Abstract
Protein-disulfide isomerase (PDI) catalyzes the formation of the correct pattern of disulfide bonds in secretory proteins. A low resolution crystal structure of yeast PDI described here reveals large scale conformational changes compared with the initially reported structure, indicating that PDI is a highly flexible molecule with its catalytic domains, a and a′, representing two mobile arms connected to a more rigid core composed of the b and b′ domains. Limited proteolysis revealed that the linker between the a domain and the core is more susceptible to degradation than that connecting the a′ domain to the core. By restricting the two arms with inter-domain disulfide bonds, the molecular flexibility of PDI, especially that of its a domain, was demonstrated to be essential for the enzymatic activity in vitro and in vivo. The crystal structure also featured a PDI dimer, and a propensity to dimerize in solution and in the ER was confirmed by cross-linking experiments and the split green fluorescent protein system. Although sedimentation studies suggested that the self-association of PDI is weak, we hypothesize that PDI exists as an interconvertible mixture of monomers and dimers in the endoplasmic reticulum due to its high abundance in this compartment.
In the extracellular oxidative environment, intra- and inter-molecular disulfide bonds are utilized to maintain the tertiary or quarternary structure of most secreted proteins. The formation of the correct pattern of disulfide bonds is essential for the function of these proteins and, in turn, the viability of the cell. This process requires two major steps: oxidation of the sulfhydryl groups of cysteines to form disulfide bonds and isomerization of mispaired disulfide bonds. In bacteria, two periplasmic enzymes catalyze these reactions, with DsbA being responsible for oxidation and DsbC for isomerization (reviewed in Ref. 1). In eukaryotes, these processes occur in the endoplasmic reticulum (ER),2 and are catalyzed by a single enzyme, protein-disulfide isomerase (PDI). PDI is an abundant ER lumenal protein that is essential for cellular viability (2, 3), which is highly conserved from yeast to human (reviewed in Ref. 4). There are several PDI paralogs in humans (5) and to a lesser extent also in yeast (6), but their detailed functions have not yet been determined.
Yeast PDI is 522 amino acids in length and contains four thioredoxin-like domains, a, b, b′, and a′, followed by an acidic C-terminal extension (supplemental Fig. S1A) containing the ER-retention signal HDEL. The a and a′ domains each contain one active site, characterized by a Cys-Gly-His-Cys thioredoxin motif, which mediates the formation and rearrangement of protein disulfide bonds via oxidation/reduction reactions involving the two cysteine residues. The substrate binding site in mammalian PDI has been localized to the b′ domain (7, 8). Besides catalyzing the formation of correct disulfide bonds, it has been reported that PDI also acts as a chaperone that can facilitate the folding of proteins that lack disulfide bonds (9). In addition, PDI constitutes the non-catalytic subunit of prolyl 4-hydroxylase (10) and is a subunit of the microsomal triglyceride transfer protein (11), however, its function as a component of these enzymes has not been fully defined.
Recently we reported the crystal structure of full-length PDI from Saccharomyces cerevisiae deduced from crystals that were grown at 4 °C (12). All four domains of yeast PDI were found to be structurally similar to thioredoxin and the acidic C-terminal extension was partially ordered. These four domains were found to be spatially organized in the shape of a twisted “U” with the a and a′ domains on the ends of the U and the b and b′ domains forming the base. The two active sites in the a and a′ domains face each other from a distance of ∼28 Å. A highly hydrophobic patch in the b′ domain is located between the two active sites, which, together with hydrophobic areas surrounding the active sites, form a continuous hydrophobic surface across the a, b′, and a′ domains. The domain arrangement, active site location, and surface features strikingly resemble the “V” shaped, homodimeric structure of DsbC (13), a functional homolog of PDI in Escherichia coli. In addition, the two active sites of PDI are structurally asymmetric in both redox state and accessibility, which mirrors their differential involvement in the activity of PDI in vitro (14-16) and in vivo (17). In this study, a distinct 3.7-Å resolution structure derived from a different crystal form of full-length PDI obtained at 22 °C is described. The large scale conformational changes and a PDI dimer observed in this crystal structure have been studied by biochemical and biophysical techniques and the relevance of these features to the function of PDI are discussed.
EXPERIMENTAL PROCEDURES
Protein Purification, Crystallization, and Data Collection—Full-length yeast PDI and all mutants were expressed and purified as described (12). Crystallization was performed utilizing published conditions (12), except that the temperature was 22 °C. The resulting crystals diffracted to 3.7-Å resolution and diffraction data were collected as previously described (12). The crystals belong to space group C2221 compared with I4 for the crystals obtained at 4 °C (Table 1).
TABLE 1.
Data collection and refinement statistics
Rsym = ∑hkl∑iIi - <I>|/∑i<I>, where Ii is the ith measurement and <I> is the weighted mean of all measurements of I. <I/sigI> indicates the average of the intensity divided by its standard deviation. Numbers in parentheses refer to the highest resolution data shell. Rcryst = ∑∥Fo| - Fc∥/∑|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes. Rfree same as R for 5% of the data randomly omitted from refinement. Ramachandran statistics indicate the fraction of residues in the favored, allowed, and outlier regions of the Ramachandran diagram as defined by MolProbity.
| Data collection statistics | |
|---|---|
| Space group | C2221 |
| Unit cell dimensions (Å) | a = 116.9, b = 123.2, c = 75.7 |
| Unique reflections | 6,049 |
| Resolution limits (Å) | 20-3.7 |
| Completeness (highest shell) | 99.9 (99.7) |
| Redundancy | 5.6 (5.5) |
| Rsym (highest shell) | 0.092 (0.696) |
| <I/σI> (highest shell) | 18.3 (2.1) |
| Refinement statistics | |
| Resolution limits | 20-3.7 |
| Number of working/test reflections | 5,738/306 |
| Number of protein/solvent atoms | 3,864/0 |
| Overall average B-factor (Å2) | 179.9 |
| (individual domains: a/b/b′/a′/c) | 180.0/179.4/180.8/179.2/181.2 |
| R-factor (Rfree) | 0.242 (0.347) |
| Different component precision index (Å) | 1.073 |
| Root mean square deviations from ideal values | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.773 |
| Torsion angles (°) | 8.6/40.9/19.9/17.8 |
| Planar groups (Å) | 0.005 |
| Ramachandran statistics | 76.8/96.3/3.7 |
Structure Determination and Refinement—The 2.4-Å resolution structure of full-length PDI (12) was used to solve the structure of PDI in the 22 °C crystals by molecular replacement with the programs MOLREP (18) and PHASER (19) at 5.0-Å resolution. After attempts with the intact molecule as the search model failed, the individual domains were used instead. The b, b′, and a′ domains could be easily located and appeared to be in a similar spatial arrangement as in the 4 °C structure. However, the a domain could only be located manually by fitting it into a region of significant difference density located near the N terminus of the b domain. The resulting model was carefully refined with REFMAC and the refinement statistics are listed in Table 1. Secondary structure elements were defined with PROMOTIF (20) and figures were generated with PYMOL (21).
Generation of Yeast PDI Variants and in Vitro Assays—Based on the pTYB12-PDI plasmid (12) point mutations were created using the QuikChange XL site-directed mutagenesis kit (Stratagene). Truncation mutants were prepared according to the domain boundaries as defined in Fig. S1A using a PCR cloning approach. All mutants were expressed and purified according to the protocol described for wild-type PDI. Enzymatic activity assays of PDI were carried out using either reduced or scrambled RNase (12).
Native PAGE and Ellmans Assay—Native PAGE was carried out as described (22) with or without a 30-min preincubation with 50 mm DTT at room temperature. A mixture of thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and bovine serum albumin (67 kDa) was used as molecular mass marker for native PAGE. Ellmans assay was carried out using the protocol recommended by the manufacturer (Pierce) except that glutathione was used instead of cysteine to generate the standard curve. The number of free -SH groups was calculated as the molar concentration of free -SH groups measured by Ellmans assay divided by the molar concentration of the protein.
Strains and General Yeast Genetics—All strains prepared during this study are listed in Table 2. MLY200 (MATa ura3-52 leu2-1 lys2-801 ade2-101 trp1-1 his3-200 Δpdi1::HIS6 pRS316-PDI) (23) was used as parent strain for transformations. Standard synthetic defined medium containing specific drop-out amino acids mixtures (24) was used for growth of cells at 30 °C unless specified otherwise. Plasmid shuffling experiments were performed as described (25), and yeast transformations were carried out by the lithium acetate procedure, enhanced by the addition of dimethyl sulfoxide (26). The growth rates of yeast strains were evaluated by spotting 3 μl of serial 10-fold dilutions of yeast cells in early log phase (A600 less than 0.5) on synthetic defined agar plates lacking tryptophan. Plates supplemented with DTT were prepared by adding DTT to hot medium immediately before pouring. The plates were stored under N2 and used within 12 h.
TABLE 2.
List of yeast strains
| Strains | Genotype |
|---|---|
| MLY200 | MATa ura3-52 leu2-1 lys2-801 ade2-101 trp1-1 his3-200 Δpdi1::HIS6 pRS316-PDI |
| TG100 | MLY200a transformed by pRS314-PDI |
| TG101 | MLY200a transformed by pRS314-PDI (E123C-S247C) |
| TG102 | MLY200a transformed by pRS314-PDI (N306C-D441C) |
| TG103 | MLY200a transformed by pRS314-PDI (E123C-S247C, N306C-D441C) |
| TG106 | MLY200a transformed by pRS314-PDI-Myc3 |
| TG107 | MLY200a transformed by pRS315-PDI-HA3 |
| TG108 | MLY200a transformed by pRS314-PDI-HA3 and pRS315-PDI-Myc3 |
| TG109 | MLY200 transformed by pRS402 |
| TG110 | TG109a transformed by pRS314-GFP-PDI |
| TG111 | TG109a transformed by pRS314-NGFP-PDI and pRS315-CGFP-PDI |
| TG112 | TG109a transformed by pRS314-PDI-NGFP and pRS315-CGFP-PDI |
| TG113 | TG109a transformed by pRS314-NGFP-PDI and pRS315-PDI-CGFP |
| TG114 | TG109a transformed by pRS314-PDI-NGFP and pRS315-PDI-CGFP |
| TG115 | TG109a transformed by pRS314-PDI-NGFP and pRS315-PDI-CGFP |
| TG116 | MLY200a transformed by pRS314-PDI (E123C) |
| TG117 | MLY200a transformed by pRS314-PDI (S247C) |
| TG118 | MLY200a transformed by pRS314-PDI (N306C) |
| TG119 | MLY200a transformed by pRS314-PDI (D441C) |
The plasmid pRS316-PDI in the parent strain was replaced with the indicated plasmid(s) by the plasmid shuffle procedure.
Cross-linking and Immunoprecipitation of PDI-HA3 and PDI-Myc3—The C terminus of PDI was tagged with either a HA3 or Myc3 epitope using the pRS314 or pRS315 vectors, respectively. Microsomes from various strains were prepared from stationary phase cells using an established protocol (27), but without reducing reagents and the sucrose gradient ultracentrifugation step. Cross-linking with dithiobis(succinimidyl)propionate and immunoprecipitation were carried out as described (28) with either microsomes prepared from 250 A600 units of yeast cells or 2 mg/ml of PDI-HA3 and PDI-Myc3 purified from E. coli.
Split GFP Construction and Measurement of GFP Reassembly—Residues 1 to 157 of GFP from pEGFP-C1 were used as the N-terminal half of GFP (NGFP) and residues 158 to 239 as the C-terminal half of GFP (CGFP). In the pRS314 and pRS315 vectors, respectively, NGFP and CGFP were attached to PDI either on the N terminus through a (Gly-Gly-Gly-Gly-Ser)2 linker, or on the C terminus through a Gly-Gly-Gly-Gly-Ser linker. The pRS402 vector (29), generously provided by Dr. Nancy Hollingsworth (Stony Brook University), was linearized with StuI and used to transform the MLY200 strain to generate strain TG109 that is devoid of autofluorescence. A set of strains co-expressing PDI tagged with NGFP and CGFP (TG111-114 in Table 2) were generated from TG109 by plasmid shuffling. The cells were grown at a low temperature, 15 °C, according to the reported protocol of the same technique in E. coli (30) and mammalian cells (31). After 72 h of cell growth at 15 °C on synthetic defined medium lacking tryptophan and leucine, colonies were isolated and resuspended in water. Images of GFP fluorescence were captured with a Zeiss LSM 510 META NLO two-photon laser scanning confocal microscope system. To evaluate the GFP reassembly statistically, the fluorescence intensity of GFP in 2 × 105 cells was measured by flow cytometry with a FAScan (BD Biosciences) at 488/530 nm. The fluorescence intensity distribution chart was reported together with the mean fluorescence intensity. PDI tagged with intact GFP at the N terminus was transfected into strain TG109 by the plasmid shuffle procedure to create strain TG110. This strain was used as the positive control, whereas strain TG109 was used as the non-GFP control. Strain 115 expressing isolated N- and C-terminal halves of GFP was used as the negative control to show that the split GFP did not self-reassemble during this experiment.
Determination of Disulfide Bond Linkages—40 μg of the quadruple E123C, S247C, N306C, and D441C yPDI mutant were proteolytically digested for 2.5 h with 2.5 μg of trypsin (sequencing grade, Promega, Madison, MA) in 100 μl of 50 mm NH4HCO3 at 37 °C. Optionally, the resulting peptides were reduced by the addition of 10 mm DTT and incubation for 10 min at 56 °C. Samples were desalted using Zip-Tip microcolumns (Millipore) according to the protocol provided by the manufacturer. Peptides were directly eluted onto a stainless steel target by the addition of saturated α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 0.1% trifluoroacetic acid. Matrix-assisted laser desorption ionization time-of-flight mass spectrometric detection was achieved using an Ultraflex TOF/TOF I (Bruker Daltonics, Bremen, Germany) at 25 kV IS1 voltage in the reflector mode.
RESULTS
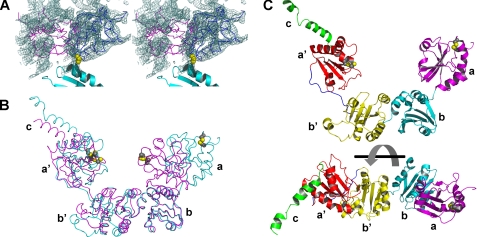
Structure of Full-length Yeast PDI Crystallized at Room Temperature—Initially, full-length yeast PDI was crystallized at 22 °C. However, crystals obtained at this temperature only diffracted to 3.7-Å resolution. At this limited resolution, the structure could not be easily solved. By reducing the temperature to 4 °C, crystal quality was significantly improved and a 2.4-Å structure could be determined (12). Armed with the 4 °C structure, the 3.7-Å dataset of the 22 °C crystals was solved by molecular replacement. Although only the temperature is different between the two crystallization conditions, a distinctly different structure was obtained. The final model was refined with Refmac, initially by rigid body refinement, followed by TLS refinement and a few cycles of restrained refinement with tight restraints to an R-factor of 0.242 (Rfree = 0.347). Despite the limited resolution and the correspondingly high free R-factor of the model the large conformational difference between the two structures are defined unambiguously in the electron density maps (Fig. 1A).
FIGURE 1.
The 22° C structure of yeast PDI. A, stereoview of a SIGMAA weighted 2Fo-Fc electron density map at a contour level of one times the root mean square deviation in the vicinity of the a domain, which is shown as a Cα trace in its new orientation in the 22 °C structure (blue) and its position in the original 4 °C structure (magenta). Parts of the adjacent b domain are shown as a cyan ribbon with the side chain of its N-terminal residue, Pro141, in yellow CPK representation. The density in the vicinity of the magenta model corresponds to the b and b′ domains of a symmetry related molecule. B, superposition of the 4 °C (magenta) and 22 °C (cyan) structures (shown as Cα traces) based on their b domains. The active site cysteines are shown in CPK representation with their S atoms in yellow. C, ribbon diagram of the 22 °C structure in two different orientations with the a domain in magenta, b domain in cyan, b′ domain in yellow, a′ domain in red, and C-terminal extension in green. The two diagrams differ by a 90° rotation around the horizontal axis. The side chains of the active site cysteines are displayed as in B.
Full-length yeast PDI contains residues 23 to 522 of the PDI1 gene and an additional vector-derived tetrapeptide, Ala-Gly-His-Met, at the N terminus with the Met numbered 22. The first three residues at the N terminus and eight residues at the C terminus are invisible, presumably due to disorder. As shown in Fig. 1B, the spatial arrangement of the b, b′, and a′ domains is very similar between the two crystal structures of full-length yeast PDI. Although the structure of the a domain does not change significantly, the relative position of this domain to the remainder of the molecule is dramatically different. The a domain is no longer in contact with the b′ domain and the center of gravity of the a domain now almost falls in the same plane as that of the other three domains (Fig. 1B). Thus, the overall structure of yeast PDI is no longer in the twisted U conformation described earlier, but instead adopts the shape of a “boat” (Fig. 1C) with the b and b′ domains at the bottom and the a and a′ domains at the bow and stern. Compared with the 4 °C structure, this structure is more extended, with a length of ∼90 Å and a height of ∼50 Å. Most importantly, the active sites of the a and the a′ domains no longer face each other.
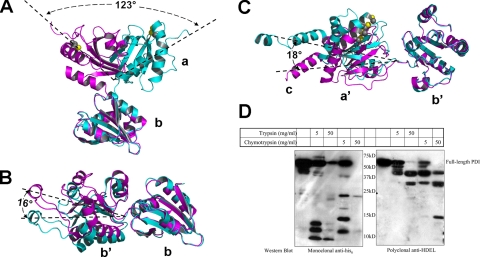
Molecular Flexibility—Based on both the 4 °C structure and this 22 °C structure, an examination of the inter-domain contacts indicates that the interaction between the b and b′ domain is relatively tight in the 4 and 22 °C structures, with a buried surface area of 650∼700 Å2. In contrast, the a and a′ domains are only loosely connected to this b-b′ base with negligibly small contact areas of ∼200 Å2 in each case. Therefore, the a and a′ domains can be viewed as two flexible arms connected to the relatively rigid base formed by the b and b′ domains. A comparison of the 4 and 22 °C structures of yeast PDI clearly confirms this molecular flexibility, especially the inter-domain mobility. As discussed earlier, the a domain exhibits the most dramatic difference between the two structures resulting in a ∼22-Å dislocation of the center of gravity of the a domain due to a ∼123° rotation around the loop connecting the a and b domains (Fig. 2A). This loop is formed by residues 139-142, which link the last α-helix of the a domain and the first β-strand of the b domain. The dihedral angles of residue Ser139 undergo the largest change, from ϕ = -77° and ψ = -11° in the 4 °C structure to ϕ = -100° and ψ = +138° in the 22 °C structure. In addition to the pronounced conformational change of the a domain, the relative orientations of the b, b′, and a′ domains also undergo small changes (Fig. 2, B and C). Keeping the b domain fixed, the b′ domain undergoes a ∼16° rotation around Ala237, resulting in a shift of the center of gravity by ∼5 Å, whereas the a′ domain is rotated around Ile364 relative to the b′ domain by ∼18°, resulting in a ∼7-Å displacement of the center of gravity.
FIGURE 2.
Domain flexibility in PDI. A, conformational change of the a domain relative to the b domain in the 4 °C (magenta) and 22 °C (cyan) structures of full-length PDI. Only the a and b domains are shown. The active site cysteines in the a domain are shown in CPK representation with the S-atoms in yellow. All angles of rotation were computed with the CCP4 program LSQKAB. B, comparison of the relative orientation between the b and b′ domains in the 4 °C (magenta) and 22 °C (cyan) structures based on a superposition of the b domains. C, comparison of the relative orientation between the b′ and a′ domains in the 4 °C and the 22 °C structures based on a superposition of the b′ domain and color-coded as in A. D, Western blot analysis following limited proteolysis of His6-PDI. The digestion of 1 mg/ml His6-PDI was carried out in buffer containing 100 mm Tris, pH 8.5, 250 mm NaCl, 1 mm glutathione with either trypsin or chymotrypsin at the indicated concentration at 4 °C for 30 min. The reaction was terminated by the addition of SDS-PAGE sample buffer and boiling for 10 min. Western blot analysis of the digestion product was carried out with anti-His6 and anti-HDEL antibodies.
Asymmetric Molecular Flexibility between the Two Arms of Yeast PDI—It has previously been demonstrated that PDI exhibits different sensitivities to protease digestion under reducing and oxidizing conditions (32) and that mammalian PDI yielded two major fragments after partial digestion with trypsin (Fig. 3, panel G, in Ref. 32). The sizes of these two fragments prompted us to speculate that the smaller fragment represents the arm of PDI that is more flexible and therefore more sensitive to trypsin digestion. Accordingly, we carried out limited proteolysis on yeast PDI with both chymotrypsin and trypsin under similar conditions. To determine which arm the smaller fragment belongs, two different markers were utilized: the N-terminal His6 tag for the a domain and the C-terminal HDEL ER retention signal for the a′ domain. As shown in Fig. 2D, the smaller fragments around 10 kDa that due to their size correspond to a single domain always contain the His6 tag, but not the HDEL sequence. On the other hand, the larger fragment around 40 kDa, which probably represents PDI with one domain trimmed off was detected by the anti-HDEL antibody, but not the anti-His6 tag antibody. This observation strongly suggests that the a domain arm was cleaved off of PDI first. Examination of the potential trypsin and chymotrypsin digestion sites in the loops connecting the a and a′ domains to the b-b′ base indicate that both of them appear to have an equal chance of being cleaved. Therefore, the observed asymmetric protease sensitivity of the a and a′ domain linkers can be attributed to differences in the molecular flexibility of the associated domains. In other words, the a arm of PDI is more flexible than the a′ arm. This was surprising because we had previously speculated that the a′ domain should be more flexible due to the long b′-a′ loop (12), however, most of this long loop is in close contact with either the b′ or a′ domain.
FIGURE 3.
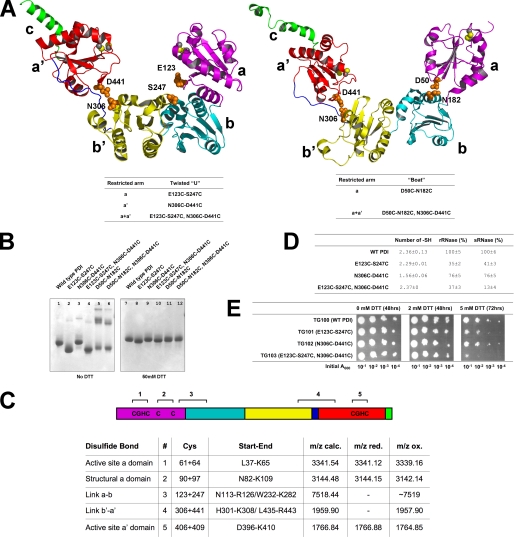
Disulfide scan mutants. A, left panel, the twisted U structure (ribbon diagram) with the same color code as in Fig. 1C. Right panel, the boat structure. Residues highlighted with their side chains in orange CPK representation were mutated to cysteines to introduce inter-domain disulfide bonds. The corresponding disulfide scan mutants are listed below the ribbon diagram. B, native PAGE (stained with Coomassie Blue) of PDI and its variants. Lanes 1-6 are in the absence and lanes 7-12 in the presence of 50 mm DTT. C, mass spectrometric analysis of the disulfide bonds of PDI. Schematic diagram of the domain architecture with possible disulfide bonds for the E123C/S247C/N306C/D441C quadruple mutant (top). SS-bonds 3 and 4 correspond to the introduced disulfide bonds. The table summarizes the mass spectrometry results under reducing (with DTT, red.) and oxidizing (without DTT, ox.) conditions. In case of the disulfide bonds existing in the wild-type (1, 2, and 5) single peptides are generated that encompass both cysteines. In case of the engineered disulfide bonds each cysteine resides in a separate peptide and for these linkages peaks only exist in the oxidized form and hence there are no entries in the m/z red. column as indicated by a minus (-). Upon reduction two separate peptides are formed for disulfide linkages 3 and 4 resulting in a total of four peptides of which only the peptide Trp232-Leu282 could not be observed. D, determination of the number of free -SH groups and enzyme activity of cross-linked PDI mutants. The number of free -SH groups was calculated as described under “Experimental Procedures.” Standard deviations are reported for two independent experiments. In vitro activity of PDI was assayed using scrambled ribonuclease (sRNase) and reduced ribonuclease (rRNase) as substrates. The indicated standard deviations result from three independent experiments. E, in vivo ability of disulfide scan mutants to support the cell growth. Cells were grown in the absence or presence of the indicated amount of DTT for the indicated time periods using the four different indicated initial cell concentrations.
Disulfide Scan Mutants—As discussed above, yeast PDI can be viewed as a molecule with two highly mobile arms, represented by the a and a′ domains. To study the relationship between molecular flexibility and enzymatic activity, inter-domain disulfide bonds were introduced to restrict the flexibility of the mobile arms. This “disulfide scan” method has been successfully utilized in studying the molecular movements of a K+ channel voltage-sensing domain (33) and the mobility of the pore plug of the Sec61 translocon (34). Based on the PDI structures, two amino acids that are spatially close to each other, but belong to different domains and are thus distant in the primary sequence were mutated to cysteine. Given their close spatial proximity, the two cysteines were expected to form a non-native inter-domain disulfide bond, thereby restricting the conformational flexibility of the two arms. As shown in Fig. 3A, left panel, for PDI in the twisted U conformation, Glu123 and Ser247 were chosen to connect the a and b′ domains, and Asn306 and Asp441 as the pair connecting the a′ and b′ domains. Therefore three sets of mutants based on the twisted U conformation were prepared as listed in Fig. 3A; none of these mutants displayed significant structural changes when examined by CD spectroscopy (Fig. S2). Moreover, activity assays with the individual E123C, S247C, N306C, and D441C variants revealed that all retained activities similar to the wild type in the in vitro rRNase refolding assay. In addition, all mutants supported the growth of yeast cells as well as wild-type PDI under normal conditions or under redox stress (Fig. S3).
The formation of the engineered disulfide bonds was confirmed by three different approaches. First, analysis by native PAGE revealed that the electrophoretic mobility of each mutant under non-reducing conditions (Fig. 3B) is either higher (lanes 2 and 4) or slightly lower (lane 3) than wild-type PDI (lane 1). However, upon reduction with 50 mm DTT, all of the mutants and the wild-type PDI migrated with essentially identical mobility (Fig. 3B, lanes 7-10). Under oxidizing conditions the introduced cysteines clearly changed the shape of PDI and, in turn, its mobility on native PAGE. Second we determined the number of free sulfyhydryl (-SH) groups in these mutants using Ellmans assay and compared them to wild-type PDI. Despite the added cysteine residues, the number of free -SH groups in the mutant should be the same as wild-type PDI under oxidizing conditions. As shown in Fig. 3C, all mutants have a similar number or fewer -SH groups than wild-type PDI. In case of the N306C/D441C mutant, the smaller number of -SH groups can possibly be attributed to the fact that it has a slight tendency to form redox state-sensitive oligomers (compare Fig. 3B, lanes 3 and 9), that may partially bury the native cysteines of PDI. Thus, the results from both approaches confirmed the formation of the engineered intra-molecular disulfide bonds. To demonstrate that the introduced cysteines do not form bonds with one of the six native cysteines, the disulfide bonds were identified by mass spectrometry following tryptic digestion.
The PDI wild-type contains 41 predicted trypsin cleavage sites, none of which were altered with the introduction of the engineered disulfide bonds. Regarding the three wild-type disulfide bonds the cysteines of each pair reside within the same tryptic peptide, and the only difference one should observe between the oxidized and reduced states is a mass increase by 2 Da resulting from the reduction of the disulfide bond. In contrast, the four introduced cysteines are located on different tryptic peptides and consequently one expects to find a larger fragment under oxidizing conditions and two separate peptides, each containing one cysteine, under reducing conditions. All three peptides containing the native disulfide bonds (Fig. 3C, peptides 1, 2, and 5) are smaller by 2 Da in the oxidized compared with the reduced state, showing that both cysteines are oxidized and connected by a intra-peptide disulfide bond. For the newly engineered disulfide bond linking the b′ and a′ domains we detected the linked peptides containing Cys306 and Cys441 (Fig. 3C, peptide 4) with a mass (m/z) of 1957.9, in addition to two signals 1 and 2 Da smaller, which can probably be attributed to deamidation products. This equals the sum of both peptides minus 2 Da, thus demonstrating that both peptides are linked by a disulfide bond. When analyzed under reducing conditions in the presence of DTT, the m/z signal at 1957.9 is no longer detectable, indicating a cleavage of the interpeptide disulfide linkage. Last, the linked tryptic fragments containing Cys123 and Cys247 were detectable by an m/z signal at ∼7519 (average mass). Although the instrument used in the analysis does not allow for isotopic resolution at this high mass range and thus a more accurate mass determination failed, the signal is no longer detectable under reducing conditions with DTT. This observation further confirms the m/z signal at 7519 belongs to the Cys123-Cys247-linked peptides. The MS data therefore account for all cysteines within the protein under oxidizing and reducing conditions and confirm that the engineered disulfides, Cys123, Cys247, Cys306, and Cys441, link the domains as designed.
To study the boat conformation, Asp50 and Asn182 were chosen to restrict the a domain in the conformation observed in the 3.7-Å structure, whereas Asn306 and Asp441 were again selected for the a′ domain, because the b-b′-a′ module is similar in the two structures (Fig. 3A, right panel). However, these two mutants were found to be prone to oligomerization as demonstrated by native PAGE (Fig. 3B, lanes 5 and 6), and these oligomers appear to be covalently linked by inter-molecular disulfide bonds as judged by their sensitivity to reducing reagents (Fig. 3B, lanes 11 and 12). It is impossible at present to determine whether the introduced cysteines or the native cysteines form the inter-molecular disulfide bond. More importantly, it is also difficult to make comparisons between these mutants and wild-type PDI because their oligomeric state is obviously different. For these reasons, the two mutants were not studied further.
Relationship between Molecular Flexibility and Activity—To study the influence of molecular flexibility on the activity of PDI, the remaining three disulfide scan mutants were either analyzed for their in vitro RNase refolding ability, or introduced into cells to examine their ability to support growth. The redox condition used for the RNase refolding assay contained a GSH/GSSG ratio of 5 to 1, which is close to the measured value in the lumen of the ER (35). To determine whether the introduced disulfide bonds were stable under these conditions, the electrophoretic mobility of these mutants on native PAGE in a redox buffer containing 1 mm GSH and 0.2 mm GSSG was compared with non-reducing conditions. The mutants retained the same eletrophoretic mobility in both buffers (data not shown), thus suggesting that the introduced disulfide bonds were preserved under the redox conditions employed in the subsequent studies.
The RNase refolding abilities of these mutants were assayed with reduced RNase (rRNase) and scrambled RNase (sRNase) as substrates. As shown in Fig. 3D, restricting the flexibility of the a domain results in a 60-70% decrease in enzymatic activity for both substrates (line 2), whereas restricting the flexibility of the a′ domain results in only a ∼24% reduction in activity (line 3). This result clearly demonstrates that the flexibility of the a domain is much more important than that of the a′ domain in catalyzing the refolding of RNase. When the flexibility of both the a and a′ domains was restricted (line 4), the PDI activity in refolding rRNase was roughly the same as when only the a domain was restricted. However, the refolding activity of sRNase was further decreased from 30-40 to 12-13%. This suggests that catalyzing disulfide isomerization during refolding of sRNase is more dependent on conformational flexibility than catalyzing disulfide oxidation during refolding of sRNase.
The ability of these disulfide scan mutants to support cell growth were analyzed in vivo by replacing wild-type PDI with the mutants using the plasmid shuffle technique. Under normal and mild redox stress conditions (2 mm DTT), these mutants supported the growth as well as the wild-type protein (Fig. 3E). However, under a more stringent stress condition (5 mm DTT), reduced growth was observed for the mutants. Decreasing the flexibility of the a domain (line 2) caused the cells to be very sensitive to redox stress, whereas restricting the a′ domain (line 3) only caused a slight increase in sensitivity. On the other hand, the mutant in which both the a and a′ domains are restricted (line 4), results in the greatest sensitivity to reducing conditions. Considering the results from the RNase refolding and cell growth assays, we conclude that the molecular flexibility of both catalytically active thioredoxin domains is important to the in vitro and in vivo activities of PDI. Moreover, both assays also demonstrate that restriction of the a domain leads to a significantly larger decrease in each activity than restricting the a′ domain.
The Elongated Helical Structure of the C-terminal Tail—In the 4 °C crystal structure, residues 490-502 of the C-terminal tail were found to form an α-helix with the remaining 18 C-terminal residues being disordered (12). In the 22 °C structure additional C-terminal residues are ordered, with residues 506-512 forming a second short α-helix. The two helices in the C-terminal tail are almost colinear, spanning a distance of about 45 Å with the last helix being completely solvent exposed. The C-terminal extension has been demonstrated to be required for full activity of yeast PDI (12), and it has been speculated that it stabilizes the structure of the a′ domain through hydrophobic interactions. This idea was confirmed by the fact that, during attempts to purify the isolated domains of yeast PDI, the a′ domain was only soluble when the C-terminal extension was attached (data not shown). A similar observation was reported recently for the a′ domain of ERp57 (36), a paralog of PDI. Besides the hydrophobic interaction with the a′ domain, the function of the highly charged region of the C-terminal extension is currently unknown. It has been proposed that it is responsible for the Ca2+ binding ability of PDI (37), however, Ba2+ ions, which were included at a concentration of 10 mm as additive during crystallization, could not be observed in either the 4 or 22 °C structure.
The 22 °C Crystal Structure Reveals a PDI Dimer—In the I4 space group of the 4 °C structure, four PDI molecules form a tetramer around a 4-fold crystallographic symmetry axis, but this oligomeric state is almost certainly a crystallization artifact because the contact area between the PDI molecules is rather limited and there is no evidence for a PDI tetramer in solution (see below). In the C2221 space group of the 22 °C structure, PDI is present as a dimer, in which a surface area of ∼2700 A2 corresponding to 9.6% of the monomer surface area is buried. The a domain contributes ∼17% to the contact interface, whereas the other three domains participate almost equally in dimerization. In the dimer, the active sites of the a domain of each PDI monomer are pointing in the same direction, whereas the planes defined by the centers of gravity of the a, b, b′, and a′ domains of the two PDI molecules intersect at an angle of ∼35° (Fig. 4A). The active site in the a′ domain and the previously identified hydrophobic pocket in the b′ domain both become buried, whereas the active site of the a domain remains solvent exposed.
FIGURE 4.
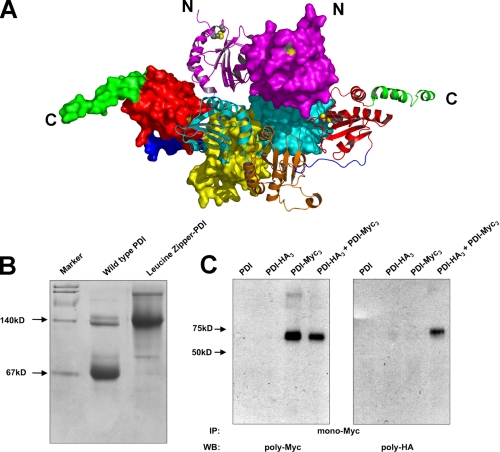
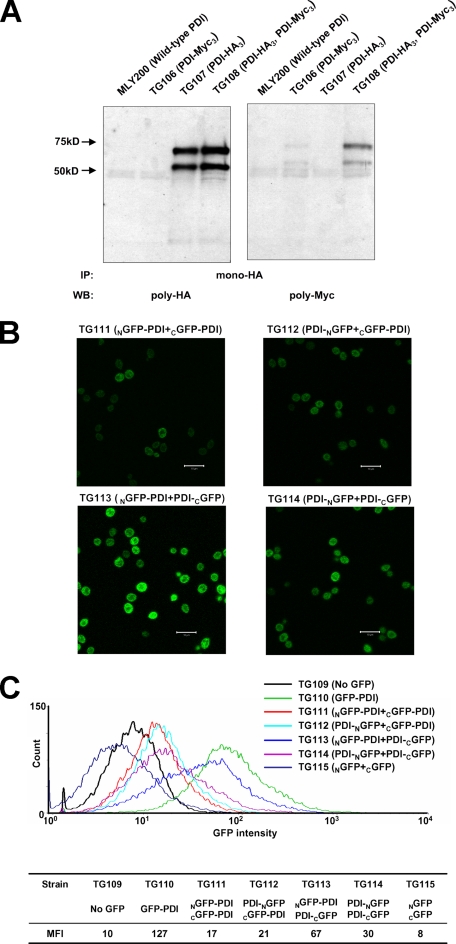
Dimerization of PDI. A, structure of the PDI dimer present in the crystals grown at 22 °C. One monomer is displayed in surface representation with its domains color coded as described in the legend to Fig. 1C, whereas the other is shown in a ribbon representation in the same color code except that the b′ domain is in orange to increase visibility. Active site cysteines are either mapped in yellow onto the surface or are shown in CPK representation (S atoms in yellow) in the ribbon diagram. B, Coomassie Blue stained native PAGE of PDI and PDI tagged with a leucine zipper. Wild-type PDI and dimeric PDI tagged with a leucine zipper on the N terminus are analyzed on native PAGE stained with Coomassie Blue. A doublet at the dimer position has always been observed and presumably represents two different forms of dimeric PDI. C, following cross-linking of purified epitope-tagged forms of PDI (PDI-HA3 and PDI-Myc3) with dithiobis(succinimidyl)propionate, PDI-Myc3 was immunoprecipitated (IP) with a monocolonal anti-Myc antibody and protein G-agarose. The resulting precipitate was analyzed by reducing SDS-PAGE and Western blot (WB) with either a polyclonal anti-Myc (left panel) or anti-HA (right panel) antibody.
Because the buried surface area is quite extensive considering the size of the protein (38), we asked whether this dimer is also present in solution. In fact, during purification of yeast PDI, a small shoulder with an apparent molecular weight twice that of the major peak was always observed during size exclusion chromatography. To investigate the existence of this possible PDI dimer, a dimeric form of PDI was created by tagging the protein on the N terminus with a leucine zipper derived from the BZIP motif (residue 220-291) of GCN4 (39). Upon native PAGE (Fig. 4B), the small fraction of purified native PDI that migrates more slowly than the major form had an electrophoretic mobility similar to the “artificial” dimeric form of PDI, thus suggesting that a minor fraction of PDI is present in solution as a dimer. To further demonstrate that PDI dimerizes in solution, it was modified with either an HA or a Myc tag at its C terminus. Tagged forms (PDI-HA3 and PDI-Myc3) were purified and equal amounts of each protein were mixed at a final concentration of 4 mg/ml protein in phosphate-buffered saline buffer. Because the dimerization of PDI appears to be relatively weak, cross-linking followed by immunoprecipitation was utilized instead of direct co-immunoprecipitation to avoid dissociation of the dimer due to the harsh conditions utilized during immunoprecipitation. As shown in Fig. 4C, PDI-Myc3 could be cross-linked to PDI-HA3, which is consistent with the existence of dimeric PDI in solution.
Analytical Ultracentrifugation Studies—The propensity of yeast PDI to dimerize in solution was further studied by analytical ultracentrifugation. Irrespective of protein concentration and temperature all sedimentation velocity data revealed two species corresponding to the monomeric and dimeric forms of PDI with the monomer as the predominant form, as also previously demonstrated for mammalian PDI (40). The standard sedimentation coefficients (s20,w) of the monomeric and dimeric forms of yeast PDI were measured as 3.4 ± 0.04 and 5.6 ± 0.31 S, respectively (Table 3, line 1). This is only slightly different from the calculated s20,w values of 3.70 and 5.86 S, based on the respective crystal structures (Table 3, line 2). Sedimentation equilibrium data collected at different speeds and concentrations (Table 3, 3-5 lines) were first fit against a single-component model. With this model, the calculated apparent molecular mass was always intermediate between the monomer and dimer, whereas the distribution of residuals indicated a poor fit (Fig. S4A). The fit could be significantly improved when the sedimentation equilibrium data were interpreted with a monomer-dimer model as indicated by the sedimentation velocity experiments (Fig. S4B). The dissociation constant (KD) of dimeric PDI was estimated to be roughly 100 μm, with values ranging from 50 to 125 μm (Table 3, 3-5 lines), thus suggesting a rather weak self-association of yeast PDI. Because the concentration of PDI used in the analytical ultracentrifugation studies is far below its KD, it is difficult to accurately measure the KD by this technique alone. Because variations in temperature did not reveal significant effects on dimerization (Table 3, 3 and 4 lines versus 5), it remains unclear why different oligomeric states of PDI were crystallized at 4 and 22 °C.
TABLE 3.
Sedimentation data of full-length PDI
| S20,w (monomer) | S20,w (dimer) | KDa | ||
|---|---|---|---|---|
| Sedimentation velocityb | ||||
| Measured S20,wc | 3.4 ± 0.04 | 5.6 ± 0.31 | 1 | |
| Predicted S20,wc | 3.70 | 5.86 | 2 | |
| Sedimentation equilibriumd | ||||
| 4 °C, 0.1 mg/ml | ∼66 μm | 3 | ||
| 4 °C, 0.2 mg/ml | ∼125 μm | 4 | ||
| 22 °C, 0.2 mg/ml | ∼50 μm | 5 |
KD is estimated based on the monomer-dimer model which represents the best fit for full-length yeast PDI.
Four sedimentation velocity experiments were carried out either at 22 °C with 0.2, 0.5, and 0.8 mg/ml PDI or at 4 °C with 0.8 mg/ml PDI.
The measured standard sedimentation coefficient (S20,w) is expressed as the mean ± S.D. of the four sedimentation velocity experiments. The predicted S20,w was calculated with Hydropro based on the 4 °C monomeric and the 22 °C dimeric structures of yeast PDI.
Three sedimentation equilibrium experiments were carried out at the indicated temperatures and concentrations. The speeds for sedimentation equilibrium experiments at 4 °C were 14,000, 17,000, and 20,000 rpm, and the speeds for sedimentation equilibrium experiment at 22 °C were 14,000, 20,500, and 26,000 rpm.
Detection of Dimeric PDI in the ER—Although the analytical ultracentrifugation studies revealed that dimerization of PDI is weak, the high abundance of PDI in the ER, with a reported concentration of ∼0.2-0.5 mm (41, 42) should promote dimerization. At these concentrations the PDI dimer would account for more than 50% of the total protein based on the estimated KD. To test whether dimeric PDI exists in the ER, a haploid PDI strain was generated in which two plasmid-borne copies of PDI exist, one with an HA3 tag and one with a Myc3 tag. From microsomes prepared from this strain, dimeric PDI was first fixed by cross-linking to avoid dissociation during immunoprecipitation. The cross-linked product was precipitated with a monoclonal anti-HA antibody, and the resulting precipitate was separated by SDS-PAGE, followed by Western blot analysis with polyclonal anti-HA and anti-Myc antibodies. Myc3-tagged PDI was found to cross-link with HA3-tagged PDI (Fig. 5A), indicating that PDI in the ER can form either a dimer or higher oligomers.
FIGURE 5.
In vivo detection of a dimeric form of PDI. A, epitope-tagged forms of PDI (PDI-HA3 and PDI-Myc3) from yeast microsomes were cross-linked by dithiobis(succinimidyl)propionate and immunoprecipitated (IP) by a monoclonal anti-HA antibody. The precipitant was probed with either a polyclonal anti-HA antibody (left panel) or a polyclonal anti-myc antibody (right panel). The additional band at 50 kDa was always present when PDI-HA3 or PDI-Myc3 were expressed in yeast. It appears to be a glycosylated degradation product because it could be detected with an anti-PDI antibody and was sensitive to EndoH digestion. B, fluorescence microscopy of yeast cells co-expressing the indicated proteins in the ER. Equal amounts of yeast cells (104-105 cells) were mounted on each slide. The scale bar represents 10 μm. C, GFP fluorescence intensity distribution curves of the combinations of split GFP reassemblies as well as the non-GFP and GFP controls. The mean fluorescence intensities (MFI) of all strains are listed. WB, Western blot.
In addition, the so-called “split GFP” approach was utilized to detect possible in situ formation of a PDI dimer in the ER. This technique has been successfully utilized to detect protein-protein interactions in E. coli (30) and mammalian cells (31). Accordingly GFP was divided into an NGFP and a CGFP, which were individually attached to PDI. Assuming that PDI dimerizes, the two halves of GFP will be brought into close spatial proximity to reconstitute intact GFP as characterized by its ability to emit fluorescence.
To minimize constraints during the reassembly of split GFP, the two halves of GFP were attached to either terminus of PDI and the following four combinations were explored: NGFP-PDI + CGFP-PDI, PDI-NGFP + CGFP-PDI, NGFP-PDI + PDI-CGFP, and PDI-NGFP + PDI-CGFP. For all four combinations, a subset of cells was observed to emit fluorescence (Fig. 5B), which indicated the successful reassembly of the GFP halves into intact GFP. However, different efficiencies of the split GFP reassemblies were clearly observed with the following order: NGFP-PDI + PDI-CGFP > PDI-NGFP + PDI-CGFP > PDI-NGFP + CGFP-PDI = NGFP-PDI + CGFP-PDI. This observation was subsequently confirmed by measuring the GFP fluorescence intensity for 2 × 105 cells using flow cytometry with the non-GFP containing TG109 strain as negative control and the GFP-containing TG110 strain as positive control (Fig. 5C). The reassembly of split GFP was confirmed by a shift of the fluorescence intensity distribution curve toward the positive control, whereas the order of reassembly efficiency was confirmed by the mean fluorescence intensity. It is important to note that the GFP reassembly efficiency depends on both the proximity and mobility of the GFP halves. Because the C terminus of PDI is more flexible than its N terminus, the order of GFP reassembly efficiency is not directly related to the distance between the termini of the two monomers. In any case, both the split GFP experiments and the cross-linking experiment are consistent with the observation that yeast PDI does indeed form a dimer in the ER.
DISCUSSION
We have solved the crystal structure of full-length yeast PDI derived from crystals grown at 22 °C and compared it to the previously reported twisted U shaped structure obtained with crystals grown at 4 °C (12). Although the new structure was only determined at 3.7-Å resolution, it nevertheless, provided unambiguous structural information on the spatial arrangement of the four domains that differed significantly from the structure reported earlier. In the new structure, the conformation of PDI can be described as a boat due to the drastic rotation and translation of the a domain. As a result of this change, the two active sites of PDI no longer face each other and the previously identified “potential” catalytic pocket formed by the a-b′-a′ domains is no longer intact. A comparison of the two conformations suggests that the 4 °C structure represents the catalytically competent state of the enzyme, because in this structure both active sites and the potential substrate binding site in the b′ domain are accessible and form a sufficiently wide cleft to accommodate protein substrates. Furthermore, the spatial arrangement of these structural elements in the 4 °C structure is strikingly similar to the homodimeric structure of DsbC, which is in an active state as demonstrated by mutagenesis experiments (43).
To accommodate its structurally diverse substrates, ranging in size from insulin to procollagen (44), PDI must be a highly flexible enzyme. An analysis of the inter-domain contacts in the two crystal structures suggests that PDI is a molecule with two flexible arms, represented by the a and a′ domains, that are connected to a relatively rigid base formed by the b and b′ domain module. The new structure provides the first direct evidence of this molecular flexibility. Of course, it should be pointed out that the two PDI structures may only represent two snapshots of many “possible” conformations of PDI as a result of its intrinsic flexibility. The loops connecting the a and a′ domains to the b-b′ base enable PDI to facilitate these conformational changes. By introducing interdomain disulfide bridges the importance of molecular flexibility for the in vitro catalytic activity and in vivo growth-supporting activity of PDI was demonstrated. However, it should be pointed out that a discrepancy has previously been observed between the in vitro and in vivo activity of PDI (14, 17). Because the reason for this discrepancy is still uncertain (15, 45), the observed similar effects of molecular flexibility on the in vitro and in vivo function of PDI might not necessary reflect the same underlying mechanism. It is possible that the molecular flexibility influences the activity of PDI in two ways. As mentioned above, PDI might alter the separation of the a and a′ domains and hence the width of the catalytic pocket formed by the a-b′-a′ domains to adjust to differently sized substrates. In addition, the molecular flexibility may also be required during the isomerization reaction to ensure that the active sites can locate incorrect disulfide bonds in the substrate.
The conformational flexibility of the yeast enzyme is mirrored in mammalian PDI for which two “extreme” conformations were reported recently (46, 47). Analytical ultracentrifugation studies revealed that the four domains of mammalian PDI are arranged in a linear fashion to form an elongated molecule with a diameter of 3.4 ± 0.1 nm and a length of 16.8 ± 0.5 nm (46). In contrast, as demonstrated by small angle x-ray scattering, the four domains of PDI can form a counterclockwise circle with the a and a′ domains close to each other (47). The molecular flexibility of PDI apparently enables mammalian PDI to adopt multiple conformations, ranging from the closed form observed in the small angle x-ray scattering experiments to the fully extended structure suggested by analytical ultracentrifugation. It is possible that these conformational changes are modulated by different redox environments (32), but the molecular details of this regulation remain unclear at present.
Mammalian PDI was originally reported to exist as a dimer in solution (48) based on its hydrodynamic properties during size exclusion chromatography. However, on the basis of the now known crystal structures it can be argued that the elution profile was caused by the non-spherical molecular shape of the PDI monomer rather than by dimerization. More recently, sedimentation velocity ultracentrifugation studies suggested that mammalian PDI exists mainly as a monomer in solution with only a small fraction forming a dimer that is stabilized by the addition of Zn2+ (40). Erp57 has also been reported to form a homodimer (36), which in this case is redox-dependent and involves its C-terminal active site. In the 3.7-Å structure of yeast PDI reported in this study, a dimeric form of yeast PDI was observed. The extensive inter-subunit contact area of this structure suggests that it might have physiological relevance.
Prompted by the observed dimeric form of PDI from the crystal obtained at 22 °C, the oligomeric status of yeast PDI was studied and the existence of a PDI dimer in solution and in the ER was confirmed by several techniques including analytical ultracentrifugation, immunoprecipitation, and the split GFP system. It should be pointed out, however, that no experimental evidence yet demonstrates that the PDI dimer in the ER adopts the same conformation as that observed in the 22 °C crystals. Based on analytical ultracentrifugation studies, the KD of the PDI dimer was estimated to be ∼100 μm and the sedimentation coefficients of monomeric and dimeric PDI were characterized by s20,w values of 3.4 S and 5.6 S, respectively. In similar sedimentation studies, the s20,w values of monomeric rat PDI (40), human PDI (47), human ERp57 (46), and dimeric human PDI (40) have been reported as 3.48 S, 3.48 S, 3.53 S, and 5.18 S, respectively. The high similarity of these values to the data derived from yeast PDI suggests that the mammalian PDI homologues may adopt conformations similar to those of yeast PDI in solution. This is not surprising because PDI is highly conserved from yeast to mammals. In addition, it has also been reported that sedimentation equilibrium data of rat and human PDI (40, 47) best fit with a mixture of monomers and dimers.
It has been speculated that the Zn2+-dependent dimerization of mammalian PDI is involved in regulating the activity of PDI or its interaction with other ER-resident chaperones and folding catalysts (40). The redox-dependent dimerization of ERp57 is required for its ability to bind DNA in the nucleus (36). In the dimer observed in the 22 °C structure, both the active site in the a′ domain and the potential substrate-binding site in the b′ domain are buried. This conformation of the dimer therefore presumably represents an “inactive” state of PDI, because most of its catalytic elements are inaccessible. On the other hand, as discussed earlier, the twisted U conformation most likely represents an “active” form of PDI. The transition between monomeric and dimeric PDI could be a regulatory mechanism whereby the ER adjusts its folding capacity to meet different cellular demands. It has been demonstrated that only 60% of the level of native PDI expression is required for normal growth of yeast (45). Therefore, it is possible that 40% of the PDI molecules in the ER are in an inactive dimeric form that can be converted to the active monomeric form under certain conditions, for instance during different stages of the cell cycle, or in quick response to cellular stress before the unfolded protein response is triggered.
Supplementary Material
Acknowledgments
We thank Dr. Robert Noiva, University of South Dakota, for assistance with the in vitro enzyme assays, Dr. Daniel Raleigh for help with CD experiments, Drs. Neta Dean and Aaron Neiman for generous assistance with the yeast fluorescence experiments, Dr. Neta Dean for the generous gift from of a polyclonal anti-HDEL antibody. Dr. Song Xiang for help with crystallographic data collection, Dr. Guowei Tian for operation of the confocal microscope, and Dr. Zhixin Chen for assistance with the analytical ultracentrifugation experiments. We thank the staff at beamlines X26C at the NSLS at Brookhaven National Laboratory and at 19ID at the Advanced Photon Source at Argonne National Laboratory.
The atomic coordinates and structure factors (code 3BOA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants GM33184 (to W. J. L.) and DK54835 (to H. S.). This work was also supported by Deutsche Forschungsgemeinschaft Grant FZ82 (to H. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4 and additional Methods.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; PDI, protein-disulfide isomerase; GFP, green fluorescent protein; DTT, dithiothreitol; NGFP, N-terminal half of GFP; CGFP, C-terminal half of GFP; HA, hemagglutinin; EndoH, endoglycosidase H; sRNase, scrambled ribonuclease; rRNase, reduced ribonuclease.
References
- 1.Nakamoto, H., and Bardwell, J. C. (2004) Biochim. Biophys. Acta 1694 111-119 [DOI] [PubMed] [Google Scholar]
- 2.Farquhar, R., Honey, N., Murant, S. J., Bossier, P., Schultz, L., Montgomery, D., Ellis, R. W., Freedman, R. B., and Tuite, M. F. (1991) Gene (Amst.) 108 81-89 [DOI] [PubMed] [Google Scholar]
- 3.LaMantia, M., Miura, T., Tachikawa, H., Kaplan, H. A., Lennarz, W. J., and Mizunaga, T. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 4453-4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson, B., and Gilbert, H. F. (2004) Biochim. Biophys. Acta 1699 35-44 [DOI] [PubMed] [Google Scholar]
- 5.Ellgaard, L., and Ruddock, L. W. (2005) EMBO Rep. 6 28-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norgaard, P., Westphal, V., Tachibana, C., Alsoe, L., Holst, B., and Winther, J. R. (2001) J. Cell Biol. 152 553-562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klappa, P., Ruddock, L. W., Darby, N. J., and Freedman, R. B. (1998) EMBO J. 17 927-935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pirneskoski, A., Klappa, P., Lobell, M., Williamson, R. A., Byrne, L., Alanen, H. I., Salo, K. E., Kivirikko, K. I., Freedman, R. B., and Ruddock, L. W. (2004) J. Biol. Chem. 279 10374-10381 [DOI] [PubMed] [Google Scholar]
- 9.Cai, H., Wang, C. C., and Tsou, C. L. (1994) J. Biol. Chem. 269 24550-24552 [PubMed] [Google Scholar]
- 10.Koivu, J., Myllyla, R., Helaakoski, T., Pihlajaniemi, T., Tasanen, K., and Kivirikko, K. I. (1987) J. Biol. Chem. 262 6447-6449 [PubMed] [Google Scholar]
- 11.Wetterau, J. R., Combs, K. A., Spinner, S. N., and Joiner, B. J. (1990) J. Biol. Chem. 265 9801-9807 [PubMed] [Google Scholar]
- 12.Tian, G., Xiang, S., Noiva, R., Lennarz, W. J., and Schindelin, H. (2006) Cell 124 61-73 [DOI] [PubMed] [Google Scholar]
- 13.McCarthy, A. A., Haebel, P. W., Torronen, A., Rybin, V., Baker, E. N., and Metcalf, P. (2000) Nat. Struct. Biol. 7 196-199 [DOI] [PubMed] [Google Scholar]
- 14.Westphal, V., Darby, N. J., and Winther, J. R. (1999) J. Mol. Biol. 286 1229-1239 [DOI] [PubMed] [Google Scholar]
- 15.Kulp, M. S., Frickel, E. M., Ellgaard, L., and Weissman, J. S. (2006) J. Biol. Chem. 281 876-884 [DOI] [PubMed] [Google Scholar]
- 16.Tsai, B., and Rapoport, T. A. (2002) J. Cell Biol. 159 207-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holst, B., Tachibana, C., and Winther, J. R. (1997) J. Cell Biol. 138 1229-1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vagin, A., and Teplyakov, A. (1997) J. Appl. Crystallogr. 30 1022-1025 [Google Scholar]
- 19.Storoni, L. C., McCoy, A. J., and Read, R. J. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 432-438 [DOI] [PubMed] [Google Scholar]
- 20.Hutchinson, E. G., and Thornton, J. M. (1996) Protein Sci. 5 212-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA
- 22.Gallagher, S. R. (1999) in Current Protocols in Molecular Biology (Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K., eds) Vol. 2, pp. 10.12B.11-10.12B.11, John Wiley & Sons, Inc., New York [Google Scholar]
- 23.LaMantia, M. L., and Lennarz, W. J. (1993) Cell 74 899-908 [DOI] [PubMed] [Google Scholar]
- 24.Sherman, F. (1991) Methods Enzymol. 194 3-21 [DOI] [PubMed] [Google Scholar]
- 25.Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987) Methods Enzymol. 154 164-175 [DOI] [PubMed] [Google Scholar]
- 26.Elble, R. (1992) BioTechniques 13 18-20 [PubMed] [Google Scholar]
- 27.Kelleher, D. J., and Gilmore, R. (1994) J. Biol. Chem. 269 12908-12917 [PubMed] [Google Scholar]
- 28.Yan, A., Ahmed, E., Yan, Q., and Lennarz, W. J. (2003) J. Biol. Chem. 278 33078-33087 [DOI] [PubMed] [Google Scholar]
- 29.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P., and Boeke, J. D. (1998) Yeast 14 115-132 [DOI] [PubMed] [Google Scholar]
- 30.Wilson, C. G., Magliery, T. J., and Regan, L. (2004) Nat. Methods 1 255-262 [DOI] [PubMed] [Google Scholar]
- 31.Nyfeler, B., Michnick, S. W., and Hauri, H. P. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 6350-6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai, B., Rodighiero, C., Lencer, W. I., and Rapoport, T. A. (2001) Cell 104 937-948 [DOI] [PubMed] [Google Scholar]
- 33.Gandhi, C. S., Clark, E., Loots, E., Pralle, A., and Isacoff, E. Y. (2003) Neuron 40 515-525 [DOI] [PubMed] [Google Scholar]
- 34.Van den Berg, B., Clemons, W. M., Jr., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C., and Rapoport, T. A. (2004) Nature 427 36-44 [DOI] [PubMed] [Google Scholar]
- 35.Hwang, C., Sinskey, A. J., and Lodish, H. F. (1992) Science 257 1496-1502 [DOI] [PubMed] [Google Scholar]
- 36.Grillo, C., D'Ambrosio, C., Consalvi, V., Chiaraluce, R., Scaloni, A., Maceroni, M., Eufemi, M., and Altieri, F. (2007) J. Biol. Chem. 272 10299-10310 [DOI] [PubMed] [Google Scholar]
- 37.Lebeche, D., Lucero, H. A., and Kaminer, B. (1994) Biochem. Biophys. Res. Commun. 202 556-561 [DOI] [PubMed] [Google Scholar]
- 38.Jones, S., and Thornton, J. M. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 13-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talanian, R. V., McKnight, C. J., and Kim, P. S. (1990) Science 249 769-771 [DOI] [PubMed] [Google Scholar]
- 40.Solovyov, A., and Gilbert, H. F. (2004) Protein Sci. 13 1902-1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zapun, A., Creighton, T. E., Rowling, P. J., and Freedman, R. B. (1992) Proteins 14 10-15 [DOI] [PubMed] [Google Scholar]
- 42.Lyles, M. M., and Gilbert, H. F. (1991) Biochemistry 30 613-619 [DOI] [PubMed] [Google Scholar]
- 43.Sun, X. X., and Wang, C. C. (2000) J. Biol. Chem. 275 22743-22749 [DOI] [PubMed] [Google Scholar]
- 44.Wilson, R., Lees, J. F., and Bulleid, N. J. (1998) J. Biol. Chem. 273 9637-9643 [DOI] [PubMed] [Google Scholar]
- 45.Solovyov, A., Xiao, R., and Gilbert, H. F. (2004) J. Biol. Chem. 279 34095-34100 [DOI] [PubMed] [Google Scholar]
- 46.Frickel, E. M., Frei, P., Bouvier, M., Stafford, W. F., Helenius, A., Glockshuber, R., and Ellgaard, L. (2004) J. Biol. Chem. 279 18277-18287 [DOI] [PubMed] [Google Scholar]
- 47.Li, S. J., Hong, X. G., Shi, Y. Y., Li, H., and Wang, C. C. (2005) J. Biol. Chem.
- 48.Lambert, N., and Freedman, R. B. (1983) Biochem. J. 213 225-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.