Abstract
Neonatal respiratory distress syndrome (RDS) is mainly the result of perturbation in surfactant production and is a common complication seen in premature infants. Normal fetal lung development and alveolar cell differentiation is regulated by a network of transcription factors. Functional loss of any of these factors will alter the developmental program and impact surfactant production and normal gas exchange. During development, the fetus is exposed to varying oxygen concentrations and must be able to quickly adapt to these changes in order to survive. Hypoxia-inducible factor 1α (HIF1α) is the primary transcription factor that is responsible for regulating the cellular response to changes in oxygen tension and is essential for normal development. Its role in lung maturation is not well defined and to address this knowledge gap, a lung-specific HIF1α knock-out model has been developed. Loss of HIF1α early in lung development leads to pups that die within hours of parturition, exhibiting symptoms similar to RDS. Lungs from these pups display impaired alveolar epithelial differentiation and an almost complete loss of surfactant protein expression. Ultrastructural analysis of lungs from HIF1α deletion pups had high levels of glycogen, aberrant septal development, and decreased expression of several factors necessary for proper lung development, including HIF2α, β-catenin, and vascular endothelial growth factor. These results suggest that HIF1α is essential for proper lung maturation and alteration in its normal signaling during premature delivery might explain the pathophysiology of neonatal RDS.
During development, an embryo is exposed to varying levels of oxygen as a balance is created between vascularization and tissue growth. Localized hypoxia, a decrease in available oxygen, is a normal part of this process. The programmed responses to these decreases in available oxygen are essential for viability (1, 2). In utero, the embryo is supplied with oxygen and nutrients through the placental barrier. Following parturition, oxygen is supplied by the neonatal lungs and therefore, proper intrauterine development of the alveolar gas exchange regions of the lung is essential for the newborn's first breath and for sustaining life outside the womb (3).
Lung morphogenesis is a complex process that is orchestrated by several transcription factors, growth factors, and extracellular cues (4). For example, thyroid transcription factor-1 regulates the expression of the genes for Clara cell secretory protein (CCSP)2 produced in Clara cells in the tracheobronchial airways and various surfactants produced by alveolar type II cells in the lung parenchyma (5, 6). CCAAT-enhancer-binding protein α (CEBPα) is essential for proper regulation of alveolar Type II cell differentiation, and Forkhead box A2 (Foxa2) controls various cellular programs involved in lung development (e.g. surfactant expression) (6-8). Ablation of any of these factors results in major structural and functional abnormalities ranging from undeveloped alveolar structure and/or improper airway branching to the faulty processing of various secretory components (7, 8). One of the extracellular cues that is important for fetal vascular growth and lung morphogenesis is the physiologically low O2 environment of the fetus. The ability to cope with this developmental “hypoxia” is not only important for lung maturation but essential for the viability of the fetus (1, 2).
Cellular responses to decreased oxygen availability are regulated by a family of proteins called hypoxia-inducible factors (HIFs). The ability of HIFs to respond to hypoxia is controlled by oxygen-dependent post-translational hydroxylation. The prolyl hydroxylases, PHDs, modify HIFs on essential residues in an oxygen-, iron-, and α-ketoglutarate-dependent manner. Once hydroxylated, the HIF is quickly degraded in a proteosomal-dependent process that involves the Von Hippel Lindau (VHL) tumor suppressor. HIF1α, the most ubiquitously expressed HIF, has been shown to play a critical role in normal development. HIF1α regulates the expression of genes important for cellular adaptation to hypoxia, including glycolytic enzymes and angiogenic factors (9). Gene deletion studies have established that HIF1α is indispensable during fetal development as HIF1α-/- mice die at midgestation, in association with defects in VEGF expression and vascularization (10-12). In addition, HIF1+/- mice developed pulmonary hypertension and pulmonary vascular remodeling under hypoxic conditions (13). PHD inhibitor-induced HIF stabilization has been shown to improve lung growth in prematurely born baboon neonates, predominantly through enhanced expression of VEGF (14). Finally, loss of HIF2α perturbs normal lung development through loss of VEGF expression and to subsequently decrease alveolar type II cell production of pulmonary surfactants (15).
Previous studies using systemic HIF1α deletions have demonstrated its central role in development and partial HIF1α deletions resulted in disrupted normal functioning of tissues such as heart and lung (13, 16, 17). The present study was aimed at elucidating the role(s) of HIF1α in the in utero differentiation of the alveolar epithelium. To this aim, mice with a lung-specific deletion of HIF1α (termed HIF1αΔ/Δ) were generated. Lung specific embryonic deletion of HIF1α earlier than 3 days prior to parturition led to HIF1αΔ/Δ neonates exhibiting cyanosis and respiratory failure soon after birth. The expression of surfactant proteins was significantly lower in HIF1αΔ/Δ pups as compared with littermate control pups. Examination of the lungs from HIF1αΔ/Δ pups via light and transmission electron microscopy confirmed defects in both alveolar epithelial differentiation and septal development that resulted in the observed respiratory distress.
MATERIALS AND METHODS
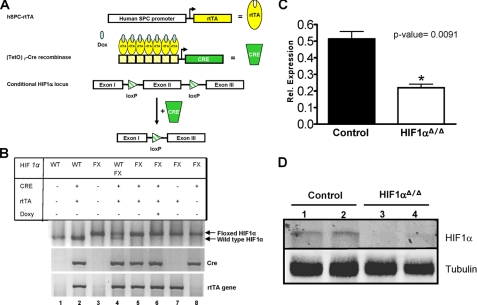
Transgenic Mice and Genotyping—HIF1αflox/flox and SP-C-rtTA-/tg/(tetO)7-CMV-Cretg/tg transgenic mice were generous gifts from Randall Johnson (University of California, San Diego) and Jeffrey Whitsett (Cincinnati Children's Hospital Medical Center), respectively (10, 17-19). HIF1αflox/flox and SP-C-rtTA-/tg/(tetO)7-CMV-Cretg/tg transgenic mice were mated to generate SP-C-rtTA-/tg/(tetO)7-CMV-Cretg/tg/HIF1αflox/flox mice capable of respiratory epithelium-specific conditional recombination in the floxed HIF1α gene. In this model, depending on the day doxycycline is administered to the dam, various cell populations within the lung undergo recombination in HIF1αflox/flox alleles (20). Genotyping of the mice progeny was performed by PCR for all the three loci using previously published primer sequences (supplemental Table S1). Genomic DNA extraction from tail clipping was performed using the Direct PCR extraction system (Viagen Biotech) per the manufacturer's instructions. PCR conditions were standardized for all the three alleles: denaturation at 94 °C for 3 min; 38 cycles of denaturation at 94 °C for 45 s, annealing at 60 °C for 45 s, and polymerization at 72 °C for 60 s followed by a 7-min extension at 72 °C. Sizes of the amplified products obtained were ∼210 bp for HIF1α (wild type); 244 bp for HIF1αflox/flox; 370 bp for Cre transgene; and 350 bp for rtTA transgene (Fig. 1B).
FIGURE 1.
Lung-specific, doxycycline-inducible deletion of HIF1α in triple transgenic mice. A, transgenic mice were generated that express three transgenes, the rtTA protein under the control of the human SP-C promoter, the (tetO)7-CMV-Cre recombinase transgene in which Cre recombinase expression is controlled by the (tetO)7-CMV promoter, and the conditional HIF1α locus. In the presence of doxycycline and rtTA, Cre recombinase is induced in a lung-specific manner and facilitates the homologous recombination of the loxP sites flanking exon 2 thus inactivating the HIF1α locus. B, agarose gel showing the genotyping results for all the combinations of the three transgenes. Floxed HIF1α size is 274 bp and wild-type HIF1α is 240 bp. C, qRT-PCR results for HIF1α mRNA from triple transgenic mice in the absence (Control) and presence (HIF1aΔ/Δ) of doxycycline (14 day in utero exposure). Values are the average of five separate animals per group. D, Western blot analysis for HIF1α from lungs of triple transgenic mice in the absence (Control) and presence (HIF1αΔ/Δ) of doxycycline (14 day in utero exposure). Blots were also probed with a tubulin-specific antibody to demonstrate equal loading. Two independent animals from both groups are shown (1-4).
Doxycycline Treatment and Animal Husbandry—Dams bearing triple (SP-C-rtTA-/tg/(tetO)7-CMV-Cretg/tg/HIF1αflox/flox) and double ((tetO)7-CMV-Cretg/tg/HIF1αflox/flox) transgenic embryos were maintained on doxycycline feed (625 mg of doxycycline/kg; Harlan Teklad, Madison, WI) and drinking water (0.8 mg/ml; Sigma Chemicals Co.). The day of parturition was taken as the reference point to calculate the duration of doxycycline exposure. Control and HIF1aΔ/Δ pups exposed to doxycycline treatment for 0-14 days in utero were sacked within 1 h of parturition. All pups were sacrificed by decapitation. Mice used in this study were kept at the animal housing facility under the strict hygienic and pathogen-free conditions approved by the University Laboratory Animal Resource regulatory unit.
Lung Harvesting and Processing—3-8 pups were analyzed from each genotype and doxycycline treatment. Pups were assessed for total body and lung weight. Lung tissues were isolated and the left lung lobe was fixed in 10% neutral-buffered formalin (NBF) or 4% glutaraldehyde for morphological analysis. The remaining lung tissue was divided; half was snap-frozen for Western blotting, half was stored in RNAlater RNA Stabilization Reagent (Qiagen) for RNA isolation and qRT-PCR analysis.
Western Blotting—Snap-frozen lung tissue (10 mg) was lysed and homogenized in radioimmune precipitation assay buffer (50 mm Tris-HCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm each of EDTA, phenylmethylsulfonyl fluoride, sodium vandate, sodium fluoride, 1 m dithiothreitol) containing protease inhibitors (1 μg/ml each of aprotinin, leupeptin, pepstatin) using a bead beater system (45 s at 30 Hz frequency). Insoluble material was removed by centrifugation (10,000 × g for 10 min.) and protein concentration was measured in supernatant using Bradford protein assay (20). Protein samples (30 μg) were separated by SDS-PAGE (NuPage 4-12% Bis-Tris gradient gel, Invitrogen) and transferred to a nitrocellulose membrane. Western blots were performed with rabbit or goat antibodies against surfactant-associated proteins (SP-A (sc 13977, Santa Cruz Biotechnology), SP-B (AB 3780, Chemicon), SP-C (sc-13979, Santa Cruz Biotechnology) and SP-D (SC 7709, Santa Cruz Biotechnology), HIF1α (NB100-479, Novus Biologicals), tubulin (T5168, Sigma), and β-actin (SC-7210, Santa Cruz Biotechnology). Proteins were visualized with horseradish peroxidase-conjugated goat anti-rabbit IgG (sc-2004, Santa Cruz Biotechnology) or rabbit anti-goat (sc-2768, Santa Cruz Biotechnology) and ECL Western blot system (Pierce) or fluorescently using Alexa fluor 680 anti-rabbit IgG (A21109, Invitrogen) or IRdye800 antimouse IgG (610-132-121, Rockland).
Histopathology and Immunohistochemistry—At least 4-6 pups of each genotype and doxycycline treatment were analyzed for histopathological changes. Formalin-fixed left lung lobe tissues were paraffin-embedded and 5-micron thick sections were mounted on glass slides and stained with hematoxylin and eosin (H&E), periodic acid Schiff (PAS), or immunostained with SP-B (1:500 dilution, AB3780, Chemicon, MA), SP-C (1:4000 dilution, antibody kindly provided by Jeffrey Whitsett), and CRE (1:100 dilution, AB24608; Abcam, Cambridge, MA). Briefly, tissue sections were deparaffinized and endogenous peroxidase activity was quenched by incubation with 6% H2O2 for 30 min. Immunostaining was performed using Rabbit Vector Elite ABC kit (Vector Laboratories), according to the manufacturer's recommendations.
Transmission Electron Microscopy—Neonatal lung sections were fixed in 4% buffered glutaraldehyde overnight at 4 °C, and stored in the 10% NBF fixative until postfixation with 1% phosphate-buffered osmium tetroxide. Tissues were dehydrated through a graded series of ethanol and propylene oxide, and embedded in Poly/Bed-Araldite resin (Polysciences, Inc., Warrington, PA). 1 μm-thick sections were cut and stained with toluidine blue for light microscopic identification of specific tissue sites for transmission electron microscopy (TEM). Ultrathin tissue sections for TEM were cut at ∼75 nm and stained with lead citrate and uranyl acetate. Sectioning was performed with an LKB Ultratome III (LKB Instruments, Inc., Rockville, MD). Ultrastructural tissue examination and photography were performed with a JEOL JEM 100CXII electron microscope (JEOL Ltd., Tokyo, Japan).
RNA Isolation and Quantitative Real-time PCR (qRT-PCR) Analysis—10 mg of lung tissue stored in RNAlater RNA Stabilization Reagent was homogenized in RLT buffer using a Retsch MM200 bead beater system (Retsch, Haan, Germany). Total RNA quantification was performed spectrophotometrically (NanoDrop ND-1000 UV-Vis Spectrophotometer). Total RNA (1 μg) was reverse-transcribed using the Superscript II reverse transcriptase kit (Invitrogen). The expression level of selected genes involved in surfactant metabolism and lung development was analyzed by qRT-PCR using SYBR green (Applied Biosystems, Foster City, CA) as previously described (21). Gene-specific primers are listed in supplemental Table S2. Copy number was determined by comparison with standard curves of the respective genes. This measurement was controlled for RNA quality, quantity, and RT efficiency by normalizing it to the expression level of the hypoxanthine guanine phosphoribosyl transferase (HGPRT) gene. Statistical significance was determined by use of normalized relative changes and analysis of variance.
Quantitative Analysis—qRT-PCR analysis was performed using unpaired two-tailed Student's t test on GraphPad Prism. Results were considered significant at the 5% level.
RESULTS
Conditional Inactivation of HIF1α in the Lung—The HIF1α gene was inactivated in the lung by mating HIF1α-conditional null mice (HIF1αflox/flox) (10) to an inducible bitransgenic mouse, SPC-rtTA, that expresses the reverse tetracycline transactivator (rtTA) under the control of the human surfactant protein C (SP-C) promoter and the Cre recombinase gene with a tetracycline operon (Fig. 1A) (18). SPC-rtTA mice have been shown to express Cre recombinase specifically in the epithelial cells of the primordial lung buds as well as in the alveolar and bronchiolar epithelium of postnatal mice in the presence of an inducer (i.e. doxycycline) (19). In the present study, compound litters from reciprocal matings between SP-C-rtTA-/tg/(tetO)7-CMV-Cretg/tg/HIF1αflox/flox and (tetO)7-CMV-Cretg/tg/HIF1αflox/flox mice were composed of pups with mixed genotypes with respect to SPC-rtTA transgene. Their genotypic ratios (data not shown) were in accordance with Mendelian inheritance as demonstrated by genotyping (Fig. 1B). Analysis of these ratios from litters studied (n = 30 litters and 190 pups) confirmed no in utero mortalities (data not shown). In the absence of doxycycline treatment, pups were phenotypically indistinguishable between litters. The functionality of the doxycycline treatment was demonstrated by the significant decrease in HIF1α mRNA as measured by qRT-PCR (Fig. 1C) and loss of HIF1α protein by Western blot analysis (Fig. 1D).
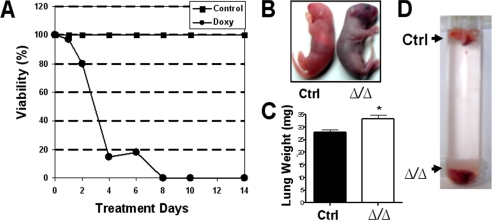
Lung-specific Deletion of HIF1α Causes Lethal Phenotype—To determine if in utero loss of HIF1α influenced litter size or the Mendelian distribution of pups, dams carrying triple transgenic embryos were exposed to doxycycline through feed and water for various lengths of time prior to parturition (i.e. 2, 4, 6, 8, 10, 12, and 14 days). Our findings indicate pups exposed to doxycycline in utero for at least 8 days prior to parturition had a 100% mortality rate, while those receiving the drug for 4-6 days prior to parturition had approximately a 15% chance of survival (Fig. 2A). Following parturition, HIF1αΔ/Δ pups (doxycycline-induced) developed respiratory distress, severe cyanosis, and died within an hour of birth (range of 10-60 min) (Fig. 2B). No observable differences were noted in survival times following parturition from pups treated with doxycycline for only 1-6 days compared with those exposed for at least 8 days. More importantly, no mortality was observed in HIF1α+fl/+fl mice lacking either the rtTA or Cre recombinase transgene in the presence or absence of doxycycline (data not shown). These data suggest that the mortality is due to loss of HIF1α and not off-target effects of the other transgenes.
FIGURE 2.
Effects of lung-specific deletion of HIF1α. A, graph of the percent viability of pups at birth versus intrauterine doxycycline treatment days. Doxycycline-induced lethality was more pronounced when the drug was delivered more than 4 days before parturition (circles). No lethality was observed in the control group (no doxycycline, squares) or other possible genotype controls (data not shown). B, at birth HIF1αΔ/Δ pups (14 day in utero exposure) were cyanotic (right) compared with normal, pink well oxygenated controls (left). C, mean lung weights of control (black bar) and HIF1αΔ/Δ pups (white bar, exposed to doxycycline for more than 8 days). The asterisk indicates a significant difference, p value < 0.01. D, vial containing excised lungs from newborn pups. Lungs from HIF1αΔ/Δ (Δ/Δ, 14 day doxycycline exposure) pups sink whereas control (Ctrl) lungs float in saline.
At necropsy, the lungs from HIF1αΔ/Δ pups were dark red and atelectic, in contrast to pink, aerated lungs from control (in the absence of doxycycline treatment) pups. Excised lung lobes from HIF1αΔ/Δ pups weighed slightly more than the lungs from control mice and sank to the bottom of a saline-filled vial in contrast to the lungs from control littermates, which floated in saline (Fig. 2, C and D). At birth, the average total body weights of control littermate and HIF1αΔ/Δ were 1.318±0.018 (S.E.) and 1.288 ± 0.024 grams, respectively.
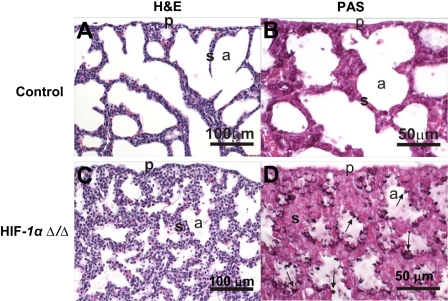
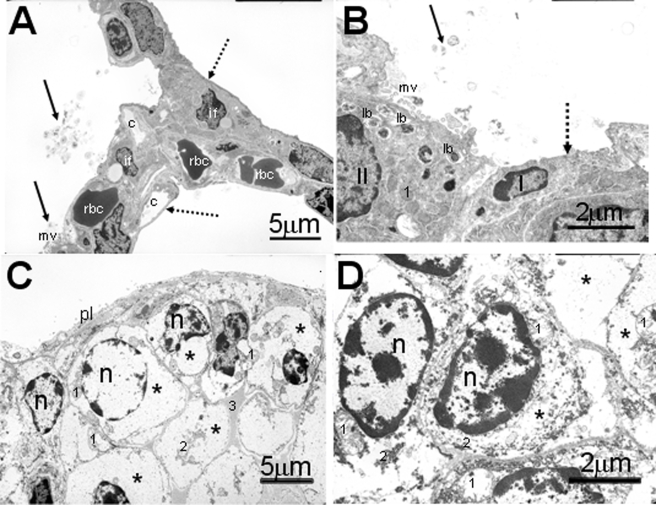
Defective Lung Morphology—The lungs of HIF1αΔ/Δ pups had histologic and ultrastructural features that indicate an impairment of normal in utero differentiation of alveolar epithelium, with a related loss of lung surfactant proteins that are essential for preventing collapse of individual alveoli and ensuring proper gas exchange to maintain life. Lungs of HIF1αΔ/Δ pups at birth had abnormally thickened alveolar septa (Fig. 3C) lined by undifferentiated cuboidal epithelial cells (immature pneumocytes) containing large amounts of intracytoplasmic periodic acid Schiff (PAS)-positive material (Fig. 3D) that was ultrastructurally identified, via transmission electron microscopy, as glycogen (Fig. 4, C and D). There was a marked reduction of alveolar airspaces in the lungs of these mice due in part to the thickened septa and to focal areas of atelectasis.
FIGURE 3.
Pulmonary histopathology. Light photomicrographs of hematoxylin and eosin-stained lung sections from control (A) and HIF1αΔ/Δ (C) pups. The lung from HIF1αΔ/Δ pup has less alveolar airspace (a) and thicker alveolar septa (s) compared with control. Large cuboidal epithelial cells line the alveolar septa of HIF1αΔ/Δ pup, whereas the alveolar septa of control pup is lined primarily by squamous epithelial cells (type 1) and only a few widely scattered cuboidal cells (type II). PAS-stained lung sections from control (B) and HIF1αΔ/Δ (D) pups. Cuboidal epithelium lining alveolar septa in HIF1αΔ/Δ pup has greater PAS-stained glycogen (arrows) than that of the control pup (D). All doxycycline treatments were at least 12 days.
FIGURE 4.
Electron photomicrographs of alveolar epithelium of newborn pups. The lung from the control pup (A and B) has a well differentiated alveolar epithelium containing both type I and II cells (B). Stippled arrow indicates thin, squamoid, type I cells, and solid arrows indicate tubular myelin figures (secreted surfactant) within the alveolar airspace of control pup. Type II cells have numerous intracytoplasmic lamellar bodies (lb) and apical microvilli (mv). In contrast the alveolar surface in HIF1αΔ/Δ pup (C and D) is lined by large undifferentiated cuboidal epithelial cells. The cytoplasm of these epithelial cells are primarily filled with glycogen (*). No lamellar bodies are present in these cells. c, capillary; rbc, red blood cells; II, type II alveolar epithelial cell; I, type I alveolar epithelial cell; if, interstitial fibroblast; *, glycogen; n, nucleus; 1, mitochondrion; 2, rough endoplasmic reticulum; pl, pleural surface; solid black arrows, tubular myelin; stippled arrow, type I cells. All doxycycline treatments were at least 12 days.
In contrast, the lungs of control mice at birth had uniformly dilated alveolar airspaces with much thinner alveolar septa that were lined by more differentiated surface epithelium containing much less PAS-positive glycogen (Fig. 3, A and B). Ultrastructurally, the luminal epithelium lining the alveolar airspace of control pups consisted of both alveolar type I and II cells (differentiated pneumocytes). Distinctive lamellar bodies were present in the apical cytoplasm of alveolar type II cells along with tubular myelin and secreted lamellar material (ultrastructural features of secreted surfactant) widely scattered along the alveolar luminal surface (Fig. 4, A and B). These distinctive cuboidal epithelial cells also had a round to ovoid nucleus, an apical luminal surface lined by short microvilli, and numerous cytoplasmic organelles consisting of mitochondria, rough endoplasmic reticulum, and lysosomes. Type I cells were identified by their thin squamous morphology containing few organelles, and a fusiform nucleus (Fig. 4, A and B). Interestingly, there were no microscopically detectable differences in the morphology of intrapulmonary conducting airways (preterminal and terminal bronchioles), lined principally by ciliated cells and nonciliated cuboidal (Clara) cells, between control and HIF1αΔ/Δ pups at birth.
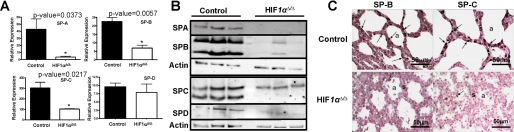
Altered Surfactant Metabolism—qRT-PCR analysis of the genes for surfactant proteins show a decreased expression of SP-A, SP-B, and SP-C in the HIF1αΔ/Δ mice compared with control pups (Fig. 5A). SP-D mRNA expression was not significantly different between the two genotypes. Western blot analysis of protein extract from HIF1αΔ/Δ and control animals was also performed to determine if surfactant protein levels were similar to gene expression patterns. All four surfactants displayed a substantial reduction in protein levels in the HIF1αΔ/Δ mice as compared with control pups (Fig. 5B). Interestingly, SP-C displayed an overall decrease in protein levels but the pre-processed form (Fig. 5B, SPC upper band) was not as affected by deletion of HIF1α as was the fully processed form (Fig. 5B, SPC lower band). This suggests that SP-C transcription is being decreased by the loss of HIF1α and processing of any translated SP-C protein is being inhibited by the aberrant differentiation of the HIF1αΔ/Δ lung. In comparison, SP-B monomer and dimer forms were equally affected by loss of HIF1α. Immunohistochemical staining on lung sections confirmed the reduced levels of SP-B and SP-C in the HIF1αΔ/Δ compared with control pups (Fig. 5C). The reduced levels of surfactant mRNA and protein expression were consistent with the lack of alveolar epithelial differentiation and respiratory distress in the HIF1αΔ/Δ pups.
FIGURE 5.
Surfactant proteins expression. A, mRNA levels for surfactant-associated proteins (SP-A, SP-B, SP-C, and SP-D). Each expression level is expressed relative to HGPRT. B, immunoblot analysis of surfactant-associated proteins (SP-A, SP-B, SP-C, and SP-D) in lung homogenates from control and HIF1αΔ/Δ pups (n = 3). β-Actin (actin) was used as loading control. C, immunohistochemical staining of SP-B (left panels) and SP-C (right panels) in lung sections from control (upper panels) and HIF1αΔ/Δ (lower panels) pups. HIF1αΔ/Δ pups show decreased expression of SP-B and SP-C as compared with control littermates. The asterisk indicates a significant difference (p value < 0.05) based on a Student's two-tailed t test for samples of unequal variance. All doxycycline treatments were for at least 12 days.
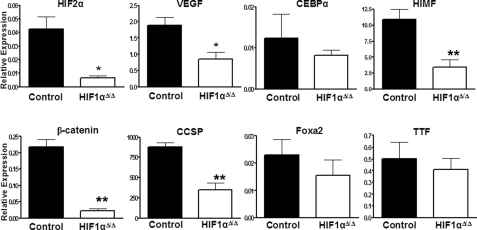
Altered Gene Expression—To begin to understand how loss of HIF1α induced changes in lung morphology, qRT-PCR was performed on several critical genes known to be involved in lung development. These genes include β-catenin, Clara cell secretory protein (CCSP), forkhead box A2 (Foxa2), forkhead box protein A1 (Foxa1, also known as HNF3α), thyroid transcription factor 1 (TTF-1), GATA-binding protein 6 (Gata6), ATP-binding cassette, subfamily A (ABC1 member 3 (ABCA3), and CCAAT/enhancer-binding protein α (CEBPα). There were no significant differences in the expression levels of Foxa2, ABCA3, CEBPα, TTF-1, Foxa1, or GATA, suggesting that HIF1α is not required for their normal expression and is most likely involved in later events in lung development (Fig. 6 and data not shown). However, lower levels of β-catenin, CCSP, HIMF, VEGF, and HIF2α mRNA in neonatal HIF1αΔ/Δ pups compared with control littermates suggest that HIF1α plays a role in controlling their expression during maturation of the lung (7, 15). Given the established role of these factors in regulating lung development, our data suggest that HIF1α is centrally located in the transcriptional network that is necessary for proper lung morphogenesis.
FIGURE 6.
Expression of developmentally important genes. qRT-PCR was used to measure the mRNA levels of various genes known to play a role in lung development. Each expression level is expressed relative to HGPRT. The asterisk indicates a significant difference (*, p value < 0.05; **, p value < 0.01) based on a Student's two-tailed t test for samples of unequal variance. All doxycycline treatments were for at least 12 days.
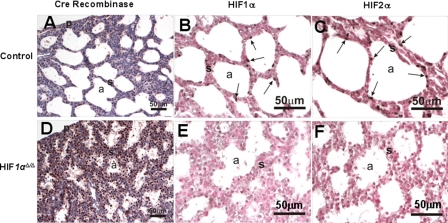
The previously described role of HIF2α in lung development, the similarity in phenotype between the HIF2α knock-out survivors and HIF1α lung deletion strains, and the loss of HIF2α expression in the HIF1αΔ/Δ pups led us to determine the level of HIF2α expression at the protein level using immunohistochemistry (15). In the presence of doxycycline there was a pronounced expression of Cre recombinase in all cell types of the lung (Fig. 7D). There was a corresponding loss in HIF1α expression in the alveolar tissue (Fig. 7E). Though HIF1α is not expressed ubiquitously throughout the lung, its cell-specific expression was abrogated upon doxycycline treatment in the triple transgenic animal (Fig. 7). Surprisingly, the increased Cre recombinase and subsequent loss of HIF1α expression also led to a drastic decline in HIF2α levels in the lung (Fig. 7, C and F).
FIGURE 7.
Immunohistochemistry for Cre recombinase, HIF1α, and HIF2α. Lung sections from control (A-C) and HIF1αΔ/Δ pups (D-F) were immunostained for Cre recombinase (A and D), HIF1α (B and E), and HIF2α (C and F). Immunohistochemistry for Cre recombinase shows strong staining of HIF1αΔ/Δ mouse alveolar epithelial cells (D, brown color). Positively stained cells for HIF1α and HIF2α are depicted by solid arrows. HIF1αΔ/Δ pups show decreased expression of HIF1α and HIF2α as compared with control littermates. a, air space; p, pleural surface; s, septa. All doxycycline treatments were for at least 12 days.
DISCUSSION
Mammalian lung development and successful transition to extrauterine respiration involves a wide battery of biochemical, cellular, and ultrastructural changes. This complex process begins at embryonic day E9.5 with most of the alveolar epithelial differentiation events starting at embryonic day E17.5. Several transcription factors control the temporal and positional epithelial differentiation process, as well as the function of these differentiated cells. When the function of these critical factors is disrupted during development, lung function is compromised, leading to respiratory distress at birth. Our results have added HIF1α to this list of transcription factors that are critical for lung development. We have also demonstrated that loss of HIF1α during development leads to changes in the expression of other critical factors, such as HIF2α, VEGF, and β-catenin (23).
HIFs are a class of transcription factors that play a critical role in oxygen-sensing and the metabolic adaptations to hypoxia. Previous studies have shown that HIF1α levels are predominant during lung development until day E13.5, at which time, HIF2α becomes the major HIF in this tissue (24, 25). Targeted gene disruption of the HIF1α locus results in the embryonic lethality due to cardiovascular defects (12). Mice partially deficient for HIF1α alleles showed normal development but were physiologically compromised as they exhibited impaired pulmonary vascular remodeling under chronic hypoxic stress. In addition, the heterozygote was used to show that partial loss of HIF1α alleles results in impaired pulmonary arterial myocyte electrophysiological responses under hypoxic stress (13, 25, 26). The study presented here has shown that the lung-specific loss of HIF1α leads to neonatal respiratory distress syndrome due to impaired alveolar epithelial differentiation. Interestingly, the lungs of HIF1αΔ/Δ pups had normal morphogenesis of conducting airways, with phenotypic alterations restricted to the alveolar parenchyma. These results suggest that HIF1α is involved in the late events of lung morphogenesis and alveolar epithelial differentiation but not in the early lung biogenesis and airway branching morphogenesis.
Respiratory distress syndrome and associated histopathology are phenotypic hallmarks of targeted deletion of several transcription factors, such as Foxa2, CEBPα, GATA-6, SMAD3, calcineurin β1, and thyroid transcription factor-1, suggesting an intricate network of transcriptional regulation governing the complex ultrastructural maturation of lungs (8, 22, 27-29). Expression analysis demonstrated that the levels of Foxa2, Foxa1, CEBPα, and GATA-6 are comparable among pups with and without HIF1α (Fig. 6 and data not shown). This suggests that HIF1α acts either downstream or independent of these transcription factors. In contrast, the loss of HIF1α led to a decreased expression of other critical factors, including CCSP, β-catenin, HIMF, VEGF, and HIF2α (Fig. 6). Recently, the loss of HIF2α and subsequent decrease in VEGF has been linked to respiratory distress syndrome (15). These experiments involved a subset of HIF2α-/- mice that were viable up to parturition and displayed similar pathology to the HIF1αΔ/Δ mice described here. Delivery of VEGF, either intrauterine or postnatal intratracheal instillation, protected the HIF2α-/- mice from symptoms of respiratory distress syndrome. Given that HIF2α expression is abrogated in the HIF1aΔ/Δ pups suggests that HIF1α regulates HIF2α early in lung development and the subsequent loss of VEGF might explain the pathology of the HIF1αΔ/Δ pups. It remains to be seen if the decrease in VEGF expression is due to direct loss of HIF1α or indirectly through the loss of HIF2α expression. More importantly, the pathophysiology of the HIF1αΔ/Δ mouse resembles that of the HIF2α-/- mouse in relation to the lack of differentiation of type II pneumocytes and increased septal thickness. In addition, the HIF1αΔ/Δ pups display large glycogen stores with concurrent loss of surfactant protein expression.
In conclusion, these studies provide evidence that HIF1α plays an important role in lung development. The respiratory failure induced by lung-specific deletion of HIF1α, suggests that hypoxia and HIF1α-regulated genes play an essential role in lung maturation, especially proper differentiation of type II alveolar cells. HIF1α potential upstream regulatory function of HIF2α and VEGF expression, directly or indirectly, offers a mechanism of action for the phenotypic observations. Finally, the results suggest that the hypoxia signaling cascade is important during the later stages of lung development but not essential in the initial patterning of the tissue. HIFs seem to play a later role in directing cell differentiation and regulating growth factors necessary for lung remodeling required for sustaining life extrauterine. Understanding the pathways necessary for proper HIF expression and function and the downstream pathways influenced by these factors will impact our ability to treat RDS and other diseases of the lung.
Supplementary Material
Acknowledgments
We thank Dr. Randall Johnson (University of California, San Diego) for the gift of the conditional HIF1α mice and the gift of SP-C-rtTA/(tetO)7-CMV-Cre transgenic from Dr. Jeffrey Whitsett (Cincinnati Children's Hospital Medical Center). We would also like to acknowledge Melinda Kochenderfer for editorial help in preparing this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-ES12186 and P42 ES04911-17. This work was also supported by the Michigan Agriculture Experimental Station and the Michigan State University Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
Footnotes
The abbreviations used are: CCSP, Clara cell secretory protein; CEBPα, CCAAT-enhancer-binding protein α; Foxa1, Forkhead box A1; Foxa2, Forkhead box A2; GATA 6, GATA-binding protein 6; HGPRT, hypoxanthine guanine phosphoribosyl transferase; HIF, hypoxia-inducible factor; HIMF, hypoxia-inducible mitotic factor; HNF3α, hepatocyte nuclear factor 3α; PAS, periodic acid-Schiff; qRT-PCR, quantitative real-time PCR; RDS, respiratory distress syndrome; rtTA, reverse tetracycline transactivator; SP(A-D), surfactant-associated proteins A-D; VEGF, vascular endothelial growth factor.
References
- 1.Webster, W. S., and Abela, D. (2007) Birth Defects Res. C Embryo Today 81 215-228 [DOI] [PubMed] [Google Scholar]
- 2.Giaccia, A. J., Simon, M. C., and Johnson, R. (2004) Genes Dev. 18 2183-2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James Metcalfe, H. B., and Waldemar Moll. (1967) Physiol. Rev. 47 782-838 [DOI] [PubMed] [Google Scholar]
- 4.Groenman, F., Unger, S., and Post, M. (2005) Biol. Neonate 87 164-177 [DOI] [PubMed] [Google Scholar]
- 5.Cassel, T. N., Suske, G., and Nord, M. (2000) Ann. N. Y. Acad. Sci. 923 300-302 [DOI] [PubMed] [Google Scholar]
- 6.deFelice, M., Silberschmidt, D., DiLauro, R., Xu, Y., Wert, S. E., Weaver, T. E., Bachurski, C. J., Clark, J. C., and Whitsett, J. A. (2003) J. Biol. Chem. 278 35574-35583 [DOI] [PubMed] [Google Scholar]
- 7.Mucenski, M. L., Wert, S. E., Nation, J. M., Loudy, D. E., Huelsken, J., Birchmeier, W., Morrisey, E. E., and Whitsett, J. A. (2003) J. Biol. Chem. 278 40231-40238 [DOI] [PubMed] [Google Scholar]
- 8.Wan, H. J., Xu, Y., Ikegami, M., Stahlman, M. T., Kaestner, K. H., Ang, S. L., and Whitsett, J. A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 14449-14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenza, G. L. (2000) J. Appl. Physiol. 88 1474-1480 [DOI] [PubMed] [Google Scholar]
- 10.Ryan, H. E., Lo, J., and Johnson, R. S. (1998) EMBO J. 17 3005-3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotch, L. E., Iyer, N. V., Laughner, E., and Semenza, G. L. (1999) Dev. Biol. 209 254-267 [DOI] [PubMed] [Google Scholar]
- 12.Iyer, N. V., Kotch, L. E., Agani, F., Leung, S. W., Laughner, E., Wenger, R. H., Gassmann, M., Gearhart, J. D., Lawler, A. M., Yu, A. Y., and Semenza, G. L. (1998) Gene Dev. 12 149-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu, A. Y., Shimoda, L. A., Iyer, N. V., Huso, D. L., Sun, X., McWilliams, R., Beaty, T., Sham, J. S., Wiener, C. M., Sylvester, J. T., and Semenza, G. L. (1999) J. Clin. Investig. 103 691-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asikainen, T. M., Chang, L. Y., Coalson, J. J., Schneider, B. K., Waleh, N. S., Ikegami, M., Shannon, J. M., Winter, V. T., Grubb, P., Clyman, R. I., Yoder, B. A., Crapo, J. D., and White, C. W. (2006) FASEB J. 20 1698-1700 [DOI] [PubMed] [Google Scholar]
- 15.Compernolle, V., Brusselmans, K., Acker, T., Hoet, P., Tjwa, M., Beck, H., Plaisance, S., Dor, Y., Keshet, E., Lupu, F., Nemery, B., Dewerchin, M., Van Veldhoven, P., Plate, K., Moons, L., Collen, D., and Carmeliet, P. (2002) Nat. Med. 8 702-710 [DOI] [PubMed] [Google Scholar]
- 16.Tomita, S., Ueno, M., Sakamoto, M., Kitahama, Y., Ueki, M., Maekawa, N., Sakamoto, H., Gassmann, M., Kageyama, R., Ueda, N., Gonzalez, F. J., and Takahama, Y. (2003) Mol. Cell Biol. 23 6739-6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan, H. E., Poloni, M., McNulty, W., Elson, D., Gassmann, M., Arbeit, J. M., and Johnson, R. S. (2000) Cancer Res. 60 4010-4015 [PubMed] [Google Scholar]
- 18.Perl, A. K., Wert, S. E., Nagy, A., Lobe, C. G., and Whitsett, J. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 10482-10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobe, C. G., Koop, K. E., Kreppner, W., Lomeli, H., Gertsenstein, M., and Nagy, A. (1999) Dev. Biol. 208 281-292 [DOI] [PubMed] [Google Scholar]
- 20.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 21.Vengellur, A., and LaPres, J. J. (2004) Toxicol. Sci. 82 638-646 [DOI] [PubMed] [Google Scholar]
- 22.Martis, P. C., Whitsett, J. A., Xu, Y., Perl, A. K. T., Wan, H. J., and Ikegami, M. (2006) Development 133 1155-1164 [DOI] [PubMed] [Google Scholar]
- 23.Maeda, Y., Dave, V., and Whitsett, J. A. (2007) Physiol. Rev. 87 219-244 [DOI] [PubMed] [Google Scholar]
- 24.Ema, M., Taya, S., Yokotani, N., Sogawa, K., Matsuda, Y., and FujiiKuriyama, Y. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 4273-4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimoda, L. A., Manalo, D. J., Sham, J. S. K., Semenza, G. L., and Sylvester, J. T. (2001) Am. J. Physiol.-Lung C 281 L202-L208 [DOI] [PubMed] [Google Scholar]
- 26.Semenza, G. L. (2005) Proc. Amer. Thor. Soc. 2 68-70 [DOI] [PubMed] [Google Scholar]
- 27.Dave, V., Childs, T., Xu, Y., Ikegami, M., Besnard, V., Maeda, Y., Wert, S. E., Neilson, J. R., Crabtree, G. R., and Whitsett, J. A. (2006) J. Clin. Investig. 116 2597-2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devriendt, K., Vanhole, C., Matthijs, G., and de Zegher, F. (1998) N. Engl. J. Med. 338 1317-1318 [DOI] [PubMed] [Google Scholar]
- 29.Liu, C., Morrisey, E. E., and Whitsett, J. A. (2002) Am. J. Physiol.-Lung C 283 L468-L475 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.