Abstract
The hormone-sensitive lipase (HSL) and adipocyte fatty acid-binding protein (AFABP/aP2) form a physical complex that affects basal and hormone-stimulated adipocyte fatty acid efflux. Previous work has established that AFABP/aP2-HSL complex formation requires that HSL be in its activated, phosphorylated form and AFABP/aP2 have a bound fatty acid. To identify the HSL binding site of AFABP/aP2 a combination of alanine-scanning mutagenesis and fluorescence resonance energy transfer was used. Mutation of Asp17, Asp18, Lys21, or Arg30 (but not other amino acids in the helix-turn-helix region) to alanine inhibited interaction with HSL without affecting fatty acid binding. The cluster of residues on the helical domain of AFABP/aP2 form two ion pairs (Asp17-Arg30 and Asp18-Lys21) and identifies the region we have termed the charge quartet as the HSL interaction site. To demonstrate direct association, the non-interacting AFABP/aP2-D18K mutant was rescued by complementary mutation of HSL (K196E). The charge quartet is conserved on other FABPs that interact with HSL such as the heart and epithelial FABPs but not on non-interacting proteins from the liver or intestine and may be a general protein interaction domain utilized by fatty acid-binding proteins in regulatory control of lipid metabolism.
Basal and hormone-stimulated triacylglycerol lipolysis and efflux of fatty acids from adipocytes is a complex process involving multiple regulatory factors, structural proteins, lipid hydrolases, and carrier proteins (1). Complete triacylglycerol hydrolysis to 3 mol of fatty acids and 1 mol of glycerol is facilitated by multiple hydrolases (triglyceride hydrolase and adipose triglyceride hydrolase), a diacylglycerol hydrolase (hormone-sensitive lipase (HSL)2), and a monoacylglycerol lipase all localized onto the surface of the lipid droplet possibly in complex with the docking protein perilipin and regulated by interaction with CGI-58 (2-6). The product fatty acids are either re-esterified by associated acyl-CoA ligases or effluxed from the cell via a trafficking system involving the adipocyte fatty acid-binding protein (AFABP/aP2) as a lipid shuttle (7-9).
AFABP/aP2 forms a physical complex with HSL leading to the original proposition that such interaction facilitated FFA efflux (10). However, more recent work using mutational analysis coupled with fluorescence resonance energy transfer between ECFP-HSL and EYFP-AFABP/aP2 has established that for interaction to occur AFABP/aP2 must have an FFA bound and HSL must be in its activated phosphorylated form suggesting that the interaction may be regulatory leading to inhibition of lipase activity or influencing complex formation on the lipid droplet surface (11).
The site of AFABP/aP2 interaction on HSL has been mapped via a combination of mutagenesis and truncation analysis to a region bounded by amino acids 193-199 containing the core sequence EHYKRNE (10). In contrast, the site of interaction on AFABP/aP2 that binds to HSL has not been mapped previously. The requirement for HSL to be in its phosphorylated form and AFABP/aP2 to have a lipid bound makes in vitro assessment of the interaction sites technically difficult. As such, we have turned to in situ methods involving mutational analysis coupled with energy transfer to map the interaction domain on AFABP/aP2. In this report we present studies defining the site of HSL interaction on AFABP/aP2 as two ion pairs linking Asp17 to Lys21 and Asp18 to Arg30. This cluster of residues on helix α1 and helix α2 are defined as a “charge quartet” and is conserved in the heart and epithelial FABPs that also associate with HSL but not in the intestinal or liver FABP (12) that do not associate with HSL. The charge quartet, components of which have previously been suggested to be a nuclear localization sequence (13), is likely to represent a protein-protein interaction motif linking AFABP/aP2 to the regulation of a variety of cellular proteins involved in lipid metabolism and homeostasis.
EXPERIMENTAL PROCEDURES
Materials—Zeocin, Geneticin, Lipofectamine, and tissue culture reagents were obtained from Invitrogen. BODIPY D 3835 (4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid), referred to as BODIPY-C12, was obtained from Molecular Probes (Eugene, OR). pEYFP-C1 and pECFP-C1 were obtained from Clontech. Pure oleic acid was purchased from NuChek Prep (Elysian, MN). Restriction and DNA-modifying enzymes were obtained from Promega, New England Biolabs, and Stratagene. 1-Anilinonaphthalene-8-sulfonic acid (1,8-ANS) was purchased from Molecular Probes. Lipidx (hydroxyalkoxypropyl dextran Type VI) and all other reagents were purchased from Sigma-Aldrich. The University of Minnesota Microchemical Facility carried out DNA sequencing and synthesis of oligonucleotides used for PCR.
Cloning and Mutagenesis—Mutations in the proposed AFABP/aP2 HSL binding site were made on the template of native AFABP/aP2 subcloned into the A206K pEYFP-C1 expression vector using the QuikChange™ site-directed mutagenesis technique of Stratagene (La Jolla, CA) as described previously (14). The A206K pEYFP-C1 vector was used for all mutants to reduce intrinsic dimerization of fluorescent fusion proteins. The mutational primers were as follows: N15A, 5′-G CTT GTC TCC AGT GAA GCC TTC GAT GAT TAC ATG-3′; F16A, 5′-G CTT GTC TCC AGT GAA AAC GCC GAT GAT TAC ATG AAA G-3′; D17A, 5′-CC AGT GAA AAC TTC GCT GAT TAC ATG AAA GAA GTG GG-3′; D18A, 5′-CC AGT GAA AAC TTC GAT GCT TAC ATG AAA GAA GTG GGA GTG GGC-3′; Y19A, 5′-CC AGT GAA AAC TTC GAT GAT GCC ATG AAA GAA GTG GGA GTG GGC-3′; M20A, 5′-CC AGT GAA AAC TTC GAT GAT TAC GCG AAA GAA GTG GGA GTG GGC-3′; K21A, 5′-C TTC GAT GAT TAC ATG GCA GAA GTG GGA GTG GGC TTT GCC-3′; E22A, 5′-C GAT GAT TAC ATG AAA GCA GTG GGA GTG GGC TTT GCC-3′; V23A, 5′-TTC GAT GAT TAC ATG AAA GAA GCG GGA GTG GGC TTT GCC-3′; and R30A, 5′-GTG GGC TTT GCC ACA GCG AAA GTG GCA GGC ATG GCC AAG CCC-3′.
To assess the heterodimerization of AFABP/aP2 and HSL using EYFP-AFABP/aP2-D18K and ECFP-HSL-K196E, the following primers were utilized: AFABP/aP2-D18K, 5′-CC AGT GAA AAC TTC GAT AAG TAC ATG AAA GAA GTG GGA GTG GGC-3′; and HSL-K196E, 5′-C TTC GGG GAA CAC TAC GAA CGC AAC GAG ACG GGC C-3′. To evaluate the interaction of ECFP-HSL with FABPs other than AFABP/aP2, epithelial FABP (EFABP), heart FABP (HFABP), intestinal FABP (IFABP), and liver FABP (LFABP) were subcloned into A206K pEYFP-C1 (14).
Live Cell Imaging and FRET—C8PA lipocytes used for FRET analyses were cultured as described previously (11, 14). Briefly C8PA cells are HEK-293 fibroblasts stably expressing fatty acid transport protein 1 and perilipin A grown and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. C8PA cells transfected for FRET analysis were grown on polylysine-coated 13-mm coverslips in 12-well plates and transfected 24 h later with the expression plasmids specific for ECFP-HSL and/or EYFP-FABP (native or mutant). 24 h later the cells were lipid-loaded for 48 h with 300 μm oleic acid, 100 μm bovine serum albumin. Essentially identical results were obtained when cells are loaded with linoleic acid (data not shown). Lipolytic conditions were initiated by the addition of 20 μm forskolin, and digital images were captured between 2 and 4 h later. During microscopy, cells were kept in medium at 37 °C with 5% CO2 until imaging at room temperature.
Normalized FRET (NFRET) Analysis—Images were captured, and real time FRET measurements were performed and calculated as previously reported where a complete description of the methods and data analysis is provided (11). Image acquisition and measurement of three types of samples were carried out on each experiment for FRET index measurements as follows: one sample with cells expressing only the donor fluorophore to evaluate the donor bleed-through coefficient, one sample with cells expressing only the acceptor fluorophore to evaluate the acceptor bleed-through coefficient, and one sample with cells co-expressing the two fluorophores in which FRET signals were measured as described in Hachet-Haas et al. (15). The FRET measurements are expressed as the NFRET index. The NFRET index value ranges from 1 to 10 and is generated using the mean bleed-through coefficients (15-17). For statistical analysis, each FRET experiment was done at least three independent times. In each experiment, cells were evaluated visually to identify typical expression patterns and to assess general transfection efficiency for each of the two fusion proteins. Monolayers of co-transfected cells were scanned using the individual EYFP and ECFP channels to identify those cells where the expression of the two reporters was comparable. Multiple fields were considered for each experiment with at least five separate cell fields assessed for localization and fluorescence measurements. Data presented in Figs. 4 and 9 represent combined results from all experiments addressing fluorescence from 15-20 cells.
FIGURE 4.
Energy transfer between ECFP-HSL and EYFP-AFABP/aP2 mutants. Values for NFRET were calculated as described under “Experimental Procedures” for various EYFP-AFABP/aP2 forms co-expressed with ECFP-HSL. For simplicity, each mutant is listed as the protein substitution, and for the co-transfection results, C:H refers to fusion proteins utilizing ECFP-HSL, whereas Y:A refers to fusion proteins in EYFP-AFABP/aP2. Gray bars represent basal NFRET, whereas black bars represent energy transfer following addition of forskolin. Error bars represent S.E. (n = 15). The * denotes statistical significance (p < 0.05) in the NFRET value (forskolin-stimulated) for the indicated isoform compared with the AFABP/aP2 control. WT, wild type.
FIGURE 9.
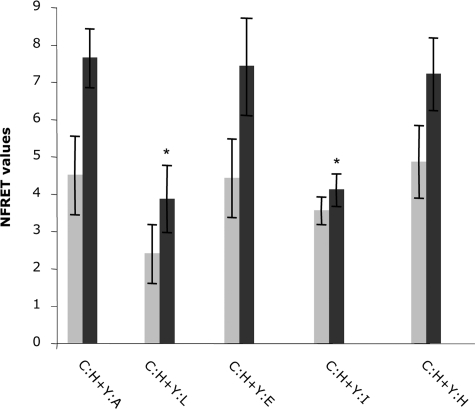
NFRET values representing relative energy transfer between ECFP-HSL and EYFP-linked fatty acid-binding proteins. Gray bars represent basal NFRET, whereas black bars represent energy transfer following addition of forskolin. Error bars represent S.E. (n = 15). C:H refers to ECFP-HSL, whereas each EYFP (Y)-fatty acid-binding protein fusion is indicated by a single letter denoting the adipocyte (A), liver (L), epithelial (E), intestinal (I), or heart (H) forms. The * denotes statistical significance (p < 0.05) in the NFRET value (forskolin-stimulated) for the indicated isoform compared with the AFABP/aP2 control.
Cells transfected with some of the mutants consistently contained aggregates under some conditions. Areas of a cell containing these aggregates were avoided when capturing images to be used for the comparative basal measurements. The forskolin-treated cells, however, were measured in the areas of HSL translocation, including any aggregates present in that area.
Expression, Purification, and Ligand Binding Analysis of His-tagged Native and Mutant Forms of AFABP/aP2—Escherichia coli BL21 DE3 pLysS cells were transformed with a plasmid encoding His-tagged native AFABP/aP2 or the indicated mutants as described previously (18). Cultures were grown, and expression of the protein was induced with 1 mm isopropyl thiogalactoside for 4 h. Following centrifugation, the cells were lysed in a French press and sonicated, and the soluble proteins were recovered following centrifugation for 1 h at 100,000 × g. The His-tagged AFABP/aP2 proteins were purified on a nickel affinity column and eluted with 250 mm imidazole. The pooled eluate was subjected to Lipidx chromatography to remove any bound ligand and subsequently dialyzed into 25 mm Tris-HCl, 50 mm NaCl (pH 7.4) buffer. The purified FABPs were verified by SDS-PAGE and quantified spectrophotometrically.
Fluorescence binding assays were performed using the probe 1-anilinonaphthalene-8-sulfonic acid as described previously (19). Briefly 1,8-ANS was dissolved in absolute ethanol and then diluted with 25 mm Tris-HCl (pH 7.4) to a final concentration of 5 μm (final ethanol concentration of 0.05%). For each assay, 500 μl of 1,8-ANS was mixed with increasing amounts of AFABP/aP2, and fluorescence was measured using a PerkinElmer Life Sciences 650-10S fluorescence spectrophotometer. Scatchard analyses were performed to calculate dissociation constants.
RESULTS
In a previous report we described studies that established that a fatty acid must be bound to AFABP/aP2 for association with HSL and that at least one amino acid, Lys21, was required for complex formation (11). In those studies, EYFP-AFABP/aP2-K21I did not exhibit FRET with ECFP-HSL, whereas EYFP-AFABP/aP2-K31I did, implying that a region on or about Lys21 of helix α1 comprises a component of the HSL binding site. As shown in Fig. 1, the side chain of Lys21 is found within a cluster of charged residues demarcated by Asp17, Asp18, Lys21, Arg30, and Lys31. Although Asp17, Asp18, and Lys21 reside on helix α1, Arg30 and Lys31 are found on helix α2 in close proximity with the hydrocarbon chain of a bound fatty acid. Analysis of crystal coordinates (20) revealed that the side chain of Asp18 forms an ion pair with that of Lys21, whereas the side chain of Asp17 forms a salt bridge to that of Arg30, forming a quartet of charges that bridge the two helices. The observation that AFABP/aP2-K21I exhibits native fatty acid binding activity but does not physically bind to HSL points toward some unique features of Lys21 that could provide an explanation for why fatty acid binding is required for interaction with HSL. Xu et al. (20) demonstrated that Lys21 is one of the few residues whose main chain atoms vary in position with fatty acid binding, and LiCata and Bernlohr (21), using computational methods to assess the accessible surface area for each of the AFABP/aP2 side chains in both the apo- and holoforms, identified Asp18 and Lys21 as residues with significantly decreased surface accessibility in the holoform. To assess the involvement of the various residues of the proposed AFABP/aP2-HSL binding site, AFABP/aP2 residues Asn15 through Val23 as well as Arg30 were sequentially changed to alanine mutants in the A206K pEYFP-C1 vector, and FRET analysis was performed between each of the mutant proteins and ECFP-HSL.
FIGURE 1.
Spatial organization of the AFABP/aP2 charge quartet. A, ribbon diagram of the AFABP/aP2 helix-turn-helix region (from Protein Data Bank code 1lib) with the side chains of Asp17, Asp18, and Lys21 on helix α1; Arg30 and Lys31 on helix α2; and oleic acid shown in space-filling mode. B, 90° rotation view showing orientation of charged residues.
Our previous studies using C8PA lipocytes (HEK-293 cells stably transfected with fatty acid transport protein 1 and perilipin A) have demonstrated that physical association between ECFP-HSL and EYFP-AFABP/aP2 occurs in the cytoplasm under basal conditions and is markedly increased in response to forskolin treatment (11, 14). Activated, phosphorylated HSL translocates to the surface of the lipid droplet and forms a complex with multiple proteins facilitating efficient lipid hydrolysis (18, 22-24). Fig. 2 shows the complete set of images and controls used for FRET analysis to determine physical association of the fusion proteins expressed by pECFP-HSL and pEYFP-AFABP/aP2 co-transfected into C8PA cells. For each mutant protein evaluated in this study the same control experiments were repeated to assure that ECFP-HSL translocated to the droplet surface and exhibited energy transfer with EYFP-AFABP/aP2.
FIGURE 2.
Energy transfer between ECFP-HSL and EYFP-AFABP/aP2 co-expressed in C8PA cells. C8PA lipocytes were co-transfected with pECFP-HSL and pEYFP-AFABP/aP2 and incubated with oleate-bovine serum albumin (3:1). After 48 h, the energy transfer between ECFP-HSL and EYFP-AFABP/aP2 in the absence (C) or presence (F) of 20 μm forskolin was determined. Images are shown for both the cyan (ECFP-HSL) and yellow (EYFP-AFABP/aP2) channels as well as energy transfer (FRET) between the two fluorophores (artificially colored as magenta). Localization of the ECFP-HSL-EYFP-AFABP/aP2 complex on the droplet surface can be compared with the BODIPY-C12 labeling of triacylglycerol. The scale bar represents 5 μm in the differential interference contrast (DIC) image.
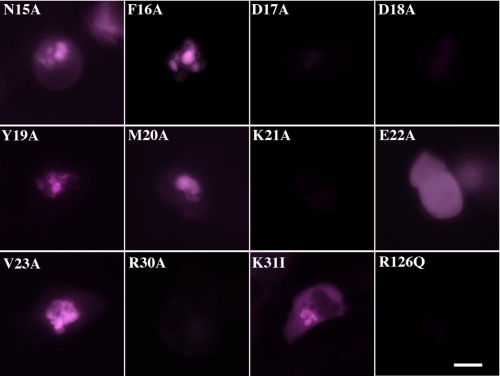
For analysis of the alanine-scanning series of mutants surrounding the potential AFABP/aP2-HSL binding site as well as for EYFP-AFABP/aP2-R126Q (non-fatty acid-binding) and EYFP-AFABP/aP2-K31I (fatty acid-binding and electrostatically equivalent to K21A) mutants, C8PA cells were grown, transfected with the appropriate cDNAs, loaded with oleic acid, and stimulated with forskolin as described previously (11, 14). Each alanine mutant fusion protein was analyzed for fluorescence energy transfer with native ECFP-HSL. Additionally because all FRET images were collected using the same acquisition settings, we were able to determine the NFRET value for HSL association with AFABP/aP2 or each mutant AFABP/aP2 protein and compare them quantitatively. Fig. 3 shows the FRET images for each of these determinations. The resulting relative values of NFRET are presented in Fig. 4 and allow each AFABP/aP2 form to be compared under both basal and lipolytic conditions. From these images and the NFRET data Asp17, Asp18, Lys21, and Arg30 were identified as the most critical residues for the binding of AFABP/aP2 to HSL. Interestingly EYFP-AFABP/aP2-E22A did not translocate to the droplet surface suggesting that this protein associates with HSL but may affect complex formation.
FIGURE 3.
FRET images for ECFP-HSL EYFP-AFABP/aP2 complex formation. Fluorescence resonance energy transfer (colored magenta as described in the legend to Fig. 2) between ECFP-HSL and the designated AFABP/aP2 mutants in forskolin-stimulated C8PA cells is shown. Each AFABP/aP2 mutant is expressed as a fusion protein with EYFP but is referred to in the figure simply by the AFABP/aP2 amino acid substitution. The scale bar represents 5 μm.
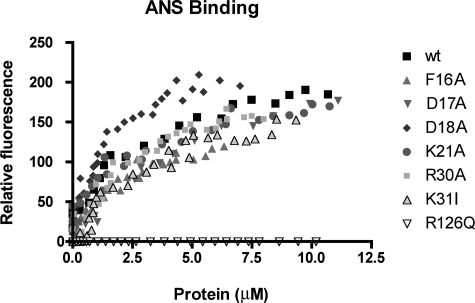
Because our prior work had established that a fatty acid must be bound to AFABP/aP2 to form a complex with HSL (11), we assessed the ligand binding activity of each mutant using the surrogate ligand 1,8-ANS. As shown in Fig. 5, although there were some differences in binding affinity, each of the alanine mutants retained fatty acid binding capacity similar to that of native AFABP/aP2. In contrast AFABP/aP2-R126Q exhibited essentially no 1,8-ANS binding and did not demonstrate energy transfer in FRET analysis with ECFP-HSL (11).
FIGURE 5.
1,8-ANS binding by selected AFABP/aP2 mutants. The indicated AFABP/aP2 mutants (expressed as His6 fusion proteins) were expressed in bacteria, purified, and evaluated for lipid binding using the surrogate ligand 1,8-ANS. Binding results for each protein shown represents one of at least three independent trials. wt, wild type.
The helix-turn-helix domain of AFABP/aP2 has been shown in some, but not all, x-ray crystal structures to be a site for homodimerization. Indeed Gillilan et al. (13) have reported that AFABP/aP2 is dimeric and, depending upon the ligand, may associate via a region encompassing the helical domain. Fig. 6 shows a sequence comparison of various FABPs with HSL and reveals that the helical region containing the charge quartet on AFABP/aP2 has some similarity to the corresponding binding site on HSL that has been identified previously (10). Of particular note are HSL Glu193 and Lys196. Although there is no x-ray or NMR structure determined for HSL, secondary structure algorithms predict that this segment of sequence would be in a helical conformation. This raises the possibility that the helical region on AFABP/aP2 interacts with a similar helical domain on HSL and that the intramolecular ion pair between Asp18 and Lys21 of AFABP/aP2 may form two intermolecular ion pairs with HSL: Asp18 of AFABP/aP2 with Lys196 of HSL and Lys21 of AFABP/aP2 with Glu193 of HSL. To test this hypothesis we carried out a charge reversal experiment. In this experiment, the EYFP-AFABP/aP2-D18K and the ECFP-HSL-K196E mutants were utilized. As shown in Fig. 7 EYFP-AFABP/aP2-D18K did not undergo significant energy transfer when co-expressed with ECFP-HSL, and conversely ECFP-HSL-K196E exhibited markedly reduced FRET when co-expressed with EYFP-AFABP/aP2. However, co-transfection of pEYFP-AFABP/aP2-D18K with the charge reversal mutant pECFP-HSL-K196E rescued energy transfer between the expressed fusion proteins to a level similar to that of the native controls. Fig. 4 provides the NFRET values obtained from these co-transfection experiments. These results indicate that during lipolytic stimulation AFABP/aP2 and HSL directly bind to each other. These results are consistent with previous titration microcalorimetry studies that had indicated a 1:1 molar stoichiometry of binding between the two proteins (18). Moreover the region of sequence similarity between AFABP/aP2 and HSL likely represents an interaction domain linking the charge quartet of AFABP/aP2 to a similar region on HSL.
FIGURE 6.
Sequence alignment of HSL with various fatty acid-binding proteins. Shaded areas show regions of identity or chemical similarity.
FIGURE 7.
Energy transfer between ECFP-HSL and EYFP-AFABP/aP2 charge reversal mutants. C8PA lipocytes were co-transfected with pECFP-HSL and pEYFP-AFABP/aP2 or the indicated mutant forms and incubated with oleate-bovine serum albumin (3:1). After 48 h FRET was evaluated in the absence (C) or presence (F) of 20 μm forskolin. For simplicity, each mutant is listed as the protein substitution, and for the co-transfection results, C:H refers to fusion proteins utilizing ECFP-HSL, whereas Y:A refers to fusion proteins in EYFP-AFABP/aP2. The scale bar represents 5 μm. WT, wild type.
The adipocyte fatty acid-binding protein is a member of the multigene family of hydrophobic lipid-binding proteins that contains several other structurally related 15-kDa polypeptides. In this family, spatial alignment of the α carbon backbones of the proteins are virtually superimposable, whereas the primary sequence varies considerably from 20 to 70% sequence identity (12, 25). Given the definition of Asp17, Asp18, Lys21, and Arg30 as primary determinants of HSL binding for AFABP/aP2, we assessed four other FABPs that have similar sequences in that region. To accomplish this epithelial FABP, heart FABP, intestinal FABP, and liver FABP were cloned into the A206K pEYFP-C1 vector, and the fluorescent fusion proteins were evaluated for binding with ECFP-HSL using FRET analyses. As shown in Figs. 8 and 9, EYFP-EFABP and EYFP-HFABP, but not EYFP-LFABP or EYFP-IFABP, exhibited energy transfer when co-expressed with ECFP-HSL with NFRET values similar to that for EYFP-AFABP/aP2. Fig. 6 shows the sequence comparison in the helical regions of the various FABPs and reveals that each of the proteins that associate with HSL contain acidic functions at positions 17 and 18 (relative to AFABP/aP2 numbering), a Lys residue at position 21 and an Arg residue at position 30. Intestinal FABP has a basic residue at position 18 and an acidic residue at position 21, whereas liver FABP contains a proline at position 18 and an isoleucine at position 30.
FIGURE 8.
Energy transfer between ECFP-HSL and other EYFP-FABP isoforms. C8PA lipocytes were co-transfected with pECFP-HSL and pEYFP fused in frame to the indicated FABP and incubated with oleate-bovine serum albumin (3:1). After 48 h FRET was evaluated in the absence (C) or presence (F) of 20 μm forskolin. For simplicity, each mutant is listed as the protein substitution, and for the co-transfection results, C:H refers to fusion proteins utilizing ECFP-HSL, whereas Y:A refers to fusion proteins in EYFP-AFABP/aP2. The scale bar represents 5 μm.
DISCUSSION
Previous work detailing the molecular and physiological interaction between AFABP/aP2 and HSL has centered upon the following components. 1) The site with which AFABP/aP2 interacts on HSL has been defined from amino acids 190 to 200 in an N-terminal docking domain discrete from the C-terminal catalytic esterase domain (10). 2) AFABP/aP2 must have a fatty acid bound to interact with HSL (11); mutation of the critical arginine residue that forms part of the H-bonding network (Arg126) to a non-H-bonding glutamine renders the protein unable to bind FFA (25) or HSL (12). 3) The interaction forms a 1:1 complex between AFABP/aP2 and HSL (18). Loss of AFABP/aP2 (AFABP/aP2-null mice) results in adipocytes with 4) an increase in intracellular FFA without activation of inflammatory pathways (26) and 5) a decrease in the efflux of FFA from the adipocyte both in situ and in vivo (7, 8). Despite the broad information base, the binding site on AFABP/aP2 that forms the cognate interaction domain with HSL has not been defined; this report addresses that identification.
Using a mutagenesis strategy in combination with fluorescence resonance energy transfer, a quartet of charges in the helix-turn-helix domain of AFABP/aP2 defined by Asp17, Asp18, Lys21, and Arg30 were identified. Examination of the AFABP/aP2 apo- and holostructures revealed that within this quartet salt bonds are formed between Asp17 and Arg30 as well as between Asp18 and Lys21 (Fig. 1). Mutagenesis of any of these four residues and subsequent analysis via FRET revealed that these mutant AFABP/aP2 forms exhibited greatly diminished interaction with HSL. The interaction is specific to these residues because mutagenesis of surrounding amino acids did not affect AFABP/aP2-HSL interaction. Included in this analysis was Lys31, a spatially adjacent residue that when mutated to alanine had no affect on HSL interaction (11) indicating that the association was not purely controlled by AFABP/aP2 electrostatics but required defined charges in a spatially concise configuration. Importantly mutagenesis of any of the quartet charges had little effect on fatty acid binding (Fig. 5) indicating that the binding determinant for HSL interaction is distinct and separable from the interaction of FFA with AFABP/aP2 and that mutants can be identified that define physical interaction with either fatty acids (e.g. R126Q), protein (e.g. D18A), or membrane phospholipids (K31I).
Previous work by LiCata and Bernlohr (21) analyzed the apo- and holoprotein structure of AFABP/aP2 and concluded that significant changes in the accessible surface area of the protein occur upon ligand binding. Of the residues that had side chains influenced by fatty acid binding, Asp17 exhibited a decrease in accessibility for several fatty acids evaluated. In contrast, both Asp18 and Lys21 demonstrated ligand-dependent changes in accessible surface area with some fatty acids increasing and others decreasing accessibility. In contrast, Arg30 did not exhibit major structural changes in response to ligand binding. These results imply a ligand-dependent change in the orientation of helix α1, particularly in the Asp18-Lys21 salt bond region.
Using a combination of in situ (FRET) (14) and in vitro (titration microcalorimetry) (18) analyses, several other, but not all, FABPs were shown to interact with HSL. Other interacting fatty acid-binding proteins include epithelial FABP and heart FABP (Fig. 8), whereas liver FABP and intestinal FABP exhibited reduced association. Comparison of the sequences in and around the helical domain of each FABP showed a consensus alignment for the binding site as F-Acidic-Acidic-YMKX2GVG (Fig. 6). Each of AFABP/aP2, EFABP, and HFABP contain this domain, whereas non-binding IFABP and LFABP forms do not, supporting the finding that this region defines the interaction region. Other FABPs that contain the binding motif but have not been evaluated for HSL association are testis and myelin FABP.
As shown in Fig. 6, the sequence of HSL from amino acids 190 to 201 also contains many of the basic core elements of the FABP interaction site. The HSL sequence FGEHYKRNETG represents a modified version of the core FABP element and the prediction that in HSL this region forms a helical interaction partner with AFABP/aP2 allowing for the acidic residue on AFABP/aP2 (Asp18) to interact with the Lys196 on HSL and Lys21 of AFABP/aP2 to ion pair with a cognate Glu193 residue on HSL. Consistent with this, Figs. 4 and 7 show that when Asp18 is mutated to a lysine residue the interaction with HSL was lost but could be rescued when the corresponding basic residue of HSL (Lys196) was converted to a glutamic acid. Although not evaluated, the extension of this is that mutation of AFABP/aP2 Lys21 to Glu would result in loss of interaction with HSL but could be restored by corresponding mutation of HSL Glu193 to Lys. This finding predicts that other FABP partners would also contain the interaction motif.
Work by Liou and Storch (27) has shown that AFABP/aP2 (and several other FABPs) donates a bound fatty acid to membranes via a collisional mechanism that requires Lys21 and Lys31. The studies herein indicated that Lys21, but not Lys31, was required for HSL interaction implying that the protein-protein interaction site overlaps with the protein-lipid interface and shares several common determinants. The structure of AFABP/aP2 (Fig. 1) shows that Lys31 is very close to Lys21 but does not form salt bonds with an adjacent amino acid. Indeed K31I served as a negative control in FRET experiments and indicates that this residue may be a discriminating factor in defining protein-protein association versus lipid trafficking.
The region on AFABP/aP2 identified as the HSL binding site overlaps with a domain suggested by Gillilan et al. (13) to be part of a nuclear localization domain involving Lys21, Arg30, and Lys31. Mutagenesis analysis of several EYFP-AFABP/aP2 fusions revealed that none of the residues affected intracellular localization when expressed into HEK-293 cells: AFABP/aP2 was found ubiquitously throughout the cell. However, in the studies of Gillilan et al. (13) the location of AFABP/aP2 in COS-7 cells was dependent upon such residues suggesting that cell-specific factors may be determining influences for localization rather than being an intrinsic property of the protein itself. Indeed in prior work we have shown in 3T3-L1 cells that early in the differentiation program (days 4-6 in the standard protocol) AFABP/aP2 is largely nuclear, whereas later (days 12-16) the protein is also found cytoplasmically (28). Given our observation that Lys21 and Arg30 are central to HSL association, translocation may represent interaction with nuclear proteins with a similar targeting sequence.
In summary, this work presents for the first time the binding site on AFABP/aP2 for HSL and suggests that association with other proteins may involve the same domain. Because the interaction with HSL requires that a fatty acid be bound to AFABP/aP2 the association is considered to be regulatory, serving as a potential negative feedback loop regulating lipid hydrolysis. Current studies are underway to evaluate this proposal.
Acknowledgments
We thank the members of the Bernlohr laboratory for helpful suggestions and comments during the development of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK053189 (to D. A. B.) and P30 DK050456 (to the Minnesota Obesity Center). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HSL, hormone-sensitive lipase; FABP, fatty acid-binding protein; AFABP/aP2, adipocyte FABP also known as aP2; FFA, free unesterified fatty acid; ECFP, enhanced cyan fluorescent protein; EYFP, enhanced yellow fluorescent protein; BODIPY, 4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid; 1,8-ANS, 1-anilinonaphthalene-8-sulfonic acid; FRET, fluorescence resonance energy transfer; NFRET, normalized FRET; EFABP, epithelial FABP; HFABP, heart FABP; IFABP, intestinal FABP; LFABP, liver FABP.
References
- 1.Greenberg, A. S., Shen, W. J., Muliro, K., Patel, S., Souza, S. C., Roth, R. A., and Kraemer, F. B. (2001) J. Biol. Chem. 276 45456-45461 [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Botas, J., Anderson, J. B., Tessier, D., Lapillonne, A., Chang, B. H., Quast, M. J., Gorenstein, D., Chen, K. H., and Chan, L. (2000) Nat. Genet. 26 474-479 [DOI] [PubMed] [Google Scholar]
- 3.Haemmerle, G., Zimmermann, R., Hayn, M., Theussl, C., Waeg, G., Wagner, E., Sattler, W., Magin, T. M., Wagner, E. F., and Zechner, R. (2002) J. Biol. Chem. 277 4806-4815 [DOI] [PubMed] [Google Scholar]
- 4.Granneman, J. G., Moore, H. P., Granneman, R. L., Greenberg, A. S., Obin, M. S., and Zhu, Z. (2007) J. Biol. Chem. 282 5726-5735 [DOI] [PubMed] [Google Scholar]
- 5.Brasaemle, D. L. (2007) J. Lipid Res. 48 2547-2559 [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi, T., Omatsu, N., Omukae, A., and Osumi, T. (2006) Mol. Cell. Biochem. 284 167-173 [DOI] [PubMed] [Google Scholar]
- 7.Baar, R. A., Dingfelder, C. S., Smith, L. A., Bernlohr, D. A. Wu, C., Lange, A. J., and Parks, E. J. (2004) Am. J. Physiol. 288 E187-E193 [DOI] [PubMed] [Google Scholar]
- 8.Scheja, L., Makowski, L., Uysal, K. T., Wiesbrock, S. M., Shimshek, D. R., Meyers, D. S., Morgan, M., Parker, R. A., and Hotamisligil, G. S. (1999) Diabetes 48; 1987-1994 [DOI] [PubMed] [Google Scholar]
- 9.Storch, J., and Thumser, A. E. (2000) Biochim. Biophys. Acta 1486 28-44 [DOI] [PubMed] [Google Scholar]
- 10.Shen, W. J., Liang, Y., Hong, R., Patel, S., Natu, V., Sridhar, K., Jenkins, A., Bernlohr, D. A., and Kraemer, F. B. (2001) J. Biol. Chem. 276 49443-49448 [DOI] [PubMed] [Google Scholar]
- 11.Smith, A. J., Thompson, B. R., Sanders, M. A., and Bernlohr, D. A. (2007) J. Biol. Chem. 282 32424-32432 [DOI] [PubMed] [Google Scholar]
- 12.Banaszak, L., Winter, N., Xu, Z., Bernlohr, D. A., Cowan, S., and Jones, T. A. (1994) Adv. Protein Chem. 45 89-151 [DOI] [PubMed] [Google Scholar]
- 13.Gillilan, R. E., Ayers, S. D., and Noy, N. (2007) J. Mol. Biol. 372 1246-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith, A. J., Sanders, M. A., Thompson, B. R., Londos, C., Kraemer, F. B., and Bernlohr, D. A. (2004) J. Biol. Chem. 279 52399-52405 [DOI] [PubMed] [Google Scholar]
- 15.Hachet-Haas, M., Converset, N., Marchal, O., Matthes, H., Gioria, S., Galzi, J. L., and Lecat, S. (2006) Microsc. Res. Tech. 69 941-956 [DOI] [PubMed] [Google Scholar]
- 16.Youvan, D. C., Silva, C. M., Bylin, E. M., Coleman, W. J., Dilworth, M. R., and Yang, M. M. (1997) Biotechnology et alia 1 1-16 [Google Scholar]
- 17.Feige, J. N., Sage, D., Wahli, W., Desvergne, B., and Gelman, L. (2005) Microsc. Res. Tech. 68 51-58 [DOI] [PubMed] [Google Scholar]
- 18.Jenkins-Kruchten, A. E., Bennaars-Eiden, A., Ross, J. R., Shen, W. J., Kraemer, F. B., and Bernlohr, D. A. (2003) J. Biol. Chem. 278 47636-47643 [DOI] [PubMed] [Google Scholar]
- 19.Kane, C. D., and Bernlohr, D. A. (1996) Anal. Biochem. 233 197-204 [DOI] [PubMed] [Google Scholar]
- 20.Xu, Z. H., Buelt, M. K., Banaszak, L. J., and Bernlohr, D. A. (1991) J. Biol. Chem. 266 14367-14370 [PubMed] [Google Scholar]
- 21.LiCata, V. J., and Bernlohr, D. A. (1998) Proteins 33 577-589 [DOI] [PubMed] [Google Scholar]
- 22.Londos, C., Brasaemle, D. L., Schultz, C. J., Segrest, J. P., and Kimmel, A. R. (1999) Semin. Cell Dev. Biol. 10 51-58 [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi, H., Souza, S. C., Zhang, H. H., Strissel, K. J., Christoffolete, M. A., Kovsan, J., Rudich, A., Kraemer, F. B., Bianco, A. C., Obin, M. S., and Greenberg, A. S. (2006) J. Biol. Chem. 281 15837-15844 [DOI] [PubMed] [Google Scholar]
- 24.Yin, W., Mu, J., and Birnbaum, M. J. (2003) J. Biol. Chem. 278 43074-43080 [DOI] [PubMed] [Google Scholar]
- 25.Storch, J., and Corsico, B. (2008) Annu. Rev. Nutr. 28 73-95 [DOI] [PubMed] [Google Scholar]
- 26.Makowski, L., Boord, J. B., Maeda, K., Babaev, V. R., Uysal, K. T., Morgan, M. A., Parker, R. A., Suttles, J., Fazio, S., Hotamisligil, G. S., and Linton, M. F. (2001) Nat. Med. 7 699-705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liou, H. L., and Storch, J. (2001) Biochemistry 40 6475-6485 [DOI] [PubMed] [Google Scholar]
- 28.Helledie, T., Antonius, M., Sorensen, R. V., Hertzel, A. V., Bernlohr, D. A., Kolvraa, S., Kristiansen, K., and Mandrup, S. (2000) J. Lipid Res. 41 1740-1751 [PubMed] [Google Scholar]