubilized membrane fractions from γ-30 cells were incubated with C100FLAG for 2 h in the presence of various concentrations of 1366 and 1367. The results shown are the average values of two independent experiments (A and B). C and D, PS1 was photolabeled with 0.5 μm III-63 (C) or 4 μm Bpa1 (D) in the presence of 500 μm indomethacin (Indo), 2 mm ibuprofen (Ibu), 2 mm aspirin (Asp), 100 μm 1366, and 100 μm 1367. Arrowhead indicates an SDS-stable NTF/CTF heterodimer. Photolabeled and immunoprecipitated PS1 was detected with anti-biotin antibody (left panels), and labeled and unlabeled immunoprecipitated PS1 was detected with anti-PS1 AB14 (right panels).
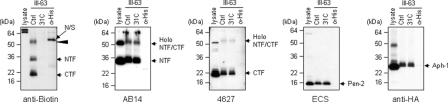
We then analyzed whether the high molecular weight band at ∼55 kDa was specifically labeled and whether other γ-secretase components were labeled or not. Solubilized membrane fractions were incubated with III-63 in the presence of 100 μm 1366 and UV-irradiated. The immunoprecipitated γ-secretase complexes with anti-HA-agarose were analyzed by Western blot, and anti-His-agarose was used as a negative control for immunoprecipitation. Labeling of PS1 proteins was completely blocked by III-31-C, the parent compound of III-63 (Fig. 8, most left panel), indicating that PS1 labeling by III-63 is specific. The uppermost ∼55-kDa band was detected with both AB14 and 4627, which recognize PS1 NTF and CTF, respectively, suggesting that this high molecular band is composed of PS1 NTF/CTF heterodimer. The other components, Pen-2 and Aph-1 were not detected at this size. These results suggest that a transition-state analogue inhibitor, which binds to the active site, can access PS1 NTF, CTF, and NTF/CTF heterodimer in the presence of naphthyl ketone inhibitors (Fig. 8); however, the substrate-binding site is no longer accessible to a helical inhibitor in the presence of naphthyl ketone inhibitors (Fig. 7D) because the naphthyl ketones directly interact with the substrate-binding site of PS or allosterically affect this site.
FIGURE 8.
Photolabeling of PS1 in the presence of an APP-selective γ-secretase inhibitor. Solubilized membrane fractions from γ-30 cells were incubated for 2 h in the presence of photoprobe III-63 and γ-secretase modulator 100 μm 1366. III-31-C (31C) (5 μm) blocked photolabeling by III-63 (0.5 μm) in the presence of 100 μm 1366. Arrowhead indicates an SDS-stable NTF/CTF heterodimer. N/S indicates nonspecific reaction from anti-His-agarose. Photolabeled and immunoprecipitated PS1 was detected with anti-biotin antibody. PS1 NTF, CTF, Pen-2, and Aph-1 were detected with AB14, 4627, ECS, and anti-HA antibodies, respectively.
DISCUSSION
In this study, we provide evidence that four different types of protease inhibitors or modulators have distinct effects on both SPP and γ-secretase. Although many γ-secretase inhibitors have been developed in industry and academia (8), the inhibitory mechanisms remain unclear in many cases. SPP and γ-secretase have the same active site motif (9), and several γ-secretase inhibitors also effectively inhibit SPP (13). On the other hand, varying degrees of selectivity have been observed for SPP or γ-secretase inhibitors; for instance DAPT (34) and compound E (35) are much more effective toward γ-secretase (data not shown), and (Z-LL)2 ketone is much more effective toward SPP (13).
Our SPP substrate-based helical peptides, designed to interact with the initial substrate-binding site (i.e. the docking site), could inhibit both SPP and γ-secretase activities, but with more potency toward SPP. One photoaffinity probe based on a helical peptide SPP-selective inhibitor, Bpa1, was able to label both SPP and γ-secretase, whereas another probe, Bpa2, only slightly labeled either SPP or γ-secretase. Helical peptide probe Bpa1 was able to bind to both SPP monomer and dimer; however, a transition-state analogue probe III-63 bound only to monomer, suggesting that although the SPP monomer and dimer can both recognize substrate, only the monomer can bring the substrate into the active site and proteolyze it. With respect to γ-secretase, Bpa1 bound only to the PS1 NTF, not the PS1 CTF or other components of the protease complex. Benzophenone photoreacts covalently with the closest C-H bond within 3 Å upon irradiation at 350 nm (29). The position and orientation of Bpa in the bound inhibitor may keep its distance from the CTF at more than 3 Å. Likewise, the distance of the Bpa moiety of Bpa2 from SPP or γ-secretase might be more than 3 Å, dramatically reducing the labeling of these proteases, even though the inhibitory potencies of Bpa1 and Bpa2 are roughly equivalent.
The mechanisms of effects of NSAIDs on SPP and γ-secretase activities are somewhat mysterious. NSAIDs shift the substrate cleavage sites of both SPP and γ-secretase, albeit in different directions (to shorter N-terminal products for γ-secretase and longer N-terminal products for SPP) (15, 31). At higher concentrations, NSAIDs inhibit overall γ-secretase activity (32, 33) but not overall SPP activity (15). The competition experiments described here using photoaffinity probes of a helical peptide inhibitor and a transition-state analogue inhibitor showed that NSAIDs at high concentrations did not block the labeling of SPP or γ-secretase by either probe (Figs. 5 and 7), suggesting that NSAIDs do not prevent the interaction of substrates with the docking site or the active site of these two proteases. How do NSAIDs shift the cleavage site by γ-secretase or SPP? A most recent study reported that the binding site NSAIDs is, surprisingly, in the juxtamembrane region of the substrate and not on the enzyme (36). NSAIDs binding to substrate may alter how the substrate interacts with γ-secretase to shift the site of cleavage. Whether NSAIDs similarly bind to SPP substrates is not known, but if so, this may explain the altered site of proteolysis by SPP as well. How then high concentrations of NSAIDs inhibit only γ-secretase activity and not SPP activity is not clear.
We found that other compounds, certain naphthyl ketone-type kinase inhibitors, also showed an inhibitory effect on both SPP and γ-secretase. Most intriguingly, SPP and γ-secretase can specifically bind to the transition-state analogue inhibitor in the presence of these naphthyl ketones (Fig. 5C and Fig. 7C), but they do not bind well to the helical peptide inhibitor in their presence. Where do these naphthyl ketones bind? After treatment with the naphthyl ketones, PS1 formed an SDS-stable NTF/CTF heterodimer (Fig. 7C), and PS1 NTF and CTF bands appear slightly shifted in mobility, suggesting that the binding site is on PS1. Identification of the target of these naphthyl ketones, however, will likely require modification into an affinity labeling reagent of their own.
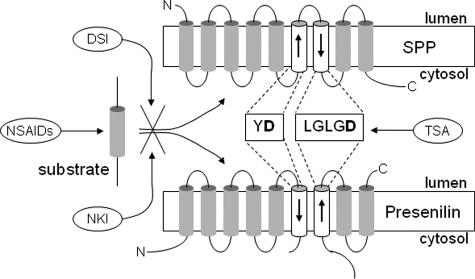
These naphthyl ketones were originally synthesized for inhibition of the Janus kinase 3 (37); however, the inhibition mechanism of γ-secretase and SPP activities by these compounds is apparently distinct from their ability to inhibit kinases, as our experiments employed cell-free protease assays. This situation is akin to that of the NSAIDs, in which the inhibition mechanism of Aβ42 production by these compounds is independent from their ability to inhibit cyclooxygenase (31). Taken together, the evidence described here suggests that the inhibitory mechanism is distinct for each of the four types of compounds (transition-state analogues, helical peptides, NSAIDs and naphthyl ketones, see Fig. 9).
FIGURE 9.
Schematic of inhibitor mechanisms. Transition-state analogue (TSA) inhibitor targets the active site, and helical peptide docking site inhibitor (DSI) prevents initial substrate interaction with the protease. NSAIDs target the substrate, and naphthyl ketone inhibitors (NKI) disrupt the initial interaction between substrate and protease (selectively for APP in the case of γ-secretase).
Understanding the nature of the inhibitor-enzyme interactions should provide clues to how structural modifications of these compounds might improve the potency and selectivity for therapeutic purposes. Inhibitors of γ-secretase are well appreciated as potential drugs for AD therapy, and a recent study suggests that malarial SPP may be a novel therapeutic target (38). Structural studies may ultimately define the details of the nature of these inhibitor interactions, facilitating the development of novel specific inhibitors for SPP or γ-secretase.
Acknowledgments
We are grateful to D. Selkoe for antibody 4627 and S. Gandy for antibody AB14.
This work was supported, in whole or in part, by National Institutes of Health Grants GM79555, NS41355, and AG17574 (to M. S. W.). This work was also supported by a research fellowship from the Uehara Memorial Foundation (to T. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AD, Alzheimer disease; Aβ, amyloid β-protein; AICD, APP intracellular domain; APP, amyloid β-protein precursor; Bpa, 4-benzoyl-l-phenylalanine; CHAPSO, 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonic acid; Cpd, compound; CRT, calreticulin; CTF, C-terminal fragment; DDM, n-dodecyl β-d-maltoside; DMSO, dimethyl sulfoxide; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; NSAIDs, nonsteroidal anti-inflammatory drugs; NTF, N-terminal fragment; Prl, prolactin; PS, presenilin; RP-HPLC, reverse phase-high performance liquid chromatography; SPP, signal peptide peptidase; HA, hemagglutinin; CHO, Chinese hamster ovary.
References
- 1.Citron, M. (2004) Nat. Rev. Neurosci. 5 677-685 [DOI] [PubMed] [Google Scholar]
- 2.Wolfe, M. S. (2006) Biochemistry 45 7931-7939 [DOI] [PubMed] [Google Scholar]
- 3.Sato, T., Diehl, T. S., Narayanan, S., Funamoto, S., Ihara, Y., De Strooper, B., Steiner, H., Haass, C., and Wolfe, M. S. (2007) J. Biol. Chem. 282 33985-33993 [DOI] [PubMed] [Google Scholar]
- 4.Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl, T. S., Kimberly, W. T., and Selkoe, D. J. (1999) Nature 398 513-517 [DOI] [PubMed] [Google Scholar]
- 5.Tanzi, R. E., and Bertram, L. (2005) Cell 120 545-555 [DOI] [PubMed] [Google Scholar]
- 6.Sato, T., Dohmae, N., Qi, Y., Kakuda, N., Misonou, H., Mitsumori, R., Maruyama, H., Koo, E. H., Haass, C., Takio, K., Morishima-Kawashima, M., Ishiura, S., and Ihara, Y. (2003) J. Biol. Chem. 278 24294-24301 [DOI] [PubMed] [Google Scholar]
- 7.Iwatsubo, T., Odaka, A., Suzuki, N., Mizusawa, H., Nukina, N., and Ihara, Y. (1994) Neuron 13 45-53 [DOI] [PubMed] [Google Scholar]
- 8.Tomita, T. (2007) Naunyn-Schmiedeberg's Arch. Pharmacol. 377 295-300 [DOI] [PubMed] [Google Scholar]
- 9.Weihofen, A., Binns, K., Lemberg, M. K., Ashman, K., and Martoglio, B. (2002) Science 296 2215-2218 [DOI] [PubMed] [Google Scholar]
- 10.Lemberg, M. K., Bland, F. A., Weihofen, A., Braud, V. M., and Martoglio, B. (2001) J. Immunol. 167 6441-6446 [DOI] [PubMed] [Google Scholar]
- 11.McLauchlan, J., Lemberg, M. K., Hope, G., and Martoglio, B. (2002) EMBO J. 21 3980-3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, J., Beher, D., Nyborg, A. C., Shearman, M. S., Golde, T. E., and Goate, A. (2006) J. Neurochem. 6 218-227 [DOI] [PubMed] [Google Scholar]
- 13.Weihofen, A., Lemberg, M. K., Friedmann, E., Rueeger, H., Schmitz, A., Paganetti, P., Rovelli, G., and Martoglio, B. (2003) J. Biol. Chem. 278 16528-16533 [DOI] [PubMed] [Google Scholar]
- 14.Lemberg, M. K., and Martoglio, B. (2002) Mol. Cell 10 735-744 [DOI] [PubMed] [Google Scholar]
- 15.Sato, T., Nyborg, A. C., Iwata, N., Diehl, T. S., Saido, T. C., Golde, T. E., and Wolfe, M. S. (2006) Biochemistry 45 8649-8656 [DOI] [PubMed] [Google Scholar]
- 16.Narayanan, S., Sato, T., and Wolfe, M. S. (2007) J. Biol. Chem. 282 20172-20179 [DOI] [PubMed] [Google Scholar]
- 17.Das, C., Berezovska, O., Diehl, T. S., Genet, C., Buldyrev, I., Tsai, J. Y., Hyman, B. T., and Wolfe, M. S. (2003) J. Am. Chem. Soc. 125 11794-11795 [DOI] [PubMed] [Google Scholar]
- 18.Bihel, F., Das, C., Bowman, M. J., and Wolfe, M. S. (2004) J. Med. Chem. 47 3931-3933 [DOI] [PubMed] [Google Scholar]
- 19.Kimberly, W. T., LaVoie, M. J., Ostaszewski, B. L., Ye, W., Wolfe, M. S., and Selkoe, D. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6382-6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esler, W. P., Kimberly, W. T., Ostaszewski, B. L., Ye, W., Diehl, T. S., Selkoe, D. J., and Wolfe, M. S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2720-2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraering, P. C., Ye, W., LaVoie, M. J., Ostaszewski, B. L., Selkoe, D. J., and Wolfe, M. S. (2005) J. Biol. Chem. 280 41987-41996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornilova, A. Y., Das, C., and Wolfe, M. S. (2003) J. Biol. Chem. 278 16470-16473 [DOI] [PubMed] [Google Scholar]
- 23.Seeger, M., Nordstedt, C., Petanceska, S., Kovacs, D. M., Gouras, G. K., Hahne, S., Fraser, P., Levesque, L., Czernik, A. J., George-Hyslop, P. S., Sisodia, S. S., Thinakaran, G., Tanzi, R. E., Greengard, P., and Gandy, S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 5090-5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Podlisny, M. B., Citron, M., Amarante, P., Sherrington, R., Xia, W., Zhang, J., Diehl, T., Levesque, G., Fraser, P., Haass, C., Koo, E. H., Seubert, P., St George-Hyslop, P., Teplow, D. B., and Selkoe, D. J. (1997) Neurobiol. Dis. 3 325-337 [DOI] [PubMed] [Google Scholar]
- 25.Kornilova, A. Y., Bihel, F., Das, C., and Wolfe, M. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3230-3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karle, I. L., and Balaram, P. (1990) Biochemistry 29 6747-6756 [DOI] [PubMed] [Google Scholar]
- 27.Klafki, H., Abramowski, D., Swoboda, R., Paganetti, P. A., and Staufenbiel, M. (1996) J. Biol. Chem. 271 28655-28659 [DOI] [PubMed] [Google Scholar]
- 28.Wolfe, M. S., Citron, M., Diehl, T. S., Xia, W., Donkor, I. O., and Selkoe, D. J. (1998) J. Med. Chem. 41 6-9 [DOI] [PubMed] [Google Scholar]
- 29.Dorman, G., and Prestwich, G. D. (1994) Biochemistry 33 5661-5673 [DOI] [PubMed] [Google Scholar]
- 30.Nyborg, A. C., Kornilova, A. Y., Jansen, K., Ladd, T. B., Wolfe, M. S., and Golde, T. E. (2004) J. Biol. Chem. 279 15153-15160 [DOI] [PubMed] [Google Scholar]
- 31.Weggen, S., Eriksen, J. L., Das, P., Sagi, S. A., Wang, R., Pietrzik, C. U., Findlay, K. A., Smith, T. E., Murphy, M. P., Bulter, T., Kang, D. E., Marquez-Sterling, N., Golde, T. E., and Koo, E. H. (2001) Nature 414 212-216 [DOI] [PubMed] [Google Scholar]
- 32.Takahashi, Y., Hayashi, I., Tominari, Y., Rikimaru, K., Morohashi, Y., Kan, T., Natsugari, H., Fukuyama, T., Tomita, T., and Iwatsubo, T. (2003) J. Biol. Chem. 278 18664-18670 [DOI] [PubMed] [Google Scholar]
- 33.Beher, D., Clarke, E. E., Wrigley, J. D., Martin, A. C., Nadin, A., Churcher, I., and Shearman, M. S. (2004) J. Biol. Chem. 279 43419-43426 [DOI] [PubMed] [Google Scholar]
- 34.Dovey, H. F., John, V., Anderson, J. P., Chen, L. Z., de Saint Andrieu, P., Fang, L. Y., Freedman, S. B., Folmer, B., Goldbach, E., Holsztynska, E. J., Hu, K. L., Johnson-Wood, K. L., Kennedy, S. L., Kholodenko, D., Knops, J. E., Latimer, L. H., Lee, M., Liao, Z., Lieberburg, I. M., Motter, R. N., Mutter, L. C., Nietz, J., Quinn, K. P., Sacchi, K. L., Seubert, P. A., Shopp, G. M., Thorsett, E. D., Tung, J. S., Wu, J., Yang, S., Yin, C. T., Schenk, D. B., May, P. C., Altstiel, L. D., Bender, M. H., Boggs, L. N., Britton, T. C., Clemens, J. C., Czilli, D. L., Dieckman-McGinty, D. K., Droste, J. J., Fuson, K. S., Gitter, B. D., Hyslop, P. A., Johnstone, E. M., Li, W. Y., Little, S. P., Mabry, T. E., Miller, F. D., and Audia, J. E. (2001) J. Neurochem. 76 173-181 [DOI] [PubMed] [Google Scholar]
- 35.Seiffert, D., Bradley, J. D., Rominger, C. M., Rominger, D. H., Yang, F., Meredith, J. E., Jr., Wang, Q., Roach, A. H., Thompson, L. A., Spitz, S. M., Higaki, J. N., Prakash, S. R., Combs, A. P., Copeland, R. A., Arneric, S. P., Hartig, P. R., Robertson, D. W., Cordell, B., Stern, A. M., Olson, R. E., and Zaczek, R. (2000) J. Biol. Chem. 275 34086-34091 [DOI] [PubMed] [Google Scholar]
- 36.Kukar, T. L., Ladd, T. B., Bann, M. A., Fraering, P. C., Narlawar, R., Maharvi, G. M., Healy, B., Chapman, R., Welzel, A. T., Price, R. W., Moore, B., Rangachari, V., Cusack, B., Eriksen, J., Jansen-West, K., Verbeeck, C., Yager, D., Eckman, C., Ye, W., Sagi, S., Cottrell, B. A., Torpey, J., Rosenberry, T. L., Fauq, A., Wolfe, M. S., Schmidt, B., Walsh, D. M., Koo, E. H., and Golde, T. E. (2008) Nature 453 925-929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown, G. R., Bamford, A. M., Bowyer, J., James, D. S., Rankine, N., Tang, E., Torr, V., and Culbert, E. J. (2000) Bioorg. Med. Chem. Lett. 10 575-579 [DOI] [PubMed] [Google Scholar]
- 38.Nyborg, A. C., Ladd, T. B., Jansen, K., Kukar, T., and Golde, T. E. (2006) FASEB J. 20 1671-1679 [DOI] [PubMed] [Google Scholar]