Abstract
The Toll-like receptors (TLR) play an instructive role in innate and adaptive immunity by recognizing specific molecular patterns from pathogens. Autophagy removes intracellular pathogens and participates in antigen presentation. Here, we demonstrate that not only TLR4, but also other TLR family members induce autophagy in macrophages, which is inhibited by MyD88, Trif, or Beclin 1 shRNA expression. MyD88 and Trif co-immunoprecipitate with Beclin 1, a key factor in autophagosome formation. TLR signaling enhances the interaction of MyD88 and Trif with Beclin 1, and reduces the binding of Beclin 1 to Bcl-2. These findings indicate TLR signaling via its adaptor proteins reduces the binding of Beclin 1 to Bcl-2 by recruiting Beclin 1 into the TLR-signaling complex leading to autophagy.
One of the mechanisms by which the innate immune system senses the invasion of pathogenic microorganisms is through Toll-like receptors (TLRs),3 which recognize specific molecular patterns that are present in microbial components. Stimulation of different TLRs induces distinct patterns of gene expression, which not only leads to the activation of innate immunity, but also instructs the development of antigen-specific acquired immunity (1, 2). TLR signaling predominantly induces interferon regulatory factors, and nuclear factor-κB (NF-κB) and AP1 activation. This elicits the production of type I interferon and inflammatory cytokines (3, 4). TLR signaling also facilitates the selection of microbial antigens for presentation by dendritic cells (5). Parasitic protozoans can also trigger TLRs enhancing resistance to them by triggering pro-inflammatory responses (6). Ten members of the TLR family have been identified in human, and 13 in mice (4). Signaling by TLRs involves five adaptor proteins known as MyD88, MyD88-adapter like (MAL), Toll/interleukin receptor (TIR) domain-containing adaptor protein inducing interferon β (TRIF), TRIF-related adaptor molecule (TRAM), and sterile α- and armadillo motifcontaining protein (SARM). MAL recruits MyD88 to TLR2 and TLR4, and TRAM recruits TRIF to TLR4 to allow interferon regulator factor 3 activation. SARM negatively regulates TRIF. MyD88 is the key signaling adaptor for all TLRs with the exception of TLR3 and a subset of TLR4 signals, which utilize Trif (7).
Autophagy is a cellular response to starvation as well as a quality control system that can remove damaged organelles and long-lived proteins from cytoplasm. Autophagy plays a role in programmed cell death, the promotion and prevention of cancer, neurodegeneration, and cellular defense to remove invading pathogens (8). Recently, it has been recognized that autophagy is involved in both innate and adaptive immunity against intracellular protozoa, bacteria, and viruses (9–11). Autophagy has been implicated in major histocompatibility complex class II antigen presentation. During autophagy cytoplasmic materials are sequestered into double membrane-coated autophagosomes, which fuse with endosomes and lysosomes to form autolysosomes where the contents are degraded by lysosomal enzymes. Autophagy is a constitutively active pathway in all major histocompatibility complex class II positive cells, where the autophagosomes continuously fuse with the multivesicular major histocompatibility complex class II-loading compartment. This provides a potential mechanism for class II positive cells to continuously present cytosolic contents to CD4+ T cells (12).
Beclin 1, the mammalian homologue of yeast Atg6, is a key component of a class III phosphatidylinositol 3-kinase complex that initiates autophagosome formation helping to localize other autophagy proteins to the pre-autophagosomal membrane (13). Bcl2 is an anti-apoptotic protein, which binds Beclin 1, and is known to inhibit autophagy (14). In this study, we use macrophages to test the potential of a panel of TLR ligands to trigger autophagy. We find that MyD88 and Trif interact with Beclin 1 and Bcl-2 and that TLR signaling alters the balance, which is likely critical in triggering autophagy.
EXPERIMENTAL PROCEDURES
Cell Culture and GPF-LC3 Stably Transduced Cell Line—RAW 264.7 cells were obtain from ATCC (American Type Culture Collection, Rockville, MD) and were maintained in Dulbecco's modified Eagle's medium, 10% fetal bovine serum. The LC3 cDNA was a gift from Dr. Noboru Mizushima (Tokyo Medical and Dental University, Tokyo, Japan). RAW 264.7 cells were transfected with the GFP-LC3 construct, using FuGENE HD Transfection Reagent (Roche Applied Science, Basel, Switzerland). The transfection protocol from the manufacturer was followed. 24 h after transfection 400 μg/ml G418 (Mediatech, Herdon, VA) was added to select positive cells for 3 weeks; after which the positive cells were enriched by cell sorting and maintained.
Plasmids—Full-length Flag-MyD88, and its C-terminal and N-terminal truncation constructs were kindly provided by Dr. A. Ding (Cornell University). The dominant-negative Trif construct was a gift from Dr. S. Akira (Osaka University, Osaka, Japan). The Trif expression vector was a gift from Dr. Douglas Golenbock (University of Massachusetts Medical School). The Beclin 1 construct was kindly provided by Dr. B. Levine (Baylor College of Medicine).
shRNAs—The shRNA constructs targeting MyD88 and Trif were gifts from Dr. J. H. Shelhamer (Critical Care Medicine Department, Clinical Center, NIH, Bethesda, MD) (15). The shRNA sequences of two different targeted sites in mouse Beclin 1 were TGGACACGAGCTTCAAGAT and TGGAGATCCTAGAGCAGAT. The annealed inserts were cloned into the RNAi-Ready pSIREN-RetroQ vector (Clontech) that had been previously digested with EcoRI-BamHI. The negative control shRNA was purchased from Clontech. The shRNAs for silencing MyD88, Trif, Beclin 1, or a negative control were transfected into RAW 267.4 cells twice separated by 24 h using FuGENE HD Transfection Reagent (Roche Applied Science, Basel, Switzerland). The cells were treated with LPS or poly(I·C) for 7 or 16 h.
Electron Microscopy—RAW 264.7 cells without treatment served as the control. RAW 264.7 cells were treated with LPS (100 ng/ml) or poly(I·C), 20 μg/ml (Apotech, Epalinges, Switzerland), for 14 h. The cells were fixed in 1% glutaraldehyde, 0.1 m sodium cacodylate buffer (pH 7.4) for 1 h. Fixed monolayers were scraped, rinsed in buffer three times, and postfixed in 1% osmium tetroxide plus 0.8% potassium ferricyanide in the above buffer for 1 h. After several rinses in the buffer, the cells were dehydrated with increasing concentrations of ethanol and gradually infiltrated with Epon-Aradite (Ted Pella, Redding, CA). Samples were polymerized at 60 °C for 1 day. Ultrathin sections (about 80 nm) were cut on Leica EM UC6 microtome (Bannockburn, IL) and collected on copper slot grids. Sections were counterstained with uranyl acetate and lead citrate, and examined using a FEI CM120 transmission electron microscope equipped with a Gatan GIF100 image filter operating at beam energy of 120 keV. Images were acquired by using a Gatan 1k × 1k cooled CCD camera.
Autophagy Assay—A PerkinElmer Ultraview spinning wheel confocal system mounted on a Zeiss Axiovert 200 microscope and equipped with an argon/krypton laser, an Orca-ERII CCD camera (Hamamatsu), and filters suitable for visualization of both fluorescein isothiocyanate and red dyes was used for the quantization of cells transfected with GFP-LC3. A minimum of 50–100 cells per sample were counted. Compared with the control, GFP-LC3 dots in autophagy cells were much more numerous and larger. All ligands except ssRNA40 used for stimulating TLRs to induce autophagosome formation were purchased from Apotech (Epalinges, Switzerland). The ligand ssRNA40 for TLR7 was purchased from Invivogen (San Diego, CA).
Western Blot Analysis and Co-immunoprecipitation Assays—The antibodies that were purchased for Western blot and co-immunoprecipitation were MyD88 and Actin (Santa Cruz Biotechnology, Santa Cruz, CA), Trif (Imgenex, San Diego, CA), Beclin 1 and IκB-α (Cell Signaling Technology, Beverly, MA), and LC3 (MBL, Woburn, MA). RAW 264.7 cells were lysed in a buffer containing 20 mm HEPES, pH 7.4, 2 mm EGTA, 50 mm β-glycerophosphate, 1 mm Na3VO4, 1% Triton X-100, 0.2% SDS, and 10% glycerol with a protease inhibitor mixture (Roche Applied Science). The cell lysates were centrifuged, and equivalent amounts of protein were loaded onto 4–20 or 10% Tris glycine/SDS-polyacrylamide gels (Invitrogen). After electrophoresis, the proteins were transferred to nitrocellulose membranes. The specific antibodies as indicated were used for immunoblots. For co-immunoprecipitation assays, a modified lysis buffer was used. Instead of 1% Triton X-100 and 0.2% SDS, 0.5% Triton and 0.5% CHAPS were used. The lysates were incubated with antibodies for 2 h at 4 °C, and then Protein G PLUS-agarose was added and incubated for 1 h at 4 °C. The immunoprecipitated beads were washed with 0.5 ml of the same buffer 8 times. The immunoprecipitates were subjected to SDS-PAGE gel electrophoresis and MyD88, Trif, and Beclin 1 detected by immunoblot analysis as described above.
RESULTS
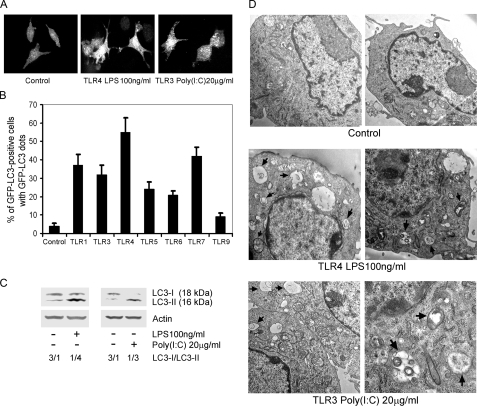
TLRs Induce Autophagy in Murine Macrophages—To detect autophagosome formation, green fluorescent protein fused to LC3 (GFP-LC3) was used as a surrogate marker. LC3 is a mammalian homologue of yeast Atg8, which is a specific constituent of the yeast autophagosomal membrane. LC3 exists in two forms, an 18-kDa cytosolic form (LC3-I) and a 16-kDa processed form (LC3-II), which localizes on the autophagosome membrane, where it functions in autophagosome enlargement (16). To determine whether TLRs induce autophagy in RAW 264.7 macrophages, we used a stably transfected cell line that expresses GFP-LC3. When autophagosomes form, GFP-LC3 is processed and recruited to the autophagosome membrane, where it can be imaged by confocal microscopy (Fig. 1A). The percentages of GFP-LC3 positive cells that have GFP-LC3 dots can be counted and are indicative of autophagosome formation. We treated the RAW 264.7/GFP-LC3 cells with different TLR ligands for 7 h and found that signaling through TLR1, TLR3, TLR4, TLR5, TLR6, and TLR7 induced significant increases in autophagosome formation. Ligands for TLR4, TLR7, TLR1, TLR3, TLR5, and TLR6 induced autophagy in 55, 42, 37, 32, 24, and 21% of the cells, respectively. Interestingly, signaling through TLR1 or TLR3 induced similar responses, despite their use of different adaptor proteins, MydD88 for TLR1 and Trif for TLR3. Ligands for TLR9 triggered autophagosome formation in 9% of the cells, only slightly above the basal level of 4% (Fig. 1B). To provide further evidence of TLR-induced autophagosome formation, we examined endogenous LC3-I and LC3-II levels by immunoblotting using either TLR4 or TLR3 ligands to treat the cells. Although the LC3-I band predominated in the untreated macrophages we could detect some LC3-II even in the absence of TLR ligand exposure. However, treatment with TLR3 or TLR4 ligands decreased the LC3-I band and increased the LC3-II band consistent with our previous assay (Fig. 1C). We also confirmed the TLR-induced autophagy at the ultrastructural level by electron microscopy. Both TLR3 and TLR4 signaling led to the appearance of autophagic vacuoles, double-membrane vacuolar structures containing cytoplasmic contents. Non-treated cells showed a few small autophagic vacuoles, some of which contained mitochondrial membranes. After TLR4 activation, we visualized large autophagic vacuoles in the cytoplasm of the RAW 264.7 cells, in which organelles did not exist. Activation of TLR3 with poly(I·C) induced some large autophagic vacuoles as well as some smaller vacuoles (Fig. 1D). Together these data indicate that TLR signaling leads to autophagosome formation.
FIGURE 1.
TLRs induce autophagy in RAW 264.7 cells. A, activated TLR4 or TLR3 leads to autophagosome formation in RAW 264.7/GFP-LC3 cells. RAW 264.7 cells stably transfected with GFP-LC3 were treated with LPS (100 ng/ml) for 7 h to activate TLR4, and with poly(I·C) (20 μg/ml) for 7 h to activate TLR3. The cells were imaged by confocal microscopy. B, TLR ligands cause autophagy in RAW 264.7/GFP-LC3 cells. Pam3CSK4·3HCl (100 ng/ml) was used to stimulate TLR1. TLR3 was activated with poly(I·C) (20 μg/ml). TLR4 was engaged by LPS (100 ng/ml). TLR5 was activated with Flagellin (50 ng/ml). TLR6 was stimulated with MALP-2 (50 ng/ml). TLR7 was activated by ssRNA40 (10 μg/ml). TLR9 was activated by CpG oligodeoxy nucleotides (5 μg/ml). All ligands were incubated with the cells for 7 h. The percentage of GPF-LC3 positive cells with GFP-LC3 dots were counted by confocal microscope. The data shown represent mean ± S.D. from three independent experiments. C, activated TLR4 or TLR3 induces autophagy in RAW 264.7 cells based on the ratio of endogenous LC3-I to LC3-II as shown by immunobloting with anti-LC3 antibodies. LPS and poly(I·C) at the above concentrations were added into the cells for 16 h; and then the cellular lysates were immunoblotted with antibodies against LC3. D, representative electron micrographics of RAW 264.7 cells treated with LPS or poly(I·C) for 16 h. Arrows denotes autophagosomes.
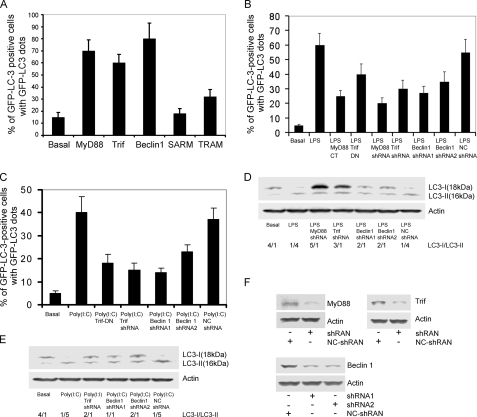
MyD88, Trif, and Beclin 1 Are Involved in Triggering Autophagy in RAW 264.7 Cells—TLR4 links to both MyD88 and Trif, TLR3 exclusively uses Trif, and the other TLRs use MyD88 as their major adaptor to transduce signals to downstream effectors (4). Beclin 1 interacts with the class III phosphoinositide-3-kinase (Vps34) to localize other autophagy proteins to the preautophagosomal membrane (17). Bcl-2 binds Beclin 1 and functions as a brake to autophagy and autophagic cell death perhaps by affecting the interaction between Beclin 1 and Vps34 (14). As an initial test of MyD88 or Trif involvement in the mediation of autophagosome formation, we expressed either MyD88 or Trif with GFP-LC3 in RAW 264.7 cells. We found that MyD88 or Trif expression led to GFP-LC3 dots in 60–70% of the GFP-LC3 positive cells, nearly similar to that achieved by the overexpression of Beclin 1. We also expressed the TIR-domain containing adapters SARM and TRAM along with GFP-LC3 in RAW 264.7. As expected, SARM, which is a negative regulator of TLR signaling (18), did not induce autophagy; however, expression of TRAM, which acts as a bridging adaptor for Trif-induced signaling (7), led to a moderate induction of autophagy (Fig. 2A). Together these data suggest that TLR signaling through MyD88 and/or Trif leads to autophagosome formation.
FIGURE 2.
Involvement of MyD88, Trif, and Beclin 1 in TLR3 and TLR4 triggered autophagy in RAW 264.7 macrophages. A, expressed MyD88, Trif, Beclin 1, or TRAM, but not SARM triggers autophagy. RAW 264.7 cells were transfected with 0.2 μg of GFP-LC3 and 0.6 μg of MyD88, Trif, Beclin 1, SARM, or TRAM plasmids for 24 h. The total amount of plasmid DNA was adjusted at 0.8 μg/transfection with pcDNA3 empty vector. The percentages of GFP-LC3 positive cells with GFP-LC3 dots were counted. The results shown represent mean ± S.D. from three independent experiments. B, a dominant negative form of MyD88 or Trif, and shRNAs targeted at MyD88, Trif, or Beclin 1 impair LPS-induced autophagy. RAW 264.7 cells were transfected with 0.8 μg of the indicated plasmids and 0.2 μg of GFP-LC3 as an autophagic marker for 24 h. The shRNA targeted at luciferase was used as a negative control. The cells were treated with LPS (100 ng/ml) for 7 h. The autophagic cells were calculated based on the percentage of GPF-LC3 positive cells with GFP-LC3 dots. The data shown are the mean ± S.D. from three independent experiments. C, dominant negative form of Trif or shRNAs targeted at Trif or Beclin 1 impairs poly(I·C)-induced autophagy. RAW 264.7 cells were transfected with 0.8 μg of the indicated plasmids and 0.2 μg of GFP-LC3 for 24 h. The shRNA that targets luciferase was used as a negative control. The cells were treated with 20 μg of poly(I·C) for 7 h. The autophagic cells were calculated as the percentage of GPF-LC3 positive cells with GFP-LC3 dots. The data shown are the mean ± S.D. from three independent experiments. D, reduced expression of MyD88, Trif, or Beclin 1 attenuates LPS-induced autophagy as determined by cleavage of endogenous LC3. RAW macrophages were transfected with 0.8 μg of the indicated shRNAs and again 24 h later. The luciferase shRNA was used as a negative control. The cells were treated with LPS (100 ng/ml) for 16 h. The levels of endogenous LC3-I and LC3-II were detected by immunobloting with anti-LC3 antibodies. Actin was used to verify equal sample loading. The data shown are one of two independent experiments performed. E, reduced expression of Trif or Beclin 1 impairs TLR3-induced autophagy as assessed by immunoblotting endogenous LC3. A double transfection of the shRNA constructs was performed. The data shown are one of two independent experiments. F, immune blots showing that the shRNAs targeting MyD88, Trif, or Beclin 1 mRNAs (two different sites) reduced expression of the corresponding protein.
MyD88 has a death domain at its NH2-terminal, which links it to downstream effectors in the TLR signaling pathways and a TIR domain at its COOH-terminal, which interacts with the cytoplasmic portion of various TLRs. An N-terminal truncated MyD88 functions as a dominant negative (19). The TIR domain of Trif has similar behavior (20). We used the dominant negative constructs or expressed shRNAs that target MyD88, Trif, or Beclin 1 to verify the roles of MyD88 and Trif in TLR4 and TLR3 induced autophagy. RAW 264.7 cells were transfected with the various constructs and GFP-LC3, treated 24 h later with either LPS or poly(I·C), and 7 h later autophagosome formation were measured by counting the percentage of GFP-LC3 positive cells that contained GFP-LC3 dots. Expression of the MyD88 or the Trif dominant negative form reduced LPS-induced autophagosome formation by 58 and 33%, respectively, whereas shRNAs targeting MyD88 or Trif reduced LPS-induced autophagosome formation by 67 and 50%, respectively. Two Beclin 1 shRNAs targeted at distinct sites in the Beclin 1 mRNA reduced autophagosome formation by 55 and 41% (Fig. 2B). Next, we examined whether Trif and Beclin 1 are required for poly(I·C)-activated TLR3-induced autophagosome formation. Using a similar experimental protocol, the expression of the Trif dominant negative form reduced TLR3-induced autophagosome formation by 55%, whereas the Trif shRNA reduced it 62%. The Beclin 1 shRNAs had similar effects as observed with TLR4-induced autophagosome formation (Fig. 2C). We also examined endogenous LC3-I and LC3-II levels following TLR3 or TLR4 signaling by immunoblotting cell lysates prepared from RAW 264.7 cells previously transfected with the Trif, MyD88, or Beclin 1 shRNAs. Consistent with the data using the RAW 264.7 cells transfected with GFP-LC3, reducing Trif, MyD88, or Beclin 1 impaired TLR4- or TLR3-induced increases in the LC3-II/LC3-I ratio (Fig. 2, D and E). The efficacy of the shRNAs in reducing MyD88, Trif, and Beclin 1 is shown (Fig. 2F).
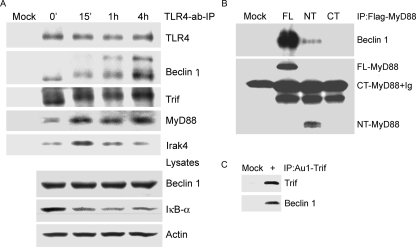
Activated TLR4 Recruits Beclin 1, MyD88, and Trif—In an attempt to link TLR signaling to autophagy, we examined whether activated TLR4 signaling recruited Beclin 1 into the TLR4 signaling complex. To do so we treated RAW 264.7 cells with an agonistic TLR4 antibody or a control antibody for 15 min, 1, and 4 h prior to cell lysis. In addition we added the same antibodies to non-treated cell lysates, which served as a time 0 point. We immunoprecipitated the TLR4 antibody or control antibody and immunoblotted for associated proteins (Fig. 3A). Beclin 1 co-immunoprecipitated with the TLR4 antibody, but not the control. The TLR4-Beclin 1 interaction increased with time and the Beclin 1 bands shifted upward suggesting a modification of Beclin 1. As expected, TLR4 adaptors MyD88 and Trif immunoprecipitated with TLR4. With the duration of exposure of the agonistic antibody, we detected more MyD88 and an altered mobility of Trif. The TLR4 antibody signaling also led to the recruitment of the protein kinase Irak4, a known member of the TLR4 signaling complex (4). Immunoblotting the cell lysates for Beclin 1 and IκBα expression revealed no change in Beclin 1 expression and degradation of IκBα indicating NF-κB activation. (Fig. 3A). To determine the relative importance of the MyD88 death and TIR domains in recruiting Beclin-1, we expressed tagged versions of full-length, TIR domaintruncated, and death domain-truncated MyD88 and examined their interaction with endogenous Beclin 1. The death domain of MyD88 pulled down Beclin 1, whereas the death domain deleted version did not (Fig. 3B). Furthermore, expression of the death domain of MyD88 induced autophagy nearly as well as full-length MyD88, whereas the death domain deleted version did not (data not shown). We also showed that Trif immunoprecipitates prepared from Trif-transfected cells contained endogenous Beclin 1 (Fig. 3C). These data strongly suggest that ligand-activated TLR4 assembles signaling complexes that contained MyD88, Trif, Irak4, and Beclin 1.
FIGURE 3.
Activated TLR4 recruits Beclin 1, MyD88, and Trif, and the death domain of MyD88 is required to associate with Beclin 1. A, an agonistic TLR4 antibody was used to stimulate RAW 267.4 cells and immunoprecipitate the TLR4-associated protein complex at the indicated time points. A matched control antibody was used for mock stimulation and immunoprecipitation. For the time 0 point non-stimulated RAW 267.4 cells were lysed, the TLR4 antibody was added and used to immunoprecipitate associated proteins. The washed immune complexes were analyzed by immunoblotting with the indicated antibodies. A portion of the cell lysates were saved and used to detect endogenous protein levels by immunoblotting with the indicated antibodies. B, MyD88 and the N terminus of MyD88 associated with Beclin 1. A FLAG-tagged MyD88, FLAG-tagged MyD88 death domain (NT), or FLAG-tagged MyD88 TIR domain (CT) was transfected into RAW 267.4 cells. An anti-FLAG monoclonal antibody was used for immunoprecipitation and immunoblotting. Anti-Beclin 1 antibodies were used for detecting the associated endogenous Beclin 1. C, Trif associates with Beclin 1. An anti-Au1 monoclonal antibody was used to immunoprecipitate expressed Au1-Trif. Endogenous Beclin 1 that co-immunoprecipitated was detected by anti-Beclin 1 antibodies. Similar results in two experiments performed.
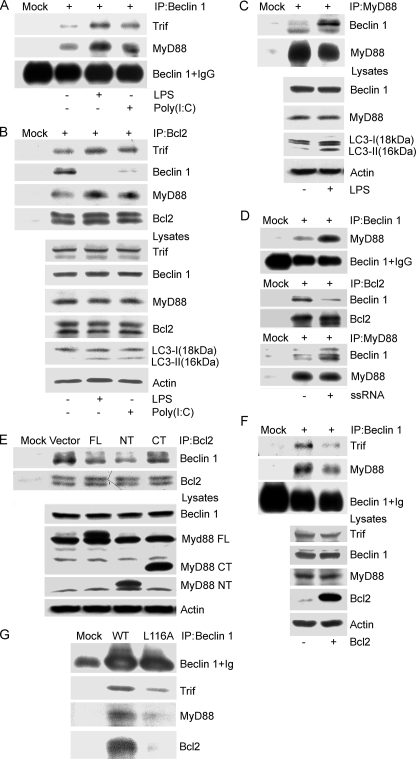
MyD88 and Trif Modulate the Interaction between Bcl2 and Beclin 1—The anti-apoptotic protein Bcl-2 inhibits starvation-induced autophagy via targeting the BH3-like domain in Beclin 1 (14, 21). Upon finding that TLR signaling leads to autophagy we sought to determine whether Bcl-2 had a role in modulating TLR-induced autophagy. However, first we verified that exposure to TLR3 and TLR4 ligands would enhance the interaction between Beclin 1 and MyD88 or Trif. The results indicated that exposure to the TLR4 ligand LPS triggered both MyD88 and Trif to recruit Beclin 1, whereas exposure to the TLR3 ligand poly(I·C) led to an increased association between Trif and Beclin 1 (Fig. 4A). Bcl-2 immunoprecipitates from lysates prepared from the same experiment were examined for the presence of MyD88, Trif, of Beclin 1. We found both MyD88 and Trif in the Bcl-2 immunoprecipitates, the levels of which increased slightly following signaling. In addition, a striking decrease in the Bcl-2 and Beclin 1 interaction occurred (Fig. 4B). Furthermore, when we immunoprecipitated MyD88 following exposure of RAW 267.4 cells to LPS we noted a large increase in co-immunoprecipitated Beclin 1 (Fig. 4C). Recently, induction of autophagy via TLR7 has been shown to depend on MyD88 and Beclin 1 expression (22). We found that similar to activated TLR4, activated TLR7 also enhanced association between MyD88 and Beclin 1, and deceased Bcl-2 binding with Beclin 1 (Fig. 4D). Next, we asked whether MyD88 or Bcl2 overexpression affected the interaction between Bcl2 and Beclin 1, or the association of Beclin 1 with MyD88 and Trif. Empty vector, MyD88, the MyD88 death domain (NT), or the death domain deleted version (CT) were transfected into RAW 264.7 cells. The expression of MyD88 or the MyD88 death domain reduced co-immunoprecipitation of Beclin 1 with Bcl-2 by 60 and 70%, respectively, whereas the death domain-deleted version did not affect it (Fig. 4E). Overexpression of Bcl-2 led to a reduced association between Beclin 1 and Trif as well as MyD88 (Fig. 4F). Because Beclin 1 has a BH-3 like domain, which is critical for binding with Bcl-2, we examined the ability of a BH-3 domain mutant, Beclin 1 L116A, to associate with MyD88 and Trif. As we had observed previously, wild type Beclin 1 readily co-immunoprecipitated Trif, MyD88, and Beclin 1, whereas Beclin 1 L116A was much less efficient (Fig. 4G). These results suggested that MyD88 and Trif may actively regulate autophagy by targeting the interaction between Beclin 1 and Bcl2.
FIGURE 4.
MyD88 and Trif modulate the interaction between Bcl2 and Beclin 1. A, LPS or poly(I·C) stimulation facilitates the association of Beclin 1 with MyD88 and Trif. Lysates from RAW 264.7 cells treated with LPS (100 ng/ml) or poly(I·C) (20 μg/ml) for 4 h were immunoprecipitated (IP) with anti-Beclin 1 antibodies. The resulting immune complexes were analyzed with the indicated antibodies. B, TLR4 and TLR3 activated by LPS and poly(I·C), respectively, promote the association of MyD88 and Trif with Bcl2, and suppress the interaction between Bcl2 and Beclin 1. The cellular lysates from the above preparation and treatment were also immunoprecipitated with a Bcl-2 antibody. The resulting immune complexes and corresponding lysates were analyzed using the indicated antibodies. The induction of autophagy was monitored by immunoblotting with anti-LC3 antibodies. C, LPS stimulation promotes MyD88 association with Beclin 1. RAW 267.4 cells were exposed to LPS (100 ng/ml) for 4 h and the cellular lysates were immunoprecipitated with anti-MyD88 antibody. They along with their corresponding lysate were analyzed by immunoblotting using the indicated antibodies. Autophagy was monitored by LC3 immunoblotting. D, ssRNA40 stimulation promotes MyD88 association with Beclin 1 and decreases Bcl-2 binding to Beclin 1. RAW 267.4 cells were treated with ssRNA40 (10 μg/ml) for 4 h and the cellular lysates were immunoprecipitated with anti-Beclin 1, Bcl-2, or a MyD88 antibody. The indicated proteins were immunoblotted. E, overexpression of FLAG-MyD88 and FLAG-MyD88 death domain (NT), but not the FLAG-MyD88 TIR domain (CT) partially suppresses the interaction between Beclin 1 and Bcl-2. The indicated constructs were transfected into RAW 267.4 cells for 24 h, and then anti-Bcl-2 antibodies were used to immunoprecipitate Bcl2. Co-immunoprecipitated Beclin 1 was detected by immunoblotting. The indicated proteins in cell lysates were detected by immunoblotting. F, overexpression of Bcl-2 attenuates the association of Beclin 1 with MyD88 and Trif. Following transfection of the Bcl-2 expression vector cell lysates were subjected to immunoprecipitation with anti-Beclin 1 antibodies. The resulting immunoprecipitates and the corresponding cell lysates were analyzed by immunoblotting with the indicated antibodies. G, FLAG-tagged Beclin 1 L116A has a decreased association with Trif, MyD88, and Bcl2. Beclin 1 or Beclin 1 L116A was transfected into RAW 267.4 cells and 24 h later immunoprecipitations were performed with anti-FLAG antibodies. The immunoprecipitates were analyzed by immunoblotting with the indicted antibodies. The above results come from one of three independent experiments that gave similar results.
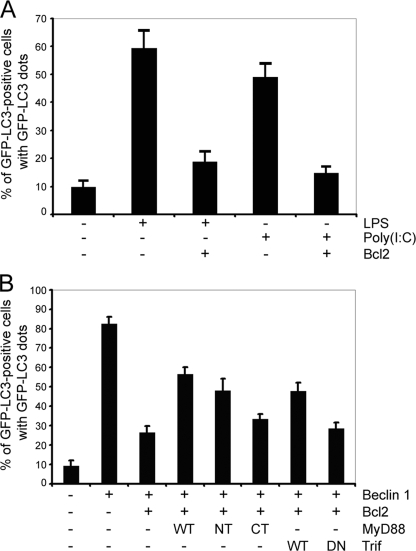
Bcl-2 Inhibits TLR-induced Autophagy and MyD88 or Trif Compete with Bcl-2 to Regulate Beclin 1-triggered Autophagy—Given that MyD88 and Trif may regulate the Beclin 1·Bcl-2 complex, we determined whether Bcl-2 is able to attenuate TLRs-induced autophagy in RAW 267.4 cells. The GFP-LC3 autophagy marker assay showed that overexpression of Bcl-2 significantly suppressed LPS and poly(I·C)-induced autophagy (Fig. 5A). Furthermore, the expression of MyD88 or Trif partially reversed the inhibition by Bcl-2 of Beclin 1-induced autophagy. Neither the death domain-deleted version of MyD88 nor the dominant negative form of Trif had a significant effect (Fig. 5B). The above results further support the concept that MyD88 and Trif are activators, whereas Bcl-2 is an inhibitor of TLR-induced autophagy and each of these proteins likely targets Beclin 1.
FIGURE 5.
Bcl2, MyD88, and Trif target Beclin 1 to regulate autophagy in RAW 264.7 macrophages. A, overexpression of Bcl-2 suppresses LPS (100 ng/ml, 7 h) and poly(I·C) (20 μg/ml, 7 h) leads to autophagy. RAW 267.4 cells were transfected with 0.2 μg of GFP-LC3 and 0.6 μg of Bcl2 plasmids for 24 h. The total amount of plasmid DNA was adjusted to 0.8 μg/transfection with the pcDNA3 vector. The percentages of GFP-LC3 positive cells with GFP-LC3 dots were counted. The results shown represent mean ± S.D. of three experiments. B, overexpression of MyD88 or Trif partially reverses the inhibition of Beclin 1-induced autophagy by Bcl2. RAW 264.7 cells were transfected with the following expression vectors: 0.2 μg of GFP-LC3, 0.3 μg of Beclin 1, 0.5 μg of Bcl-2, and MyD88, MyD88 death domain (NT), MyD88 TIR domain (CT), Trif, or Trif dominant negative. The total amount of plasmid DNA was adjusted at 1.5 μg/transfection with the pcDNA3 vector. The percentages of GFP-LC3 positive cells with GFP-LC3 dots were counted. The results shown are representative of one of three independent experiments performed.
DISCUSSION
The activation of innate immune systems is based on sensing the invasion of pathogenic microorganisms through pattern recognition receptors like Toll-like receptors, which recognize specific molecular signatures that are present in microbial components. Much attention has been focused upon TLR-mediated activation of NF-κB, mitogen-activated protein kinases, and interferon regulatory factors (4, 23). Here, we provide a link between TLR receptor signaling and the signaling pathway that leads to autophagy. We have focused on the signaling pathway downstream of TLR4, which leads to induction of autophagy and have shown that TLR4 adapter proteins MyD88 and Trif either directly or indirectly recruit Beclin 1 into the TLR4 signaling complex. This reduces the interaction between Beclin 1 and Bcl-2 and results in a biochemical modification of Beclin 1. The release of Beclin 1 from Bcl-2 biases the cell toward autophagy. We have also shown that TLR7-induced autophagy uses a similar mechanism.
Autophagy has been shown to play a role in macrophage antimicrobial activity. It is a defense mechanism that inhibits the survival of Mycobacterium tuberculosis in infected macrophages (24) and CD40 triggers autophagy in macrophages to help eliminate Toxoplasma gondii infection (25). Mice lacking MyD88 display profound defects in host resistance and immune responses to Mycobacterium avium infection not exhibited by Toll-like receptor 2 (TLR2)- and TLR4-deficient animals (26, 27). Because our investigation demonstrates defective TLR-induced autophagy from multiple Toll receptors, a failure to induce autophagy in MyD88-deficient mice could help explain the defects in host resistance in mice lacking MyD88.
Recently, TLR4 has been reported to be a sensor for autophagy in RAW 264.7 macrophages. However, in contrast to our results a Trif-dependent, but MyD88 independent, the TLR4 signaling pathway was found responsible. Receptor-interacting protein (RIP1) and p38 mitogen-activated protein kinase were downstream components of the pathway (28). This conclusion was based on silencing the expression of Trif, RIP1, and MyD88, by the use of a p38 inhibitor, and by the expression of a dominant negative MyD88. Data in this study that supports a role for MyD88 include the following: multiple TLRs can trigger autophagosome formation including TLRs that do not use Trif as an adapter, knock-down of MyD88 impairs TLR4-induced autophagosome formation, expression of the dominant negative version of MyD88 impaired LPS-induced autophagosome formation, overexpression of either full-length MyD88 or the N terminus of MyD88 triggered autophagosome formation, and finally that MyD88 co-immunoprecipitates with Beclin 1 and vice versa. Furthermore, it has been recently reported that TLR7-induced autophagy in RAW 264.7 depends upon MyD88 and Beclin 1 (22). This report clearly supports our results. Recently, genes that encode TIR domain containing proteins have been identified in Escherichia coli CFT073 (TcpC) and Brucella melitensis (TcpB). These proteins impeded MyD88-mediated TLR signaling, and thus can suppress innate immunity and increase bacterial virulence (29). Data from recent preliminary experiments indicate that the expression of TcpC or TcpB impedes LPS-induced autophagy in macrophages.4 The reason why our study implicated MyD88 and that of Xu et al. (28) did not is enigmatic. Both studies used the same cell line, similar LPS concentrations, and the same MyD88 dominant negative although the present study largely relied on transient transfections, whereas Xu et al. (28) used cells stably transfected with GFP-LC3 or GFP-LC3 and the MyD88 dominant negative.
Similar to the Xu et al. (28) study we found that Trif can trigger autophagy in RAW 264.7 cells. Although Xu et al. (28) relied on TLR4 signaling to trigger autophagy we also showed that TLR3 signaling, which uses Trif, and not MyD88, triggers autophagosome formation. Linking the Trif signaling pathway to autophagy Xu et al. (28) showed that LPS signaling led to the increase of VPS34 in the particulate fraction, which suggests an enhanced incorporation of the VSP34 complex into membranes. VPS34 is a class III phosphatidylinositol 3-kinase that functions in autophagy as a catalytic subunit in a complex with Beclin 1 (13). We found that similar to MyD88, Trif either directly or indirectly associates with Beclin 1. The data with the TLR4 agonistic antibody indicates that Beclin 1 directly associates with the TLR4 signaling complex and by analogy the TLR3 signaling complex. This data also revealed modification(s) of Beclin 1 that slowed its mobility on SDS-PAGE. Because we did not detect a change in the mobility of Beclin 1 in cell lysates, the Beclin 1 modification likely occurs following recruitment to the TLR4 signaling complex. An initial slight shift in the mobility was followed by a more dramatic shift. Prime candidates to cause such changes would be phosphorylation followed by ubiquitination.
It has been reported that the anti-apoptotic protein, Bcl-2, interacts with Beclin 1 and inhibits Beclin 1-dependent autophagy (14). Recently, a BH3-like domain has been recognized in Beclin 1, which binds Bcl-2 and Bcl-XL. The pharmacological BH3 mimetic ABT737 competitively inhibits the interaction and antagonizes autophagy, whereas inhibition of Bcl-2 stimulates autophagy. The BH3 domain mutant L116A of Beclin 1 loses binding to Bcl-2 and Bcl-XL (21). In our study the same mutant lost most of its ability to co-immunoprecipitate MyD88 and Trif. Overall our results suggest that a similar mechanism regulates TLR-induced autophagy in macrophages. We found that Bcl-2 overexpression attenuated TLR-dependent autophagy. MyD88 and Trif function as autophagic activators. TLR signaling enhances the presence of Beclin 1 in the signaling complex while reducing the interaction between Bcl-2 and Beclin 1, thereby promoting autophagy. Another interesting finding is that the death domain of MyD88 is required for the association with Beclin 1, and that overexpression of this domain by itself can induce autophagy. This result suggests a possible cross-talk pathway between autophagy and apoptosis, in which other death domain proteins might also interact with Beclin 1 to regulate autophagy.
In our study we have predominantly relied on two methods for monitoring the induction of autophagosome formation by TLR signaling both of which depend upon LC3 cleavage. LC3 is a homologue of Atg8 in yeast, which during autophagy is cleaved to become LC3-I and LC3-II. Although LC3-I is cytosolic, LC3-II is enriched on the autophagic vacuole. When GFP-LC3 expressing cells undergo autophagy LC3-II appears on newly formed autophagosomes visible as cytoplasmic puncta, which can be quantified by fluorescence microscopy. Alternatively, Western blotting of LC3-I and LC3-II can be performed and the ratio between the two monitored. These two methods have been extensively using for detecting autophagosome formation (30). Although electron microscopy can be used to visualize autophagy, it is difficult to quantify and impractical for signaling experiments.
In conclusion, not only TLR4 but also engagement of TLR1, TLR3, TLR5, TRL6, and TLR7 leads to autophagy in macrophages. Activated TLR4 recruited a protein complex that contained MyD88, Trif, and Beclin 1. MyD88 or Trif likely serve as activators for autophagy in macrophages along with Beclin 1 and triggers a reduction in the interaction between Bcl-2 and Beclin 1. Autophagy has emerged as a novel effector for TLRs facilitating innate as well as acquired immunity against a variety of pathogens.
Acknowledgments
We thank Guofeng Zhang (Division of Bioengineering & Physical Sciences, National Institutes of Health) for preparing samples and taking the electron microscope images. We also thank Mary Rust for excellent editorial assistance, and Anthony S. Fauci along with the NIAID for continued support.
This work was authored, in whole or in part, by National Institutes of Health staff. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TLR, Toll-like receptor; MAL, MyD88-adapter like; TRIF, TIR domain-containing adaptor protein inducing interferon β; TRAM, TRIF-related adaptor molecule; SARM, and sterile α- and armadillo motif-containing protein; GFP, green fluorescent protein; LPS, lipopolysaccharide; shRNA, short hairpin RNA; TIR, Toll/interleukin receptor; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; ssRNA, single strand RNA.
C. S. Shi, unpublished data.
References
- 1.Akira, S., Takeda, K., and Kaisho, T. (2001) Nat. Immunol. 2 675–680 [DOI] [PubMed] [Google Scholar]
- 2.Trinchieri, G., and Sher, A. (2007) Nat. Rev. Immunol. 7 179–190 [DOI] [PubMed] [Google Scholar]
- 3.Akira, S., and Takeda, K. (2004) Nat. Rev. Immunol. 4 499–511 [DOI] [PubMed] [Google Scholar]
- 4.Kawai, T., and Akira, S. (2006) Cell Death Differ. 13 816–825 [DOI] [PubMed] [Google Scholar]
- 5.Blander, J. M., and Medzhitov, R. (2006) Nature 440 808–812 [DOI] [PubMed] [Google Scholar]
- 6.Gazzinelli, R. T., and Denkers, E. Y. (2006) Nat. Rev. Immunol. 6 895–906 [DOI] [PubMed] [Google Scholar]
- 7.O'Neill, L. A., and Bowie, A. G. (2007) Nat. Rev. Immunol. 7 353–364 [DOI] [PubMed] [Google Scholar]
- 8.Shintani, T., and Klionsky, D. J. (2004) Science 306 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deretic, V. (2005) Trends Immunol. 26 523–528 [DOI] [PubMed] [Google Scholar]
- 10.Schmid, D., and Munz, C. (2007) Immunity 27 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine, B., and Deretic, V. (2007) Nat. Rev. Immunol. 7 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid, D., Pypaert, M., and Munz, C. (2007) Immunity 26 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kihara, A., Kabeya, Y., Ohsumi, Y., and Yoshimori, T. (2001) EMBO Rep. 2 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattingre, S., Tassa, A., Qu, X., Garuti, R., Liang, X. H., Mizushima, N., Packer, M., Schneider, M. D., and Levine, B. (2005) Cell 122 927–939 [DOI] [PubMed] [Google Scholar]
- 15.Qi, H. Y., and Shelhamer, J. H. (2005) J. Biol. Chem. 280 38969–38975 [DOI] [PubMed] [Google Scholar]
- 16.Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., Kominami, E., Ohsumi, Y., and Yoshimori, T. (2000) EMBO J. 19 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie, Z., and Klionsky, D. J. (2007) Nat. Cell Biol. 9 1102–1109 [DOI] [PubMed] [Google Scholar]
- 18.Carty, M., Goodbody, R., Schroder, M., Stack, J., Moynagh, P. N., and Bowie, A. G. (2006) Nat. Immunol. 7 1074–1081 [DOI] [PubMed] [Google Scholar]
- 19.Medzhitov, R., Preston-Hurlburt, P., Kopp, E., Stadlen, A., Chen, C., Ghosh, S., and Janeway, C. A., Jr. (1998) Mol. Cell 2 253–258 [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto, M., Sato, S., Mori, K., Hoshino, K., Takeuchi, O., Takeda, K., and Akira, S. (2002) J. Immunol. 169 6668–6672 [DOI] [PubMed] [Google Scholar]
- 21.Maiuri, M. C., Le Toumelin, G., Criollo, A., Rain, J. C., Gautier, F., Juin, P., Tasdemir, E., Pierron, G., Troulinaki, K., Tavernarakis, N., Hickman, J. A., Geneste, O., and Kroemer, G. (2007) EMBO J. 26 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delgado, M. A., Elmaoued, R. A., Davis, A. S., Kyei, G., and Deretic, V. (2008) EMBO J. 27 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783–801 [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez, M. G., Master, S. S., Singh, S. B., Taylor, G. A., Colombo, M. I., and Deretic, V. (2004) Cell 119 753–766 [DOI] [PubMed] [Google Scholar]
- 25.Andrade, R. M., Wessendarp, M., Gubbels, M. J., Striepen, B., and Subauste, C. S. (2006) J. Clin. Investig. 116 2366–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scanga, C. A., Aliberti, J., Jankovic, D., Tilloy, F., Bennouna, S., Denkers, E. Y., Medzhitov, R., and Sher, A. (2002) J. Immunol. 168 5997–6001 [DOI] [PubMed] [Google Scholar]
- 27.Feng, C. G., Scanga, C. A., Collazo-Custodio, C. M., Cheever, A. W., Hieny, S., Caspar, P., and Sher, A. (2003) J. Immunol. 171 4758–4764 [DOI] [PubMed] [Google Scholar]
- 28.Xu, Y., Jagannath, C., Liu, X. D., Sharafkhaneh, A., Kolodziejska, K. E., and Eissa, N. T. (2007) Immunity 27 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cirl, C., Wieser, A., Yadav, M., Duerr, S., Schubert, S., Fischer, H., Stappert, D., Wantia, N., Rodriguez, N., Wagner, H., Svanborg, C., and Miethke, T. (2008) Nat. Med. 14 399–406 [DOI] [PubMed] [Google Scholar]
- 30.Klionsky, D. J., Abeliovich, H., Agostinis, P., Agrawal, D. K., Aliev, G., Askew, D. S., Baba, M., Baehrecke, E. H., Bahr, B. A., Ballabio, A., Bamber, B. A., Bassham, D. C., Bergamini, E., Bi, X., Biard-Piechaczyk, M., Blum, J. S., Bredesen, D. E., Brodsky, J. L., Brumell, J. H., Brunk, U. T., Bursch, W., Camougrand, N., Cebollero, E., Cecconi, F., Chen, Y., Chin, L. S., Choi, A., Chu, C. T., Chung, J., Clarke, P. G., Clark, R. S., Clarke, S. G., Clave, C., Cleveland, J. L., Codogno, P., Colombo, M. I., Coto-Montes, A., Cregg, J. M., Cuervo, A. M., Debnath, J., Demarchi, F., Dennis, P. B., Dennis, P. A., Deretic, V., Devenish, R. J., Di Sano, F., Dice, J. F., Difiglia, M., Dinesh-Kumar, S., Distelhorst, C. W., Djavaheri-Mergny, M., Dorsey, F. C., Droge, W., Dron, M., Dunn, W. A., Jr., Duszenko, M., Eissa, N. T., Elazar, Z., Esclatine, A., Eskelinen, E. L., Fesus, L., Finley, K. D., Fuentes, J. M., Fueyo, J., Fujisaki, K., Galliot, B., Gao, F. B., Gewirtz, D. A., Gibson, S. B., Gohla, A., Goldberg, A. L., Gonzalez, R., Gonzalez-Estevez, C., Gorski, S., Gottlieb, R. A., Haussinger, D., He, Y. W., Heidenreich, K., Hill, J. A., Hoyer-Hansen, M., Hu, X., Huang, W. P., Iwasaki, A., Jaattela, M., Jackson, W. T., Jiang, X., Jin, S., Johansen, T., Jung, J. U., Kadowaki, M., Kang, C., Kelekar, A., Kessel, D. H., Kiel, J. A., Kim, H. P., Kimchi, A., Kinsella, T. J., Kiselyov, K., Kitamoto, K., Knecht, E., Komatsu, M., Kominami, E., Kondo, S., Kovacs, A. L., Kroemer, G., Kuan, C. Y., Kumar, R., Kundu, M., Landry, J., Laporte, M., Le, W., Lei, H. Y., Lenardo, M. J., Levine, B., Lieberman, A., Lim, K. L., Lin, F. C., Liou, W., Liu, L. F., Lopez-Berestein, G., Lopez-Otin, C., Lu, B., Macleod, K. F., Malorni, W., Martinet, W., Matsuoka, K., Mautner, J., Meijer, A. J., Melendez, A., Michels, P., Miotto, G., Mistiaen, W. P., Mizushima, N., Mograbi, B., Monastyrska, I., Moore, M. N., Moreira, P. I., Moriyasu, Y., Motyl, T., Munz, C., Murphy, L. O., Naqvi, N. I., Neufeld, T. P., Nishino, I., Nixon, R. A., Noda, T., Nurnberg, B., Ogawa, M., Oleinick, N. L., Olsen, L. J., Ozpolat, B., Paglin, S., Palmer, G. E., Papassideri, I., Parkes, M., Perlmutter, D. H., Perry, G., Piacentini, M., Pinkas-Kramarski, R., Prescott, M., Proikas-Cezanne, T., Raben, N., Rami, A., Reggiori, F., Rohrer, B., Rubinsztein, D. C., Ryan, K. M., Sadoshima, J., Sakagami, H., Sakai, Y., Sandri, M., Sasakawa, C., Sass, M., Schneider, C., Seglen, P. O., Seleverstov, O., Settleman, J., Shacka, J. J., Shapiro, I. M., Sibirny, A., Silva-Zacarin, E. C., Simon, H. U., Simone, C., Simonsen, A., Smith, M. A., Spanel-Borowski, K., Srinivas, V., Steeves, M., Stenmark, H., Stromhaug, P. E., Subauste, C. S., Sugimoto, S., Sulzer, D., Suzuki, T., Swanson, M. S., Tabas, I., Takeshita, F., Talbot, N. J., Talloczy, Z., Tanaka, K., Tanaka, K., Tanida, I., Taylor, G. S., Taylor, J. P., Terman, A., Tettamanti, G., Thompson, C. B., Thumm, M., Tolkovsky, A. M., Tooze, S. A., Truant, R., Tumanovska, L. V., Uchiyama, Y., Ueno, T., Uzcategui, N. L., van der Klei, I., Vaquero, E. C., Vellai, T., Vogel, M. W., Wang, H. G., Webster, P., Wiley, J. W., Xi, Z., Xiao, G., Yahalom, J., Yang, J. M., Yap, G., Yin, X. M., Yoshimori, T., Yu, L., Yue, Z., Yuzaki, M., Zabirnyk, O., Zheng, X., Zhu, X., and Deter, R. L. (2008) Autophagy 4 151–17518188003 [Google Scholar]