Abstract
To test the hypothesis that factor Xa (fXa) interacts with protein S, fXa was labeled active-site specifically with a dansyl (D) dye via a Glu-Gly-Arg (EGR) tether to yield DEGR-fXai. When protein S was added to phosphatidylcholine/phosphatidylserine (PC/PS, 4:1) vesicle-bound DEGR-fXai, the anisotropy of the dansyl moiety was altered from 0.219 ± 0.002 to 0.245 ± 0.003. This change in dansyl anisotropy was not observed when DEGR-Xai was titrated with protein S in the absence of PC/PS vesicles, or in the presence of 100% PC vesicles, or when PC/PS vesicle-bound DEGR-fXai was titrated with thrombin-cleaved protein S. The protein S-dependent dansyl fluorescence change was specific for fXa because it was not observed for two homologous and similarly labeled DEGR-fIXai and DEGR-fVIIai. Furthermore, protein S specifically and saturably altered the fluorescence anisotropy of PC/PS-bound active site-labeled LWB-FPR-fXai (Kd = 33 nm) and was photocross-linked to PC/PS-bound LWB-FPR-fXai analog, independently confirming the above results. Chemically synthesized microprotein S, comprising residues 1–116 of protein S and including the γ-carboxyglutamic-rich domain, the thrombin-sensitive region (TSR), and the first epidermal growth factor-like domain (EGF1) of protein S, altered the anisotropy of PC/PS-bound DEGR-fXai from 0.219 to 0.242, similar to the effect of the protein S titration (Kd = 303 nm), suggesting that microprotein S binds to DEGR-fXai. To identify individual protein S domain(s) that binds DEGR-fXai, the EGF1 and TSR domains were chemically synthesized and studied. The TSR altered the anisotropy of DEGR-fXai by ∼16% (Kd = 3.9 μm), but the EGF1 domain had no effect on the signal. In controls, the TSR domain did not alter the anisotropy of DEGR-fIXai and DEGR-fVIIai, respectively. These data demonstrate that membrane-bound fXa binding to protein S involves the TSR of protein S.
Protein S is known as a non-enzymatic cofactor of activated protein C in the inactivation of factors Va (fVa)2 and VIIIa, as part of a negative feedback loop to regulate coagulation (1). Plasma coagulation assays in the absence of activated protein C suggest that protein S may have other anticoagulant role(s) in the absence of activated protein C (2, 3). Consistent with this notion, other anticoagulant functions have been ascribed to protein S that are independent of activated protein C and that we will term here as “protein S direct.” For example, it has been proposed that protein S can inhibit the generation of thrombin by the prothrombinase (fXa·fVa ·phospholipid) complex (4) by forming a complex with fXa and fVa. Recently, it has been suggested that protein S down-regulates thrombin generation by stimulating fXa inhibition by the tissue factor pathway inhibitor (5, 6). The molecular interactions responsible for these effects are as yet not understood.
Protein S is a multi-modular vitamin K-dependent plasma protein of 635 amino acids (1). The N-terminal end of the mature protein is rich in γ-carboxyglutamic acid (Gla) residues and is termed the Gla domain. The Gla module mediates the interaction of protein S with negatively charged phospholipids in the presence of calcium ions. The Gla domain is followed by four epidermal growth factor-like (EGF) repeats and a C-terminal structure that resembles the sex hormone binding globulin. Between the Gla domain and the first EGF (EGF1) repeat is a loop that is sensitive to proteolysis and is commonly referred to as the TSR. Thrombin cleaves at two sites in this loop, at Arg49 and Arg70, releasing a 21-amino acid peptide. This leaves the Gla domain of protein S attached to the rest of the protein through a disulfide linkage. Thrombin cleavage of protein S results in loss of activated protein C cofactor activity of protein S. Additionally, fXa also cleaves at the TSR of protein S at Arg60, a site distinct from the cleavages by thrombin (7). This fXa-cleaved protein S no longer possesses activated protein C-dependent or direct anticoagulant activities. FXa-cleavage of protein S requires the presence of phospholipids and Ca2+ ions.
Because the proposed mechanisms of protein S directly involve fXa, we tested the hypothesis that specific domains in protein S interact with fXa using fluorescence spectroscopy. Furthermore, the formation of a fXa ·protein S complex on a PC/PS vesicle surface was also demonstrated by photocross-linking technique. To determine which domain(s) of protein S interact with fXa, various domains of protein S were synthesized using solid-phase peptide synthesis strategies. These domains were tested for binding to fluorescent fXa. The results of our investigations are presented.
EXPERIMENTAL PROCEDURES
Proteins and Reagents—Protein S was obtained from Enzyme Research Laboratory (South Bend, IN) for some assays. For some assays, protein S was purified from human plasma according to published protocols (4). Protein S preparations ranged from 50–90% activated protein C cofactor activity compared with normal pooled plasma in a Staclot protein S calibrator. Thrombin cleaved protein S was prepared as described previously (8). fXa was activated from fX using Russells viper venom-X. For some assays fXa was obtained from Hematologic Technologies (Essex Junction, VT). 5-dimethylaminonaphthalene-1-sulfonyl-glutamyl-glycyl-arginyl chloromethylketone was purchased from Calbiochem. Nα-fluorescein-p-benzoylphenylalanyllysyl(Nε bromoacetyl) amide (LWB) was synthesized as described previously (9). fXa was active site-specifically labeled with LWB via a Phe-Pro-Arg tether and purified as described (9). Dioleoylphosphatidylcholine and dioleoylphosphatidylserine were obtained from Avanti Polar Lipids (Alabaster, AL). 1, 2-Di[1-14C]oleoyl-l-3-phosphatidylcholine was purchased from Amersham Biosciences. Chromogenic substrate N-benzoylisoleucylglutamylglycylarginyl-p-nitroaniline (S2222) was obtained from Chromogenix (Milano, Italy).
5-Dimethylaminonaphthalene-1-sulfonyl-(1,5, dansyl-) Glu-Gly-Arg factor Xa (DEGR-fXai) and 1,5, dansyl-Glu-Gly-Arg factor IXa (DEGR-fIXai) were obtained from Hematologic Technologies Inc. For some experiments, DEGR-fXai was prepared as described previously (10). DEGR-fVIIai was obtained as a gift from Dr. Wolfram Ruf, Department of Immunology, The Scripps Research Institute, La Jolla, CA. Protein concentrations were determined using the molecular masses and ε1%1 cm at 280 nm of 45,000 and 14.5 for fXa, respectively. Protein S concentrations were determined by a enzyme-linked immunosorbent assay kit for protein S, Asserachrom protein S, from Diagnostica Stago (Asnieres, France) and using a molecular mass of 75,000.
Phospholipid Vesicles—Unilamellar vesicles of phosphatidylcholine/phosphatidylserine (PC/PS; 4:1 molar ratio) and 100% PC were prepared by sonication and ultracentrifugation according to procedures described previously (11). A [14C]PC tracer was added to each preparation to facilitate determination of phospholipid recovery post-centrifugation.
Synthetic Domains of Protein S—Domains of protein S were synthesized on a 0.4-mmol scale by manual solid-phase peptide synthesis using in situ neutralization/O-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate activation procedure of t-butoxycarbonyl chemistry (12). Syntheses of EGF1, TSR, and microprotein S were done as described (8, 13).
Spectral Measurements—Steady state fluorescence intensity and anisotropy measurements were made using either an SLM AB2 or an SLM 8100 photon-counting spectrofluorometer (SLM-Aminco, Rochester, NY) according to procedures described previously (9). Dansyl or fluorescein emissions were detected by using excitation wavelengths of 340 and 490 nm and an emission wavelength of 540 and 525 nm, respectively. The band pass used was 8 nm on the excitation and 16 nm on the emission beam for experiments involving DEGR-fXai and 8 nm on both the excitation and emission beams for experiments involving Light-with-a-Bite (LWB) FPR-fXai. All experiments were performed using 5 × 5 mm quartz cuvettes. Samples were mixed and adsorption of proteins to cuvette walls was minimized according to procedures described previously (14, 15).
Steady state anisotropy was measured using Glan-Thompson prism polarizers on both the excitation and emission beams. The emission intensity measured when the sample was excited by vertically plane-polarized light and the emission detected through a horizontal polarizer is termed IVH. IHH, IHV, and IVV are defined analogously. The component intensities of a dye-free blank were subtracted from the component sample intensities to give the net emission intensities. Anisotropy (r) was then calculated from the net intensities using r = (IVV – GIHV)/(IVV + 2GIVH), where the grating factor, G, equals IHV/IHH.
Each point in the titration is an average of 10 anisotropy measurements. Fitting of the saturation binding curve to determine Kd was performed according to procedures described previously (16, 17).
Photocross-linking—Photocross-linking experiments were performed using a CL-1000 ultraviolet cross-linker (Ultraviolet Products, Upland, CA). Adventitious cross-linking was prevented by storing the samples in the dark until photocross-linking experiments were performed. Samples were placed in microcentrifuge tubes ∼15 cm from the light source on ice and then irradiated with 254 nm ultraviolet light (5 × 8 watt discharge bulbs) for 0–30 s. Immediately following the photocross-linking experiments, samples were analyzed by SDS-PAGE (4–12% gradient; Novex, San Diego, CA).
Immunoblotting—After irradiating the PC/PS-bound LWB-FPR-fXai and protein S mixtures with UV light, the photocross-linked proteins were separated from the reactants by SDS-PAGE and visualized by both anti-fXa and anti-protein S Western blotting. Briefly, after SDS-PAGE, the protein bands were transferred to a polyvinylidene difluoride membrane (Bio-Rad) using a semi-dry transfer apparatus (Bio-Rad). After incubating in 50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 2 mm CaCl2, and 1% casein buffer overnight, the membrane was incubated with anti-fX monoclonal ascites (BG-X2, 1:1000 dilution; Elcatech Inc., Winston-Salem, NC) or with rabbit anti-human protein S (2.5 μg/ml; Dako, Glostrup, Denmark). The membrane was then treated with a biotin-conjugated secondary antibody directed against the primary antibody. Finally, alkaline phosphatase-conjugated streptavidin was added to the membrane for a time period of 30 min. This step was followed by the addition of alkaline phosphatase substrate, namely a pre-prepared mixture of 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (Pierce). Color development was terminated by washing the substrates off the membrane with water.
RESULTS
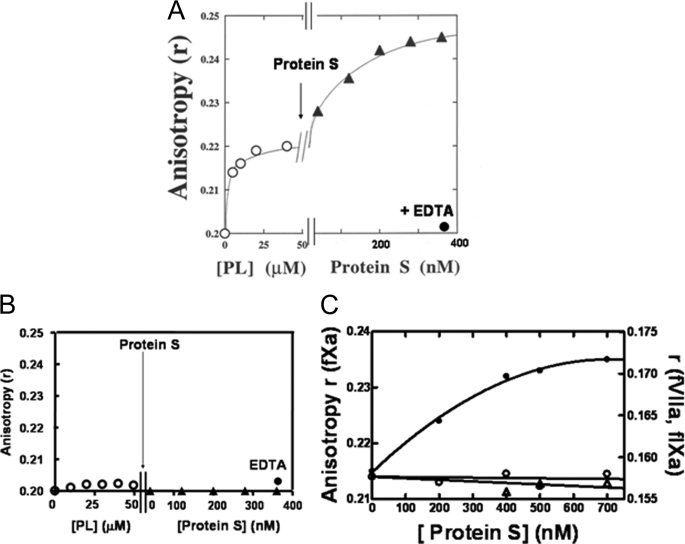
Interaction of DEGR-fXai with Protein S—The hypothesis that protein S interacts with membrane-bound fXa was tested. First, DEGR-fXai was titrated with PC/PS vesicles until essentially all the DEGR-fXai was membrane-bound. As illustrated in Fig. 1A, the fluorescence anisotropy (r) of DEGR-fXai increased from a value of 0.201 ± 0.002 for the unbound form to 0.219 ± 0.002 upon addition of PC/PS vesicles (n = 5). At this point in the titration, increasing concentrations of protein S were added. The r value of DEGR-fXai increased and reached a plateau at 0.245 ± 0.003 at ∼500 nm protein S. When EDTA was added at the end of the titration, the signal changed in the dansyl moiety because PC/PS and protein S additions were reversed, indicating that the PC/PS-dependent interaction between protein S and fXa is calcium ion-dependent.
FIGURE 1.
Interaction of protein S with DEGR-fXai in the presence of phospholipid vesicles. Samples containing DEGR-fXai (initially 200 nm) in 50 mm Hepes (pH 7. 4), 150 mm NaCl, 2 mm CaCl2 (buffer A) were titrated with phospholipids (open circles) with either PC/PS (4:1 mole ratio) (A) or 100% PC (B) vesicles prior to the addition of protein S (filled triangles). At the end of the titration, 5 mm EDTA (filled circles) was added to reverse the fluorescence changes. The anisotropy of the dansyl moiety in DEGR-fXai was monitored as described under “Experimental Procedures.” C, factor Xa dependence of dansyl anisotropy change. Three parallel titrations were performed. Equimolar concentrations of DEGR-fXai (filled circles), DEGR-fIXai (open circles), and DEGR-fVIIai (open triangles) (initially 200 nm) were each titrated with PC/PS vesicles as described (31, 32) prior to the addition of protein S. Anisotropy (r) was measured as described above.
The protein S- and phospholipid-dependent signal changes were lost upon the addition of 20 μm unlabeled fXa, suggesting that unlabeled fXa competes with DEGR-fXai for interaction with protein S and PC/PS. Here one notes that only the anisotropy of the dansyl probe in PC/PS-bound DEGR-fXai was sensitive to the addition of protein S, and no change in fluorescence emission intensity of the probe was observed.
Specificity of DEGR-fXai Spectral Change—The calcium ion-dependent protein S-induced signal change in DEGR-fXai was not observed in the absence of PC/PS vesicles (not shown). Additionally, this signal change was also not observed when DEGR-fXai was titrated with 100% PC vesicles before the addition of protein S. These data show the PC/PS dependence of the fXa-protein S interaction (Fig. 1B).
To determine the specificity of the fXa ·protein S complex, two homologs of fXa, fVIIa and fIXa, were labeled with a DEGR probe. When samples containing PC/PS-bound DEGR-fIXai or DEGR-fVIIai were titrated with protein S, no significant change in dansyl r value was observed (Fig. 1C).
Although the absence of spectral changes in Fig. 1 (B and C) does not necessarily indicate an absence of interaction, these data clearly establish the specificity of the spectral change when protein S is added to PC/PS-bound DEGR-fXai.
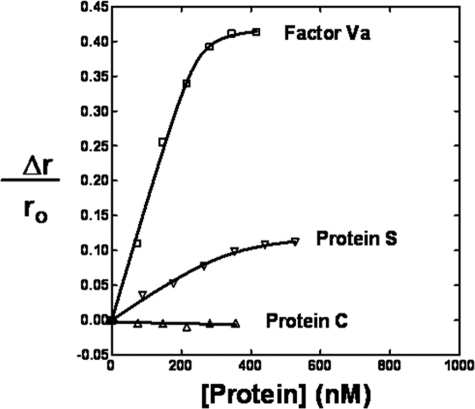
Interaction of PC/PS-bound Light-with-a-Bite-labeled-FPR-fXai with Protein S—The interaction of protein S with PC/PS-bound fXai was independently monitored using the fluorescence properties of Light-with-a-Bite (LWB)-labeled fXai. LWB is a bimodal probe that can monitor enzyme-effector interactions using both fluorescence and photocross-linking techniques (9). Initially, fXa was active site specifically labeled via a Phe-Pro-Arg (FPR) tether with LWB. When protein S was added to PC/PS-bound LWB-FPR-fXai, the fluorescence anisotropy, but not the fluorescence intensity of the fluorescein moiety was altered saturably by ∼14% (Fig. 2), suggesting that PC/PS-bound fXa interacts with protein S. Two additional controls were performed in parallel: a) the titration of fVa to PC/PS-bound LWB-FPR-fXai and b) the titration of protein C to PC/PS-bound LWB-FPR-fXai. Although both fVa and protein C are known to bind PC/PS surfaces, only the former binds fXa. Thus they are appropriate positive and negative controls for our assay. Fig. 2 shows that the fluorescence anisotropy of LWB-FPR-fXai is altered by the addition of fVa (∼42%), whereas the addition of protein C does not have any effect on LWB-FPR-fXai anisotropy. These data suggest that the fluorescein moiety in LWB is sensitive to the binding of various effectors of fXa. Additionally, the data indicate that the anisotropy of LWB is not altered by a protein binding to the lipid surface alone.
FIGURE 2.
Interaction of PC/PS-bound Light-with-a-Bite-FPR-fXai with protein S. Three parallel titrations were performed. Samples containing equimolar concentrations of LWB-FPR-fXai (initially 200 nm) containing PC/PS vesicles were titrated with protein S (open inverted triangles), fVa (open squares), or protein C (open triangles), and the anisotropy (r) of the fluorescein moiety measured as described in the text.
The protein S-dependent binding isotherms of LWB-FPR-fXai were fit to determine Kd and stoichiometry for the fXa-protein S interaction. The Kd for this interaction was determined at two different concentrations of LWB-FPR-fXai (200 and 50 nm). The Kd values were determined to be 29 and 37 nm, respectively, very similar for the two concentrations of LWB-FPR-fXai. This suggests that the Kd is independent of the concentration of starting material of LWB-FPR-fXai.
To determine the stoichiometry of the fXa-protein S interaction, the fluorescence data were analyzed according to Stinson and Holbrook (18, 19). Plots of protein S/θ versus (1/1–θ) were constructed, where θ = fractional saturation =Δr/Δrmax. Our analysis showed that the stoichiometry for LWB-FPR-fXai-protein S interaction is 1.0:0.7. Therefore, the data are consistent with a 1:1 interaction for these two proteins.
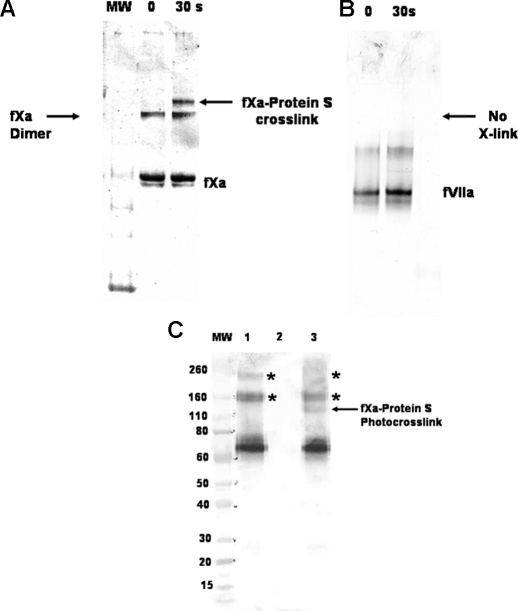
Photocross-linking of PC/PS-bound LWB-FPR-fXai to Protein S—The interaction of protein S with PC/PS-bound fXai was independently monitored using photocross-linking techniques. LWB-FPR-fXai was incubated with excess PC/PS vesicles prior to the addition of protein S and subsequent photoirradiation with UV light. Samples were analyzed using SDS-PAGE and bands were visualized by immunoblotting using anti-fXa monoclonal antibody as described previously (9). Fig. 3A (lane 3) shows that a new band of ∼120,000 Da appears after irradiating the reaction mixture for 30 s with UV light. This band corresponds in approximate molecular mass to a protein S ·fXa covalent complex. It is noteworthy that we also observed fXa dimers in our cross-linking experiments. These results are in accordance with observations that fXa can dimerize in the presence of PS (20).
FIGURE 3.
A, the protein S ·LWB-FPR-fXai complex was detected using photocross-linking. LWB-FPR-fXai (2 μm) in buffer containing buffer A was incubated with PC/PS (25 μm) and protein S (6 μm) prior to exposure to 30 s of 254 nm irradiation. Subsequently, samples were analyzed using SDS-PAGE and visualized using anti-fXa monoclonal as primary antibody in immunoblotting as described under “Experimental Procedures.” Lane 2 contains the reaction mixture prior to UV irradiation and is designated as zero time. Lane 3 contains the reaction mixture after 30 s of exposure to UV light. B, LWB-FPR-fVIIai (2 μm) in buffer A was incubated with PC/PS (25 μm) and protein S (6 μm) prior to exposure to UV light as described above. Samples were analyzed by SDS-PAGE and visualized using anti-fVIIa polyclonal (Enzyme Research Laboratories) as primary antibody in immunoblotting. C, LWB-FPR-fXai (2 μm) in buffer containing buffer A was incubated with PC/PS (25 μm) and protein S (50 nm) prior to exposure to 30 s of 254 nm irradiation. Subsequently, samples were analyzed using SDS-PAGE and visualized using anti-protein S polyclonal (Dako) as primary antibody in immunoblotting as described under “Experimental Procedures.” Lane 1 contains protein S ·PC/PS complex irradiated with UV light, lane 2 contains LWB-FPR-fXaI ·PC/PS complex irradiated with UV light, and lane 3 contains the reaction mix irradiated with UV light. Photocross-link between LWB-FPR-fXai and protein S is indicated by the arrow.
Analysis of the SDS-polyacrylamide gels with anti-protein S showed that the new 120,000-Da band stains positive for protein S (Fig. 3C, compare lanes 1 and 3), suggesting that the band is a complex between fXa and protein S. Protein S can form multimers, as reported by Seré et al. (21). We have mostly used commercial protein S preparations, which contain predominantly monomers of protein S (see supplemental Fig. 1S), for our photocross-linking studies. Although multimers of protein S were detected both on native gels and on SDS-polyacrylamide gels (Fig. 3C, lanes 1 and 3 asterisks), only one new band, corresponding to ∼120,000 Da, appears on irradiation with UV light. This suggests that fXa interacts with monomeric protein S.
When similar photocross-linking experiments were performed with mixtures containing LWB-FPR-fVIIai, excess PC/PS, and protein S, no high molecular mass cross-links were detected after irradiation with UV light (Fig. 3B). These data independently suggest that fXa and protein S (and not fVIIa and protein S) are in close proximity to each other (<15 Å) on a PC/PS vesicle surface. This is likely due to complex formation of protein S and fXa on PC/PS vesicles.
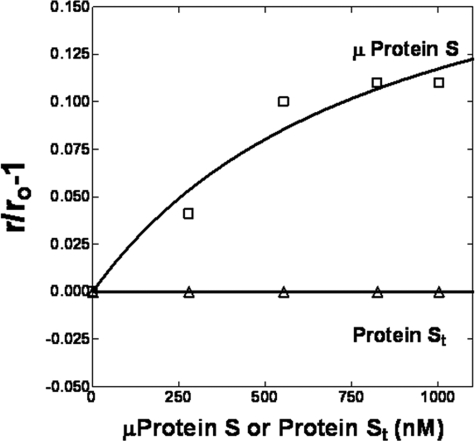
Interaction of DEGR-fXai with Microprotein S—When thrombin-cleaved protein S was titrated into samples containing PC/PS-bound DEGR-fXai, no significant change in the r value of the dansyl probe in DEGR-fXai was observed (Fig. 4, open triangles). This suggests that the N terminus of protein S may be important for the interaction with fXa. To test this hypothesis, the first 116 residues of protein S, consisting of the Gla, TSR, and EGF1 domains of protein S, were synthesized chemically and correctly folded according to procedures published previously (22). This protein termed “microprotein S” had ∼50% activated protein C cofactor activity as judged by an activated partial thromboplastin time clotting assay. When PC/PS-bound DEGR-fXai was titrated with microprotein S, the anisotropy of the dansyl probe increased from 0.219 to 0.242 (Kd = 303 nm), suggesting that the fXa binding domain was expressed in microprotein S (Fig. 4). Thus, it is highly likely that one or more domains in microprotein S (EGF1, TSR, and/or Gla) bind to fXa.
FIGURE 4.
Interaction of microprotein S with PC/PS-bound DEGR-fXai. Samples containing DEGR-fXai (initially 200 nm) in buffer A were titrated with PC/PS vesicles (not shown) up to a concentration of 50 μm before the addition of microprotein S (open squares) or thrombin-cleaved protein S (open triangles). rO is the anisotropy of PC/PS-bound DEGR-fXai, whereas r is the anisotropy of the dansyl moiety in DEGR-fXai at any point in the subsequent protein titration.
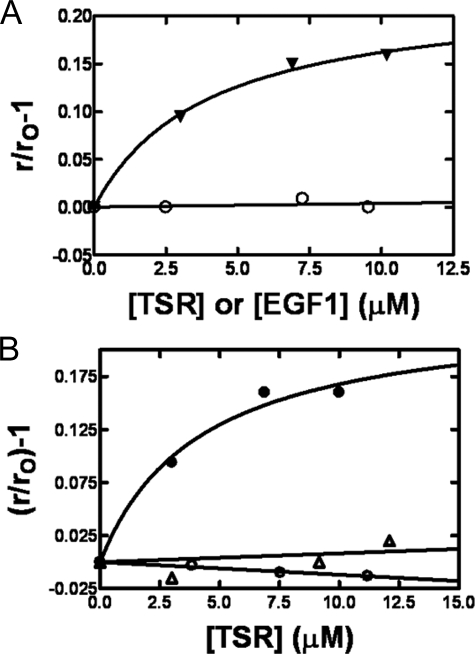
Interaction of DEGR-fXai with TSR Module of Protein S—Chemically synthesized EGF1 (residues 76–116 in protein S) and TSR (residues 47–75) of protein S were folded to native-like conformation as described (13). These modules were then tested for binding to DEGR-fXai. When the TSR of protein S was titrated into DEGR-fXai, the r value of the dansyl dye increased from 0.200 ± 0.003 to a value of 0.232 ± 0.003 (Fig. 5A), showing that the module binds to and alters the active site of DEGR-fXai (Kd = 3.9 μm). On the other hand, when the EGF1 module of protein S was titrated into samples containing DEGR-fXai, no significant change in r values was observed, suggesting the specificity of the DEGR-fXai-TSR interaction.
FIGURE 5.
Interaction of PC/PS-bound DEGR-fXai with domains of protein S. A, DEGR-fXai (initially 200 nm) was titrated with chemically synthesized domains of TSR (filled inverted triangles) or EGF1 (open circles) of protein S. rO and r are as in the legend to Fig. 3. B, the factor Xa dependence of TSR-dependent change in dansyl anisotropy. Three parallel titrations were performed. Equimolar concentrations of DEGR-fXai (closed circles), DEGR-fIXai (open triangles), and DEGR-fVIIai (open circles) (initially 200 nm) were each titrated with TSR of protein S, and the dansyl anisotropy monitored as described under “Experimental Procedures.”
To further demonstrate the specificity of TSR for fXa, samples containing either DEGR-fIXai or DEGR-fVIIai were titrated with the TSR of protein S (Fig. 5B). No significant changes in dansyl r values were observed with these two similarly labeled homologs of DEGR-fXai. These data suggest that the interaction of TSR with DEGR-fXai is specific for fXa.
DISCUSSION
The present study demonstrates that DEGR-fXai and LWB-FPR-fXai, two different analogs of fXa, form a complex with protein S via the TSR of protein S and that interaction requires negatively charged phospholipid vesicles and calcium ions. The interaction of protein S with fXa was specific because two similarly labeled homologs of fXa, fIXa, and fVIIa, did not exhibit protein S-induced spectral changes. Importantly, the protein S-dependent spectral changes in fXa were observed only when DEGR-fXai was bound to PC/PS vesicles, and signal changes were not observed in the absence of lipid or in the presence of 100% PC vesicles. These data suggest that the interaction between fXa and protein S is phospholipid-dependent and requires PS.
To confirm independently these spectroscopic findings, we employed photocross-linking techniques. fXa and fVIIa were labeled via the active site with a photocross-linkable probe to yield LWB-FPR-fXai and LWB-FPR-fVIIai, respectively. Energy-minimized molecular modeling of LWB shows that the photoreactive carbonyl in the benzophenone of LWB is ∼15 Å away from the bromoacetyl group on the Lys of LWB. Because LWB was linked to the active site of fXa and fVIIa via a FPR tether, much of the LWB is expected to be outside the active site pocket. Thus we estimate the cross-linking radius to be ∼15 Å. When irradiated with UV light, PC/PS-bound LWB-FPR-fXai, but not LWB-FPR-fVIIai, was cross-linked to protein S, yielding a new band at ∼120,000 Da under denaturing conditions. This molecular mass corresponds to a covalent complex between fXa and protein S. The formation of this complex was confirmed when the new band stained positive for both fXa and protein S in immunoblots. Specificity of the fXa-protein S photocross-linking was shown by the failure of protein S to cross-link to PC/PS-bound LWB-FPR-fVIIai.
Unlike native protein S, thrombin-cleaved protein S did not alter lipid-bound DEGR-fXai signal. This suggests that the N-terminal region of protein S, e.g. the TSR, may be important for the interaction of fXa with protein S. To determine the fXa binding loci on protein S, various N-terminal fragments of protein S were chemically synthesized and tested for binding to protein S. Microprotein S and TSR both altered similarly the fluorescence anisotropy of DEGR-fXai, whereas the EGF1 of protein S did not, suggesting that the N-terminal TSR in protein S binds to DEGR-fXai. TSR did not alter the anisotropy of DEGR-fIXai and DEGR-fVIIai, showing that this interaction was specific for fXa. These data strongly suggest that protein S interacts with lipid-bound fXa via the TSR.
In our fluorescence binding assays, an order of magnitude in binding affinity was lost in going from protein S to microprotein S to TSR (∼30 nm to 300 nm to 4 μm). This suggests that other domains of protein S could provide additional binding energy for the fXa-protein S interaction. Consistent with this notion, previous studies of the mutant protein S Tokushima demonstrated that a Lys155 → Glu mutation in the EGF2 of protein S diminished fXa binding to protein S (23). This mutation also abolished the protein S inhibition of prothrombinase on thrombin-stimulated platelets. Another study using a tandem construct containing two EGF-like domains of protein S, namely EGF3–4, showed that these domains inhibited the binding of fX to protein S (24). Thus our findings and these two reports support the concept that the TSR and EGF domains of protein S mediate direct binding of protein S to fXa that is responsible for the activated protein C-independent anticoagulant activity of protein S.
The functional consequence(s) of this protein S ·fXa interaction are as yet not clearly understood. It is possible that this interaction plays a role in the anticoagulant activity of protein S that is independent of activated protein C, known as “protein S direct” or in the fXa-dependent proteolytic inactivation of protein S (7). Heeb et al. (4) proposed that protein S direct was brought about because of protein S binding to fXa and fVa and disrupting the IIase complex assembly. However, in different studies, large variations in protein S direct have been reported (25, 26). Seré et al. contend that protein S inhibition of prothrombinase correlates with the content of protein S multimers in different preparations of protein S (21). However, they observed that protein S multimers were not observed in plasma and so concluded that these forms of protein S are unlikely to contribute to protein S direct in plasma. Whether protein S multimers are present in plasma or generated during purification is also a matter of debate (27–30). In our studies, we have used commercial protein S preparations that essentially have only protein S monomers. Thus, our results indicate that monomeric protein S binds to fXa. Recently, it has been described that protein S can down-regulate fIIa generation by stimulating fXa inhibition by tissue factor pathway inhibitor (5). This observation was confirmed and extended recently in a report that protein S enhances the tissue factor pathway inhibitor-mediated inhibition of fXa but not its inhibition of fVIIa-tissue factor (6). These recent findings are in accordance with our observations here that protein S binds to fXa but not to fVIIa. It is likely that protein S-fXa interactions play a part in these functions of protein S. Our results are also compatible with observations of Long et al. (7), who showed that fXa cleaves protein S at Arg60 of the TSR and that this reaction requires negatively charged phospholipids and calcium ions.
In conclusion, we have shown a specific interaction between the TSR of protein S and fXa that is phospholipid-dependent. Whether this interaction is relevant to protein S-cofactor activity for tissue factor pathway inhibitor, protein S-dependent inhibition of prothrombinase, or protein S proteolysis by fXa is as yet unclear. Specific experiments are needed to address these issues.
Supplementary Material
Acknowledgments
We thank Phuong M. Nguyen for technical assistance and Dr. Andrew J. Gale for reading through the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL 21544 (to J. H. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: fVa, factor Va; DEGR-fIXai, 5-dimethylaminonaphthalene-1-sulfonyl-(1,5, dansyl-) Glu-Gly-Arg factor Xa; TSR, thrombin-sensitive region; EGF1, epidermal growth factor-like; Gla, γ-carboxyglutamic acid; LWB, Nα-fluorescein-p-benzoylphenylalanyllysyl(Nε-bromoacetyl) amide; PC, phosphatidylcholine; PS, phosphatidylserine.
References
- 1.Dahlback, B. (2007) Thromb. Haemostasis 98 90–96 [PubMed] [Google Scholar]
- 2.van Wijnen, M., van't Veer, C., Meijers, J. C., Bertina, R. M., and Bouma, B. N. (1998) Thromb. Haemostasis 80 930–935 [PubMed] [Google Scholar]
- 3.Hackeng, T. M., van't Veer, C., Meijers, J. C., and Bouma, B. N. (1994) J. Biol. Chem. 269 21051–21058 [PubMed] [Google Scholar]
- 4.Heeb, M. J., Rosing, J., Bakker, H. M., Fernandez, J. A., Tans, G., and Griffin, J. H. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 2728–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackeng, T. M., Seré, K. M., Tans, G., and Rosing, J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3106–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ndonwi, M., and Broze, G., Jr. (2008) J. Thromb. Haemostasis 6 1044–1046 [DOI] [PubMed] [Google Scholar]
- 7.Long, G. L., Lu, D., Xie, R. L., and Kalafatis, M. (1998) J. Biol. Chem. 273 11521–11526 [DOI] [PubMed] [Google Scholar]
- 8.Hackeng, T. M., Yegneswaran, S., Johnson, A. E., and Griffin, J. H. (2000) Biochem. J. 349 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yegneswaran, S., Fernandez, J. A., Griffin, J. H., and Dawson, P. E. (2002) Chem. Biol. 9 485–494 [DOI] [PubMed] [Google Scholar]
- 10.Husten, E. J., Esmon, C. T., and Johnson, A. E. (1987) J. Biol. Chem. 262 12953–12961 [PubMed] [Google Scholar]
- 11.Yegneswaran, S., Wood, G. M., Esmon, C. T., and Johnson, A. E. (1997) J. Biol. Chem. 272 25013–25021 [DOI] [PubMed] [Google Scholar]
- 12.Schnolzer, M., Alewood, P., Jones, A., Alewood, D., and Kent, S. B. H. (1992) Int. J. Pept. Protein Res. 40 180–193 [DOI] [PubMed] [Google Scholar]
- 13.Hackeng, T. M., Dawson, P. E., Kent, S. B., and Griffin, J. H. (1998) Biopolymers 46 53–63 [DOI] [PubMed] [Google Scholar]
- 14.Dell, V. A., Miller, D. L., and Johnson, A. E. (1990) Biochemistry 29 1757–17563 [DOI] [PubMed] [Google Scholar]
- 15.Ye, J., Esmon, N. L., Esmon, C. T., and Johnson, A. E. (1991) J. Biol. Chem. 266 23016–23021 [PubMed] [Google Scholar]
- 16.Koppaka, V., and Lentz, B. R. (1996) Biophys. J. 70 2930–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koppaka, V., Talbot, W. F., Zhai, X., and Lentz, B. R. (1997) Biophys. J. 73 2638–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stinson, R. A., and Holbrook, J. J. (1973) Biochem. J. 131 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stinson, R. A., and Holbrook, J. J. (1973) Biochem. J. 131 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder, R., Wang, J., and Lentz, B. R. (2003) Biophys. J. 84 1238–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seré, K. M., Janssen, M. P., Willems, G. M., Tans, G., Rosing, J., and Hackeng, T. M. (2001) Biochemistry 40 8852–8860 [DOI] [PubMed] [Google Scholar]
- 22.Hackeng, T. M., Fernandez, J. A., Dawson, P. E., Kent, S. B., and Griffin, J. H. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 14074–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi, T., Nishioka, J., and Suzuki, K. (1995) Biochim. Biophys. Acta 1272 159–167 [DOI] [PubMed] [Google Scholar]
- 24.Stenberg, Y., Muranyi, A., Steen, C., Thulin, E., Drakenberg, T., and Stenflo, J. (1999) J. Mol. Biol. 293 653–665 [DOI] [PubMed] [Google Scholar]
- 25.van Wijnen, M., Stam, J. G., van't Veer, C., Meijers, J. C., Reitsma, P. H., Bertina, R. M., and Bouma, B. N. (1996) J. Thromb. Haemostasis 76 397–403 [PubMed] [Google Scholar]
- 26.Van't Veer, C., Butenas, S., Golden, N. J., and Mann, K. G. (1999) J. Thromb. Haemostasis 82 80–87 [PubMed] [Google Scholar]
- 27.Heeb, M. J., Radtke, K. P., Fernandez, J. A., and Tonnu, L. (2006) Thromb. Haemostasis 4 2215–2222 [DOI] [PubMed] [Google Scholar]
- 28.Heeb, M. J., Schuck, P., and Xu, X. (2006) Thromb. Haemostasis 4 385–391 [DOI] [PubMed] [Google Scholar]
- 29.Mann, K. G. (2005) Blood 106 1884–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pauls, J. E. D., Hockin, M. F., Long, G. L., and Mann, K. G. (2000) Biochemistry 39 5468–5473 [DOI] [PubMed] [Google Scholar]
- 31.Mutucumarana, V. P., Duffy, E. J., Lollar, P., and Johnson, A. E. (1992) J. Biol. Chem. 267 17012–17021 [PubMed] [Google Scholar]
- 32.McCallum, C. D., Hapak, R. C., Neuenschwander, P. F., Morrissey, J. H., and Johnson, A. E. (1996) J. Biol. Chem. 271 28168–28175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.