Abstract
We previously showed that a 9-nucleotide sequence from the 5′ leader of the Gtx homeodomain mRNA facilitates translation initiation by base pairing to 18S rRNA. These earlier studies tested the Gtx element in isolation; we now assess the physiological relevance of this element in the context of two natural mRNAs that contain this sequence in their 5′ leaders, Gtx itself and FGF2 (fibroblast growth factor 2). 2′-O-Methyl-modified RNA oligonucleotides were employed to block mRNA-rRNA base pairing by targeting either the Gtx-binding site in 18S rRNA or Gtx elements in recombinant mRNAs containing the Gtx or FGF2 5′ leaders linked to a reporter cistron. Studies in cell-free lysates and transfected COS-7 cells showed that translation of mRNAs containing the Gtx or FGF2 5′ leaders was decreased by >50% when oligonucleotides targeting either the rRNA or mRNA were used. Specificity was demonstrated by showing that translation of the recombinant mRNAs was unaffected by control oligonucleotides. In addition, the specific oligonucleotides did not affect the translation of recombinant mRNAs in which the Gtx elements were mutated. Experiments performed using constructs containing Gtx and FGF2 5′ leader and coding sequences ruled out possible effects of the reporter cistron. Furthermore, two-dimensional gel electrophoresis revealed that the oligonucleotides used in this study had little overall effect on the proteomes of cells transfected with these oligonucleotides. This study demonstrates that mRNA-rRNA base pairing affects the expression of two cellular mRNAs and describes a new approach for investigating putative mRNA-rRNA base pairing interactions in mammalian cells.
Numerous mRNAs contain complementary matches to 18S rRNA, and it has been suggested that such sequences may affect translation by base pairing to each other (1–3). In earlier studies, we noted that the 5′ leader of the Gtx homeodomain mRNA contains a 9-nucleotide sequence with complementarity to sequences contained within helix 26 of mouse 18S rRNA (4). Biochemical, functional, and genetic studies showed that this sequence element could mediate specific binding to 40S ribosomal subunits by base pairing to 18S rRNA and function as an internal ribosome entry site and as a translational enhancer element (TEE)2 and ribosomal shunt site (3, 5–8). We showed that this element could function in isolation and as a modular component of larger assemblages of elements, with additive or synergistic enhancements of activity depending on the nucleotide context and message configuration (3). These properties of the Gtx element facilitated our analysis of its mechanism because the ability of the individual element to affect translation is relatively weak. However, the relatively weak activity of a single Gtx element raised questions regarding its physiological relevance. It was therefore important to perform studies to determine whether an individual Gtx element affects translation efficiency in the context of natural mRNAs that contain this sequence. In this study, we investigate the physiological contribution of the Gtx element to the translation of two such mRNAs: Gtx and FGF2 (fibroblast growth factor 2).
The murine Gtx mRNA, also termed Nkx 6-2, is expressed in differentiated oligodendrocytes and testis germ cells (9, 10). The 5′ leader is 201 nucleotides in length, and the TEE is located 56 nucleotides upstream of the initiation codon. This mRNA encodes a homeobox protein that binds to DNA sequences within the promoters of the myelin basic protein and proteolipid protein genes (9) and is thought to have a role in regulating myelin gene transcription in oligodendrocytes. The 5′ leader of the human FGF2 mRNA also contains a sequence matching the Gtx-TEE (11, 12). Inasmuch as the sequence of the Gtx-binding site in rodent 18S rRNA is also found in humans, we chose the FGF2 mRNA for further analysis. The 5′ leader of this mRNA is 471 nucleotides in length and expresses five FGF2 isoforms from different initiation codons, including four noncanonical CUG codons and an AUG codon (13–15). The Gtx element is located at nucleotides 119–127, upstream of three CUG codons and the AUG codon. It is downstream of the first CUG codon, which is cap-dependent (16). In this mRNA, the complementary match to 18S rRNA (13 of 14 nucleotides) is even more extensive than in the Gtx mRNA. The various FGF2 isoforms differ in subcellular localization, function, and relative abundance in different cells and tissues (15, 17–23) and are involved in fundamental cellular processes, which include cell proliferation, differentiation, and angiogenesis (23).
This study investigates the extent to which the Gtx element contributes to the translation of the Gtx and FGF2 mRNAs by using antisense oligonucleotides to block either the Gtx-binding site in 18S rRNA or the Gtx element in either the Gtx or the FGF2 mRNAs. We expect that this antisense masking approach can also be used to identify other natural mRNAs that employ mRNA-rRNA base pairing, either via the Gtx element or through other binding sites in the 18S rRNA.
EXPERIMENTAL PROCEDURES
DNA Constructs—Monocistronic constructs included those containing the Gtx and FGF2 5′ leaders linked to the Photinus luciferase cistron, as well as those containing these 5′ leaders linked to the corresponding Gtx or FGF2 coding sequences. Luciferase reporter constructs were generated using the pGL3c (Promega) plasmid, which expresses the mRNA via the SV40 promoter. The Gtx and FGF2 5′ leader sequences were synthesized by PCR amplification of oligonucleotide templates and cloned upstream of the Photinus luciferase cistron. Briefly, constructs expressing the Gtx 5′ leader and either the Gtx or luciferase coding sequences were cloned using AatII and NcoI restriction sites. The AatII site was introduced at the transcription start site to avoid inclusion of vector-derived sequence in the recombinant transcripts; the NcoI site contains the initiation codon. The Gtx coding sequences were obtained by PCR amplification of mouse genomic DNA. The PCR-generated fragment was digested with NcoI and XbaI, which was introduced through the PCR primer, and cloned into the vector containing the Gtx 5′ leader, as described above. Constructs containing the FGF2 5′ leader and luciferase or FGF2 coding sequences were similarly constructed except that the FGF2 coding sequences were synthesized by PCR amplification of oligonucleotide templates. Mutations in the Gtx element were introduced into the Gtx 5′ leader using annealed oligonucleotides that generated a 43-nt fragment with ApaI/BssHII sticky ends. For FGF2, annealed oligonucleotides generated a 38-nt fragment with SacII sticky ends. In both cases, the annealed oligonucleotides were used to replace the corresponding fragments from the 5′ leader.
Oligonucleotides—2′-O-Methyl-modified RNA oligonucleotides were obtained from Thermo Scientific (Dharmacon RNAi Technologies). Oligonucleotide sequences are as follows, with the noncomplementary nucleotides in the Gtx-m1 and α-Gtx-m1 oligonucleotides underlined: Gtx, 5′ CCGGCGGGU 3′; Gtx-m1, 5′ CCGGAGGGU 3′; α-Gtx, 5′ ACCCGCCGG 3′; α-Gtx-m1, 5′ ACCCACCGG 3′; and Gtx-scrambled, 5′ CGGCGCUGG 3′.
In Vitro Translation—Capped mRNAs were generated from plasmids linearized using ClaI, which is located downstream of the luciferase cistron, and were transcribed by using the mMessage mMachine (Ambion). RNAs were quantified by UV absorption at 260 nm. In vitro translation reactions were performed in rabbit reticulocyte lysate as specified by the manufacturer (Promega) with the following modification: translation reactions were performed in 10 μl and incubated for 15 min at 37 °C instead of the proposed 50 μl reactions incubated at 37 °C for 90 min. These conditions were determined to be in the linear phase of the translation reaction.
Transfection—FuGENE 6 (Roche Diagnostics) was used to transfect 1–3 × 105 COS-7 cells with 0.5 μg of reporter construct according to the manufacturer's instructions. Cells were cotransfected either with a control construct p(SI/SIII)5βP (6), which expresses Photinus luciferase, or pCMVβ (Clontech), which expresses the lacZ gene. Neither of the control mRNAs contain sequences similar to the Gtx element. Transfected cells were harvested 18–24 h after transfection, and luciferase and β-galactosidase activities were determined as described previously (6). 2′-O-Methyl-modified RNA oligonucleotides were cotransfected with plasmids or were transfected alone using the abovementioned conditions at concentrations indicated in the figures.
Analyses of Reporter Gene Activity and mRNA Levels—Luciferase activities were determined as described previously (6). Recombinant mRNA levels were determined by using ribonuclease protection assays (RPAIII kit; Ambion) with 1 μg of DNase-treated total RNA. Protected fragments were size-fractionated on 6% polyacrylamide-urea gels, visualized on a Storm 860 PhosphorImager (GE Healthcare), and quantified by using ALPHAEASEFC stand-alone software (Alpha Innotech, San Leandro, CA).
Western Immunoblotting—Cell lysates were prepared 18–24 h after transfection; cell monolayers were scraped, resuspended in lysis buffer (RIPA Buffer: 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, and 1× complete mix of protease inhibitor), and sonicated. Total protein in cell lysates was quantified using the Bio-Rad DC protein assay kit. For Western blots, samples containing 25 μg of protein were heated for 10 min at 72 °C in SDS and dithiothreitol-containing sample buffer and separated in a 4–12% gradient BisTris polyacrylamide gel (Invitrogen). Proteins were transferred to polyvinylidene difluoride membranes and probed with goat anti-Gtx or anti-FGF2 polyclonal IgG 1° antibodies (Santa Cruz Biotechnology) and visualized using the CDP Star chemiluminescent Western blot immunodetection kit (Roche Applied Science) with donkey anti-goat IgG 2° antibody conjugated to alkaline phosphatase (Promega). For control experiments, an anti-β-actin monoclonal mouse IgG 1° antibody (Sigma) and an anti-mouse IgG 2° antibody (Sigma) conjugated to alkaline phosphatase were used to detect the β-actin protein.
Two-dimensional Gel Electrophoresis—Cell lysates were prepared 18–24 h after transfection by scraping the cell monolayers, resuspending in lysis buffer (50 mm Tris-HCl, pH 7.4, 20 mm NaCl, and 1% IGEPAL detergent), and sonicating. Total protein in cell lysates was quantified as above. Protein samples (50 μg per gel) were diluted to 125 μl in rehydration buffer (8 m urea, 2% CHAPS, 50 mm dithiothreitol, 0.2% Bio-Lyte ampholytes, and 0.002% bromphenol blue). The indicated volume of each sample was applied to a gel strip (17 cm, pH 3–10, Bio-Rad), rehydrated for 16–20 h, and focused using a PROTEAN isoelectric focusing cell and in three steps as follows: 1) 250 V, 1 h linear; 2) 1000 V, 2 h linear; and 3) 10,000 V, 45,000 V-h rapid. Prior to running the second dimension, gel strips were either stored at –70 °C or equilibrated in 2 ml of equilibration buffer 1 (6 m urea, 0.375 m Tris-HCl, pH 8.8, 2% SDS, 20% glycerol, 2% w/v dithiothreitol) for 10 min. Isoelectric focusing strips were then equilibrated in equilibration buffer 2 (6 m urea, 0.375 m Tris-HCl, pH 8.8, 2% SDS, 20% glycerol, 2.5% w/v iodoacetamide) for 10 min and run on 12% acrylamide SDS-polyacrylamide gels.
Protein spots were stained using the Flamingo fluorescent gel stain (Bio-Rad), visualized at 532 nm using the Typhoon Trio Variable Mode Imager (GE Healthcare), and analyzed using the Progenesis SameSpots computer software (Nonlinear Dynamics, Durham, NC). Protein spots were reduced in-gel using 10 mm dithiothreitol, alkylated with 55 mm iodoacetamide, and digested with trypsin in an estimated 1:30 enzyme to substrate ratio. The samples were then analyzed using the Agilent MSD 1100 nano-electrospray LC/MS/MS. MS/MS analysis was performed on the LTQ linear ion trap mass spectrometer (Thermo) using 2 kV at the tip. One MS spectrum was followed by four MS/MS scans on the most abundant ions after the application of the dynamic exclusion list. Tandem mass spectra were subjected to peak picking using Xcalibur (version 2.0 SR2, Thermo Electron Corp.). Protein identification was performed using Mascot (version 2.1.04, Matrix Science, London, UK).
Proteins were identified using a Mascot search (version 2.1.04, Matrix Science, London, UK) at a 95% confidence level using a data base of tryptic peptides derived from the NCBI annotation of primate genomes (June 6, 2008). Scores were adjusted to give a false-positive rate of <1% based on a decoy data base search. An ion score threshold of 35 gave an acceptable and low false-positive rate of 0.398 ± 0.57% using a dataset of 20 searches. All searches were done using this threshold, which included single peptide assignments that were included in the analysis.
RESULTS
Oligonucleotide Masking of Gtx-binding Site in 18S rRNA Blocks Activity of Isolated Gtx Elements—The feasibility of using oligonucleotides to block the base pairing interaction between the Gtx TEE and 18S rRNA was tested using synthetic mRNA constructs containing five linked copies of the Gtx TEE in the 5′ leader of the Photinus luciferase mRNA. Comparable constructs were used in our earlier work to study this element in isolation (3, 6–8). Constructs were tested both in rabbit reticulocyte lysate using mRNAs transcribed with T7 RNA polymerase and in transfected cells, with transcription driven by the SV40 promoter.
For these studies, we used 2′-O-methyl-RNA oligonucleotides, which contain a methyl group at the 2′-OH residue of the ribose molecule. These modified oligonucleotides base pair with RNA more stably than DNA or unmodified RNA oligonucleotides; they are also more resistant to nuclease degradation than unmodified RNA oligonucleotides and do not target the hybridized region of the target RNA to digestion by RNase H, which is a potential limitation associated with DNA oligonucleotides (24–26).
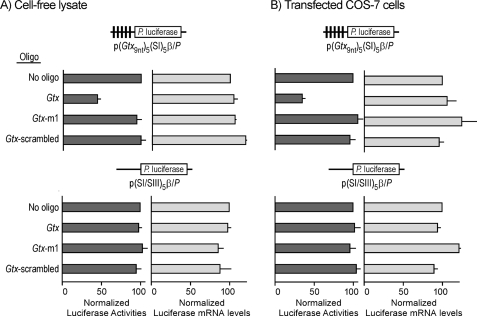
The results showed that increasing concentrations of the Gtx oligonucleotide, which targets the Gtx-binding site in 18S rRNA (nucleotides 1124–1132), led to progressively reduced luciferase activities from a construct containing five Gtx TEEs (p(Gtx9nt)5(SI)5β/P) either in an in vitro translation assay or in COS-7 transfected cells (supplemental Fig. 1A). Construct pGtx/P containing the full-length Gtx 5′ leader exhibited a similar trend both in vitro and in transfected COS-7 cells (supplemental Fig. 1B). Maximal inhibition was obtained using 1 μm both in the cell-free lysate and in cells. Cell-free studies were performed using in vitro transcribed Gtx reporter mRNAs in the absence or presence of the Gtx, scrambled, and Gtx-m1 oligonucleotides. Fig. 1A shows results obtained in the cell-free lysate with the reporter construct containing five Gtx TEEs (p(Gtx9nt)5(SI)5β/P) and 1 μm of the Gtx oligonucleotide. Luciferase expression was reduced by ≈50% compared with samples containing the Gtx reporter construct alone (no oligonucleotide). Control experiments using a scrambled oligonucleotide (Gtx-scrambled) and an oligonucleotide with a point mutation in the Gtx sequence (Gtx-m1) had no effect on luciferase expression, suggesting that the Gtx oligonucleotide specifically blocked the activity of the Gtx elements. Expression of a control construct (p(SI/SIII)5β/P) lacking Gtx elements was unaffected by any of the oligonucleotides, suggesting that the Gtx oligonucleotide affected translation of the Gtx reporter mRNA by blocking the activity of Gtx TEEs. These results are consistent with direct inhibition by the Gtx oligonucleotide of mRNA-rRNA base pairing. RNA analyses indicated that the decreased luciferase levels observed in samples incubated with the Gtx oligonucleotide were not because of decreased reporter mRNA levels but to a decrease in translation efficiency.
FIGURE 1.
Oligonucleotide masking of Gtx-binding site in 18S rRNA blocks activity of isolated Gtx elements in rabbit reticulocyte cell-free lysate (A) and transiently transfected COS-7 cells (B). The constructs used in this study are indicated schematically. Vertical black bars in the 5′ leader of the p(Gtx9nt)5(SI)5β/P mRNA represent 9-nt Gtx elements. Control construct p(SI/SIII)5β/P is a size-matched control that lacks Gtx elements. The absence or presence of specific oligonucleotides (oligo) is indicated. A and B, oligonucleotide concentration is 1 μm, and luciferase activities or luciferase mRNA levels for each construct are normalized to 100 for the activity of the no oligonucleotide control. The data points represent the results of three independent experiments; the error bars indicate S.E.
Fig. 1B shows that comparable results were obtained when COS-7 cells were cotransfected with the reporter construct containing five Gtx TEEs and 1 μm of the Gtx oligonucleotide. Luciferase expression was reduced by ≈60% compared with cells transfected with the reporter construct alone (no oligonucleotide). The control oligonucleotides did not affect luciferase expression. In addition, control construct p(SI/SIII)5β/P was unaffected by any of the oligonucleotides. RNA analysis indicated that decreased luciferase levels in cells cotransfected with the Gtx oligonucleotide were not because of decreased reporter mRNA levels but to a decrease in translation efficiency.
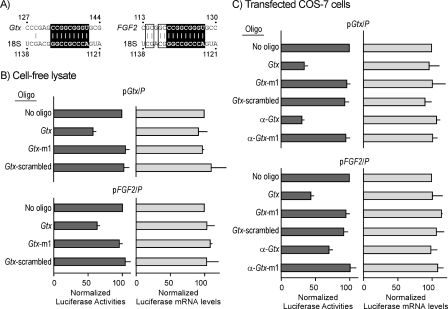
Antisense Masking Establishes Contribution of Gtx Elements in Gtx and FGF2 5′ Leaders—To begin to assess the contribution of an individual Gtx element in the context of a natural mRNA, we investigated the Gtx 5′ leader, which was the source of this element. In addition, we investigated the FGF2 5′ leader, which contains the nucleotide sequence of the Gtx element as well as a more extended complementary match to 18S rRNA (Fig. 2A). The contributions of these individual Gtx elements were initially tested using Photinus luciferase reporter constructs as follows: pGtx/P and FGF2/P, containing the full-length Gtx 5′ leader (201 nt), or FGF2 5′ leader (471 nt). The contribution of the Gtx element to the translation mediated by these two 5′ leaders was assessed using the Gtx oligonucleotide to block the Gtx-binding site in 18S rRNA in both a rabbit reticulocyte cell-free lysate and transfected COS-7 cells. The results showed that this oligonucleotide blocked expression mediated by the Gtx 5′ leader by ≈45% in the cell-free lysate (Fig. 2B) and ≈65% in transfected cells (Fig. 2C). In contrast, expression remained unaffected in both the cell-free lysate and transfected cells upon exposure to the mutated Gtx-m1 oligonucleotide. In addition, in both systems, expression from a control RNA lacking Gtx elements was unaffected by the Gtx oligonucleotide. The control Gtx-m1 and Gtx-scrambled oligonucleotides had no effect on the expression of any of these constructs. Experiments performed using constructs containing full-length FGF2 5′ leader yielded similar results, with the Gtx oligonucleotide blocking expression by ≈40% in the cell-free lysate (Fig. 2B) and 55% in the transfected cells (Fig. 2C). Furthermore, expression of both pGtx/P and FGF2/P constructs was blocked by approximately the same amount by an oligonucleotide targeting the Gtx element in the mRNA (α-Gtx). In contrast, expression was unaffected by a mutated α-Gtx oligonucleotide (α-Gtx-m1; Fig. 2C). The control construct was unaffected by any of the oligonucleotides in both systems (data not shown). Ribonuclease protection assays showed that the reporter mRNA levels were unaffected in cells transfected with the various oligonucleotides, suggesting that the effects observed were not due to altered mRNA levels (Fig. 2, B and C).
FIGURE 2.
Oligonucleotide masking of rRNA or mRNA sequences blocks translation of luciferase reporter mRNAs containing full-length Gtx and FGF2 5′ leaders. A, complementary matches between nucleotides in mouse 18S rRNA and Gtx 5′ leader or human 18S rRNA and FGF2 5′ leader. B, expression in rabbit reticulocyte lysate. C, transiently transfected COS-7 cells. Constructs pGtx/P and pFGF2/P contain the Gtx and FGF2 5′ leaders, respectively, upstream of the Photinus luciferase cistron. The absence or presence of oligonucleotides (oligo)(1 μm) is indicated. For each construct, luciferase activities or luciferase mRNA levels for each construct are normalized to 100 for the activity of the no oligonucleotide control. The data points represent the results of three independent experiments; the error bars indicate S.E.
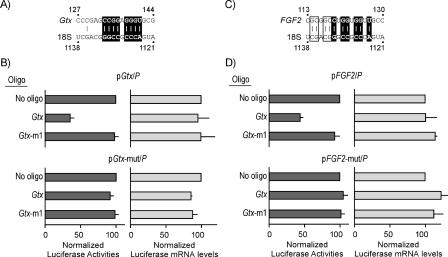
To further evaluate the specificity of the results, Gtx elements in the two 5′ leaders were disrupted by point mutations within the Gtx element (Fig. 3, A and C). The point mutation in the Gtx 5′ leader (in pGtx-mut/P) was known from earlier studies to block the activity of the Gtx-TEE (8). In these studies, this mutation reduced luciferase expression by ≈40% in transfected cells. For the FGF2 5′ leader, we introduced three point mutations to disrupt possible base pairing interactions to 18S rRNA because this 5′ leader contains a more extensive complementary match. These point mutations in the FGF2 5′ leader (in pFGF2-mut/P) reduced expression by 25%. The Gtx oligonucleotide did not affect expression from the mutated Gtx construct, similar to the controls (No oligonucleotide and Gtx-m1; Fig. 3B). Likewise, the Gtx oligonucleotide did not affect expression from the mutated FGF2 construct (Fig. 3D). Ribonuclease protection assays showed that the reporter mRNA levels were relatively unaffected in cells transfected with the various oligonucleotides, suggesting that the effects observed were not due to altered mRNA levels (Fig. 3, B and D).
FIGURE 3.
Mutation of Gtx TEE in Gtx or FGF2 5′ leaders prevents inhibition by antisense masking. Constructs pGtx/P and pFGF2/P contain the Gtx or FGF2 5′ leaders, respectively, linked to Photinus luciferase. Constructs pGtx-mut/P and pFGF2-mut/P are identical except for point mutations in the Gtx elements, which are shown in A for the Gtx 5′ leader and in B for the FGF2 5′ leader. Constructs were expressed in transiently transfected COS-7 cells. The results obtained with the Gtx constructs are shown in C and those with the FGF2 5′ leader in D. The absence or presence of oligonucleotides (oligo)(1 μm) is indicated. Luciferase activities or luciferase mRNA levels for each construct are normalized to 100 for the no oligonucleotide control. The data points represent the results of three independent experiments; the error bars indicate S.E.
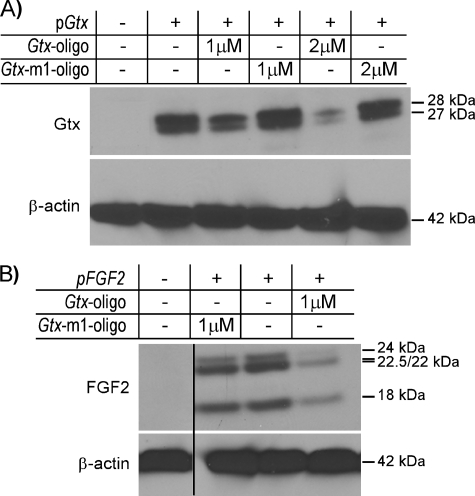
Evaluating Effects of Coding Sequences—To rule out possible nonspecific effects of the luciferase reporter cistron, we tested constructs containing both the Gtx and FGF2 5′ leader and coding sequences in COS-7 cells and evaluated expression by immunoblot analyses using anti-Gtx and anti-FGF2 antibodies, respectively. The results showed that each of these mRNAs produced several protein bands (Fig. 4, A and B). The two Gtx bands correspond to the full-length protein and a shorter product initiating from an in-frame AUG codon in the coding region. The FGF2 bands correspond to initiation from the initiation codon and three upstream CUG codons. In cells cotransfected with 1 μm of the Gtx oligonucleotide, levels of the Gtx and FGF2 proteins decreased. For Gtx, this decrease was even greater in the presence of 2 μm oligonucleotide. In contrast, the levels of these proteins were similar to those in control cells when cells were exposed to the mutated (Gtx-m1) oligonucleotide. Immunoblot analyses using an anti-β-actin antibody showed that the levels of endogenous β-actin were unaffected by the Gtx oligonucleotide, further suggesting that the effects of this oligonucleotide were specific.
FIGURE 4.
Oligonucleotide masking of Gtx-binding site in 18S rRNA blocks activity of recombinant mRNAs. Plasmids containing Gtx (pGtx)(A) or FGF2 (pFGF2)(B)5′ leader and coding sequences were transiently transfected into COS-7 cells and cell lysates analyzed by Western blot analysis using anti-Gtx, anti-FGF2, or anti-β-actin antibodies as indicated. Histogram above Western blots indicates plasmids and oligonucleotides (oligo) in the various samples. Molecular weights of Gtx, FGF2, and β-actin protein bands are indicated to the right. This experiment was repeated with similar results three times.
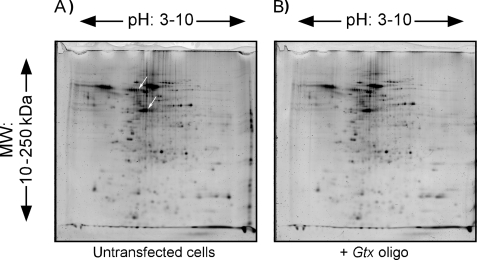
Assessing the Effects of Gtx and α-Gtx Oligonucleotides on the Proteome—The specificity of the Gtx and α-Gtx oligonucleotides was further evaluated by looking at their effects on the proteome. These effects were monitored by two-dimensional gel electrophoresis. COS-7 cells were transiently transfected with the Gtx oligonucleotide. Cells were lysed, and proteins in these lysates were compared with those from untransfected COS-7 cells using two-dimensional gel electrophoresis (Fig. 5). Comparison of the gels showed that the overall pattern of proteins, as well as the relative levels of proteins, was highly similar. The SameSpots computer software was used to match and quantify protein spots between gels. When the gel containing proteins from untransfected COS-7 cells (Fig. 5A) was compared with the gel with protein from COS-7 cells transfected with the Gtx oligonucleotide (Fig. 5B), 12 of 196 spots showed a significant change in expression. In addition, when two-dimensional gels containing proteins from untransfected COS-7 cells were compared with those from cells transiently transfected with the Gtx-m1 and α-Gtx oligonucleotides (not shown), we found that five of these spots changed only with the Gtx and α-Gtx oligonucleotides, suggesting that the expression of these five proteins may involve base pairing interactions mediated by Gtx elements. The intensity (volume) of these spots was inhibited by 30–55% compared with the same spots from untransfected cells. These spots were picked from the two-dimensional gels, digested, and analyzed using a nano-LC/MS/MS method. We were able to identify proteins from two spots with >95% confidence as follows: vimentin (gi/75075845), an intermediate filament protein, and stomatin (gi/7305503), a component of lipid rafts. The 5′ leaders of the mRNAs encoding these proteins were analyzed for the presence of a Gtx element or elements. This analysis revealed that both the 186-nt vimentin 5′ leader and the 63-nt stomatin 5′ leader contain 7- and 6-nt contiguous matches to the Gtx element, respectively (supplemental Fig. 2). Our earlier studies showed that comparable matches were functional and could be as active or more active than the 9-nt element (6, 8).
FIGURE 5.
Two-dimensional gel electrophoresis of COS-7 proteins in the absence or presence of masking or control oligonucleotides. Proteins were electrophoresed from COS-7 cells that were either untransfected (A) or transfected (B) with 1 μm Gtx oligonucleotide (oligo). The arrows in A are spots that were subjected to mass spectrometry and correspond to vimentin (upper arrow) and stomatin (lower arrow).
DISCUSSION
Base pairing between mRNA and rRNA underlies the well established Shine-Dalgarno interaction in bacteria (27, 28). Our earlier analysis of the Gtx TEE indicated that similar base pairing interactions were not limited to bacteria but could also occur in eukaryotes (6, 8). These earlier studies required testing the Gtx TEE as an isolated element, i.e. outside of the Gtx 5′ leader from which it was identified. This study now provides the first evidence that mRNA-rRNA base pairing can affect the translation of some mammalian mRNAs.
In this study, we showed that oligonucleotides targeting either the Gtx-binding site in 18S rRNA or the Gtx TEE in the Gtx and FGF2 5′ leaders reduced translation from mRNAs containing these 5′ leaders. In addition, mRNAs in which the Gtx TEEs were mutated were translated less efficiently, suggesting that the Gtx TEE enhances translation in the Gtx and FGF2 mRNAs. Moreover, the mutated mRNAs were no longer affected by the Gtx oligonucleotide, indicating that the inhibitory effect of this oligonucleotide involved blocking the activity of the TEE.
Antisense masking of the binding site in 18S rRNA enabled us to assess the contribution of the Gtx TEE in mRNAs without mutating, deleting, or otherwise modifying the mRNAs. This is an important advantage of the approach because any of these manipulations may themselves have nonspecific effects on expression, e.g. by affecting RNA conformation or altering the accessibility of other mRNA elements, including initiation codons (7). Such effects may underlie the results of a study in which deletion of a segment of the FGF2 5′ leader containing the Gtx TEE led to significantly decreased translation (11), which is consistent with our findings, but various other mutations of the Gtx TEE or flanking nucleotides yielded inconsistent results.
A limitation of the antisense masking approach is that it cannot distinguish between an mRNA element that binds to 18S rRNA or to another RNA. This is not a serious concern for the Gtx TEE because there is both biochemical as well as genetic evidence indicating that this element binds specifically to helix 26 of 18S rRNA and not to other factors (4, 6, 8). However, for a different mRNA element, additional evidence would be required to establish that the element is base pairing to rRNA. Ideally, one would perform experiments involving mutation of both mRNA and rRNA sequences to assess the requirement for an intact complementary match. Such a genetic approach was used to confirm the Shine-Dalgarno interaction in Escherichia coli (28). We used a similar approach in yeast to establish the Gtx interaction (8). Although a suitable system for performing comparable studies in mammalian cells does not yet exist, we have undertaken studies in our laboratory to develop such a system.
The oligonucleotides used in our studies did not appear to have global effects on the proteomes of cells transfected with them, as monitored by two-dimensional gel electrophoresis. However, the levels of a small number of proteins did change specifically with the Gtx and α-Gtx oligonucleotides, suggesting that the translation of mRNAs encoding these proteins is affected by base pairing to the Gtx-binding site in 18S rRNA. Indeed, mass spectrometry of two proteins showing such a change revealed that the corresponding mRNAs contain matches to the Gtx element in their 5′ leaders, which provides a plausible explanation for the effects of Gtx and α-Gtx oligonucleotides. The changes observed in the levels of proteins affected by both the Gtx and α-Gtx oligonucleotides in COS-7 cells is consistent with the total number of mRNAs containing sequences complementary to the Gtx-binding site in 18S rRNA (<1%).
The observation that numerous eukaryotic mRNAs contain sequences with complementarity to different sites in the 18S rRNA (e.g. see Refs. 1, 2, 29, 30) raises the possibility that the translation of a reasonably large number of eukaryotic mRNAs may be affected by base pairing to ribosomes. In contrast to the Shine-Dalgarno sequence in prokaryotes, which is located upstream of and in close proximity to the initiation codon, sequences with complementarity to rRNA in eukaryotic mRNAs are not restricted to the region located immediately upstream of the initiation codon but are found in all parts of mRNAs. Likewise, putative binding sites in 18S rRNA are not restricted to the 3′ end but are found throughout the molecule.
This study provides the first evidence indicating that the translation efficiency of some natural mRNAs is affected by base pairing to the Gtx-binding site in 18S rRNA. In addition, this study provides an experimental approach for investigating putative mRNA-rRNA base pairing interactions in mammalian cells via the Gtx-binding site in 18S rRNA as well as through other possible binding sites.
Supplementary Material
Acknowledgments
We thank Luke Burman for excellent technical assistance and Dr. Gerald M. Edelman for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM078071 (to V. P. M.). This work was also supported by the Skaggs Institute for Chemical Biology (to P. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: TEE, translational enhancer element; nt, nucleotide; LC/MS/MS, liquid chromatography/tandem mass spectrometry; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
References
- 1.Scheper, G. C., Voorma, H. O., and Thomas, A. A. M. (1994) FEBS Lett. 352 271–275 [DOI] [PubMed] [Google Scholar]
- 2.Mauro, V. P., and Edelman, G. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chappell, S. A., Edelman, G. M., and Mauro, V. P. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu, M. C.-Y., Tranque, P., Edelman, G. M., and Mauro, V. P. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chappell, S. A., Dresios, J., Edelman, G. M., and Mauro, V. P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 9488–9493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappell, S. A., Edelman, G. M., and Mauro, V. P. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9590–9594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chappell, S. A., Edelman, G. M., and Mauro, V. P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 18077–18082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dresios, J., Chappell, S. A., Zhou, W., and Mauro, V. P. (2006) Nat. Struct. Mol. Biol. 13 30–34 [DOI] [PubMed] [Google Scholar]
- 9.Awatramani, R., Scherer, S., Grinspan, J., Collarini, E., Skoff, R., O'Hagan, D., Garbern, J., and Kamholz, J. (1997) J. Neurosci. 17 6657–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., Volkert, T. L., Wilson, C. J., Bell, S. P., and Young, R. A. (2000) Science 290 2306–2309 [DOI] [PubMed] [Google Scholar]
- 11.Bonnal, S., Schaeffer, C., Creancier, L., Clamens, S., Moine, H., Prats, A. C., and Vagner, S. (2003) J. Biol. Chem. 278 39330–39336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vagner, S., Gensac, M. C., Maret, A., Almaric, F., Prats, H., and Prats, A. C. (1995) Mol. Cell. Biol. 15 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florkiewicz, R. Z., and Sommer, A. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 3978–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prats, H., Kaghad, M., Prats, A. C., Klagsbrun, M., Lelias, J. M., Liauzun, P., Chalon, P., Tauber, J. P., Amalric, F., Smith, J. A., and Caput, D. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 1836–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnaud, E., Touriol, C., Boutonnet, C., Gensac, M. C., Vagner, S., Prats, H., and Prats, A. C. (1999) Mol. Cell. Biol. 19 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu, P. J., Ferrari, G., Galloway, A. C., Mignatti, P., and Pintucci, G. (2007) J. Cell. Biochem. 100 1100–1108 [DOI] [PubMed] [Google Scholar]
- 17.Bugler, B., Amalric, F., and Prats, H. (1991) Mol. Cell. Biol. 11 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couderc, B., Prats, H., Bayard, F., and Amalric, F. (1991) Cell Regul. 2 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quarto, N., Finger, F. P., and Rifkin, D. B. (1991) J. Cell. Physiol. 147 311–318 [DOI] [PubMed] [Google Scholar]
- 20.Bikfalvi, A., Klein, S., Pintucci, G., Quarto, N., Mignatti, P., and Rifkin, D. B. (1995) J. Cell Biol. 129 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coffin, J. D., Florkiewicz, R. Z., Neumann, J., Mort-Hopkins, T., Dorn, G. W., II, Lightfoot, P., German, R., Howles, P. N., Kier, A., and O'Toole, B. A. (1995) Mol. Biol. Cell 6 1861–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vagner, S., Touriol, C., Galy, B., Audigier, S., Gensac, M. C., Amalric, F., Bayard, F., Prats, H., and Prats, A. C. (1996) J. Cell Biol. 135 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bikfalvi, A., Klein, S., Pintucci, G., and Rifkin, D. B. (1997) Endocr. Rev. 18 26–45 [DOI] [PubMed] [Google Scholar]
- 24.Hogrefe, H. H., Hogrefe, R. I., Walder, R. Y., and Walder, J. A. (1990) J. Biol. Chem. 265 5561–5566 [PubMed] [Google Scholar]
- 25.Cummins, L. L., Owens, S. R., Risen, L. M., Lesnik, E. A., Freier, S. M., McGee, D., Guinosso, C. J., and Cook, P. D. (1995) Nucleic Acids Res. 23 2019–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein, D., Foster, E., Huang, S. B., Weller, D., and Summerton, J. (1997) Antisense Nucleic Acid Drug Dev. 7 151–157 [DOI] [PubMed] [Google Scholar]
- 27.Shine, J., and Dalgarno, L. (1974) Proc. Natl. Acad. Sci. U. S. A. 71 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hui, A., and De Boer, H. A. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 4762–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matveeva, O. V., and Shabalina, S. A. (1993) Nucleic Acids Res. 21 1007–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mignone, F., and Pesole, G. (2002) Appl. Bioinformatics 1 145–154 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.