Abstract
Covalent modification of cullins by the ubiquitin-like protein NEDD8 (neddylation) regulates protein ubiquitination by promoting the assembly of cullin-RING ligase E3 complexes. Like ubiquitination, neddylation results from an enzymatic cascade involving the sequential activity of a dedicated E1 (APPBP1/Uba3), E2 (Ubc12), and an ill-defined E3. We show that SCCRO (also known as DCUN1D1) binds to the components of the neddylation pathway (Cullin-ROC1, Ubc12, and CAND1) and augments but is not required for cullin neddylation in reactions using purified recombinant proteins. We also show that SCCRO recruits Ubc12∼NEDD8 to the CAND1-Cul1-ROC1 complex but that this is not sufficient to dissociate or overcome the inhibitory effects of CAND1 on cullin neddylation in purified protein assays. In contrast to findings in cellular systems where no binding is seen, we show that SCCRO and CAND1 can bind to the neddylated Cul1-ROC1 complex in assays using purified recombinant proteins. Although neddylated (not unneddylated) Cul1-ROC1 is released from CAND1 upon incubation with testis lysate from SCCRO+/+ mice, the addition of recombinant SCCRO is required to achieve the same results in lysate from SCCRO–/– mice. Combined, these results suggest that SCCRO is an important component of the neddylation E3 complex that functions to recruit charged E2 and is involved in the release of inhibitory effects of CAND1 on cullin-RING ligase E3 complex assembly and activity.
Post-translational modification of proteins by ubiquitin (Ub)4 regulates diverse cellular functions including protein turnover, differentiation, apoptosis, cell cycle, and transcription (1–5). Given its essential role, ubiquitination is a highly regulated process that involves the sequential action of three enzymes termed as E1, E2, and E3. In this enzymatic cascade, E1 initiates the process by forming a high energy thioester bond with Ub in an ATP-coupled reaction. The Ub is then transferred to E2 as a thioester intermediate. Finally, E3s serve as the targeting arm in the ubiquitination process, mediating the transfer of Ub from E2 to the target protein to create an isopeptide bond between the C-terminal glycine in Ub and a lysine residue on the substrate protein. Once attached, the Ub itself can be modified to generate polyubiquitin chains on the target protein (6). The functional effects of ubiquitination are influenced by the chain length and the residue on the Ub to which the chain is attached. Polyubiquitination promotes translocation to the 26 S proteasome for degradation. Other functional effects of mono- and polyubiquitination include protein translocation, interaction, and activation.
Although there is only one known E1 (except in plants) and relatively few E2s, E3s exist in multiple forms to allow for specific protein targeting (6). In general, E3s are modular multiprotein complexes that can be divided into two broad categories based on the presence of either a HECT (homologous to E6-AP C terminus) or RING (Really Interesting New Gene)-finger domain-containing protein at their core. Although HECT E3s form a thioester intermediate with the Ub before its transfer, RING-containing complexes serve as scaffolds to facilitate the direct transfer of Ub from E2 to the target protein.
Cullin RING ligases (CRLs) constitute the largest class of E3s in mammals (7, 8). All CRLs are anchored by cullins, a highly conserved protein family with seven known isoforms in humans (Cul1, Cul2, Cul3, Cul4a, Cul4b, Cul5, and Cul7). A small RING protein (ROC1) and a variable substrate recognition subunit bind to the cullin core to form the CRL complex. The SCF (SKP1 cullin1 F-box) complex is the prototypic CRL E3 complex and is made up of ROC1 bound to the C terminus and SKP1 adaptor protein bound to the N terminus of Cul1. SKP1 in turn binds to a host of different F-box containing proteins to confer target specificity, and ROC1 binds to E2 to form the catalytic core of the SCF complex.
Assembly of CRL complexes serves as a key regulatory step for ubiquitination. Cullins normally exist as part of two mutually exclusive complexes in cells. The majority of cullins are in complex with CAND1 (Cullin-associated and neddylation-dissociated 1), which sterically inhibits assembly of CRL complexes. The covalent modification of cullins with the ubiquitin-like protein (Ublp) NEDD8, in a process termed neddylation, dissociates CAND1 and promotes assembly of CRL complexes (9–12). In addition, cullin neddylation also enhances CRL activity through the recruitment of ubiquitin E2s to the complex and possibly facilitating cullin heterodimer formation (13–15). Conversely, deneddylation of cullins, principally by the COP9 signalosome, promotes dissociation of CRL complexes and binding to CAND1. Several studies suggest that cycling of neddylation and deneddylation is required for normal CRL function (8, 16, 17).
Neddylation occurs by mechanisms analogous to Ub or Ublp conjugation, involving the sequential activity of a dedicated E1, E2, and E3. Although APPBP1/Uba3 functions as the E1 and Ubc12 as the E2, the precise components of the neddylation E3 remain to be established. Using a positional cloning strategy, we identified SCCRO (squamous cell carcinoma-related oncogene) within a recurrent amplification peak at 3q26.3 in squamous cell carcinomas (18). Activation of SCCRO by amplification is associated with malignant transformation in vitro and in vivo and an aggressive clinical course in human cancers. Moreover, cancer cell lines carrying amplification are addicted to high SCCRO levels, rapidly undergoing apoptosis with SCCRO suppression using RNAi (18). Here we show that SCCRO interacts with known cullin isoforms as well as ROC1, Ubc12 and CAND1. SCCRO preferentially binds to Ubc12∼NEDD8 thioester and augments cullin neddylation in both lysate and purified systems. Although SCCRO is not essential in a purified system, activated neddylation is reduced in extracts made from SCCRO–/– mice. In addition, these mice have reduced viability and severe developmental defects, suggesting that SCCRO plays an important role in vivo. Details of the SCCRO knock-out mouse construction and characterization will be published elsewhere.5 Although SCCRO does not release CAND1 or overcome its inhibition of cullin neddylation in assays using purified protein, Cul1-ROC1 dissociates from CAND1 when incubated in lysate from SCCRO+/+ mice but not SCCRO–/– mice. These findings suggest that SCCRO plays an essential role in neddylation and supports its inclusion as a component of the E3 complex for neddylation.
EXPERIMENTAL PROCEDURES
Alignment and Sequence Analyses—Data base and BLAST searches were carried out at NCBI (www.ncbi.nlm.nih.gov). Multiple sequence alignments were performed using the ClustalW program.
Reagents—All constructs were generated by standard PCR ligation-based methods and verified by automated sequencing. Proteins were expressed as GST fusions in Escherichia coli, induced overnight at 18 °C with the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside, and purified by passing through glutathione-Sepharose 4B beads (GE Healthcare) followed by thrombin cleavage where required. Cul1-ROC1 was expressed and purified from E. coli essentially as described previously (19). APPBP1/Uba3, Ubc12, and NEDD8 were obtained from a commercial source (Boston Biochem).
cDNAs for mammalian transfection were cloned into pUSE-amp or pCMV-HA vector (Clontech). Anti-SCCRO RNAi and scrambled RNAi were generated, validated, and used as previously described (18). Transfection was carried out with Lipofectamine 2000 (Invitrogen) or FuGENE (Roche Applied Science) using manufacturer's specifications.
All cell lines used in this study were obtained from ATCC (American Type Culture Collection, Manassas, VA) and grown at 37 °C in 5% CO2. HeLa cells were maintained in minimal essential media supplemented with 10% fetal calf serum-containing antibiotics.
The following antibodies were used in this study: anti-Cul1 (Zymed Laboratories Inc.), anti-Cul2 (Abcam), anti-Cul3 (BD Biosciences), anti-Cul4, anti-Cul5, anti-SKP2 (Santa Cruz), anti-ROC1 (Abcam), anti-SKP1 (Abnova), anti-Ubc12 (Rockland, MA), anti-NEDD8 (Invitrogen), anti-CAND1 (BD Biosciences), anti-tubulin (Calbiochem), anti-HA (Abcam), and anti-FLAG (Sigma). Anti-SCCRO (rabbit polyclonal) antibody was produced and utilized as described previously (18). Anti-SCCRO monoclonal antibody was raised against an N-terminal region of the protein in SCCRO–/– mice. This antibody was found to be highly sensitive and specific in control experiments (data not shown). Secondary antibodies conjugated to horseradish peroxidase (Santa Cruz, CA) were used according to the manufacturers' specifications.
GST Pulldown Assay—GST tagged proteins were bound to glutathione-Sepharose beads by gentle rocking at 4 °C for 30 min. The beads were washed 3 times with EBC buffer (50 mm Tris-HCl, pH 7.5, 2.5 mm MgCl2, 150 mm NaCl, and 0.5% Nonidet P-40) at 20× bead volume. The beads were incubated with 500 μg of HeLa cell lysate or purified proteins as indicated at 4 °C for 1 h followed by 3 washes with EBC buffer at 20× bead volume. Bound proteins were eluted by the addition of 6× Laemmli buffer, resolved on SDS-PAGE gels, and analyzed by Western blot. For mass spectrometric analysis, the resolved proteins were stained with Coomassie R250. Bands were excised and subjected to matrix-assisted laser desorption/ionization reflectron time-of-flight (MALDI-reTOF) mass spectrometric analysis.
Immunoprecipitations—Immunoprecipitations were performed essentially as described earlier (18). In brief, all cells were lysed using mammalian cell lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 μg/ml leupeptin) (Cell Signaling). One mg of lysate was incubated with antibody (αHA, αSCCRO, αROC1, or preimmune sera) bound to agarose beads by gentle rocking at 4 °C for 2 h. The same wash and detection sequence was used as for the GST pulldown assay.
Thioester Reactions—Reactions were performed at room temperature in a buffer (50 mm Tris-HCl, pH 7.6, 50 mm NaCl, 10 mm MgCl2, 0.5 mm DTT) with 4 mm ATP, 80 nm APPBP1/Uba3, 800 nm Ubc12, and 9 μm NEDD8. Reactions were quenched with 3× non-reducing Laemmli buffer (without DTT), and the presence of Ubc12∼NEDD8 thioester complexes were verified by Western blot for Ubc12 and/or NEDD8 in the presence and absence of 50 mm DTT. Time course experiments showed that incubation times greater that 10 min resulted in the formation of a small fraction of NEDD8 bound to Ubc12 that was not reducible by DTT (supplemental Fig. 1). Accordingly, all thioester reactions were carried out for 10 min or less. For use in binding assays, the reaction mix was quenched with the addition of EDTA to a final concentration of 50 mm and purified on a G-50 micro spin desalting column. For competition experiments, reaction conditions were identical except for the addition of a gradient of free Ubc12 to the purified thioester reaction mix. Reaction conditions for HeLa transfected with HA-Ubc12 or HA-Ubc12Δ1–26 was the same, except a longer incubation time (30 min) was employed for HA-Ubc12Δ1–26.
In Vitro Neddylation—The source of Cullin-ROC1 substrate for in vitro neddylation reactions was either HeLa lysates or mouse tissue extracts or was bacterially derived (see above). Lysates from testes were used, as the SCCRO–/– mouse had developmental defects restricted to the testis.5 For reactions using HeLa or tissue-derived Cullin-ROC1 complexes, 100 or 10 μg, respectively, of lysates (Cul1 concentration ∼20 fmol) were added to reactions containing 0.2–2 pmol of APPBP1/Uba3, 1–12.5 pmol of Ubc12, and 150 pmol of cold NEDD8 or 32P-labeled NEDD8 in neddylation buffer (50 mm Tris-HCl, pH 7.6, 55 mm NaCl, 5 mm MgCl2, and 1 mm DTT). Reactions were incubated at 30 °C and stopped with the addition of 6× Laemmli buffer. Reactions with bacterially derived substrate were performed at 30 °C in neddylation buffer containing 20 nm Cul1-ROC1, 4 mm ATP, 10 nm of APPBP1/Uba3, 20 nm Ubc12, and 0.9 μm NEDD8. In reactions containing CAND1, Cul1-ROC1 was preincubated with the indicated amounts of CAND1 for 15 min at room temperature. Proteins were resolved on an SDS-PAGE gel and subjected to autoradiography and/or Western blot analysis. For quantification Western blots were imaged using MultiImage Light Cabinet and quantified using Alpha-EaseFC v3.2.1 (Alpha Innotech Corp.).
Assay for Binding to Neddylated and Unneddylated Cullins—Neddylation reactions were performed on purified Cul1-ROC1 at 30 °C for the indicated times. The reactions were stopped by the addition of EDTA to a final concentration of 50 mm, and the products were purified on a G-50 micro spin desalting column. A small aliquot of the flow-through was subjected to Western blot analysis for Cul1 to establish the level of cullin neddylation. The remaining flow-through was incubated with GST-SCCRO and/or GST-CAND1 (at limiting concentrations) at room temperature for 20 min followed by the addition of glutathione-Sepharose beads and incubation with gentle rocking at 4 °C for 45 min. After three 20× bead volume washes with EBC wash buffer, 6x Laemmli buffer was added to the beads, and bound proteins were resolved on an SDS-PAGE gel and analyzed by Western blot for Cul1. To assess the effects of lysate on release of cullins from CAND1, neddylated or unneddylated Cul1-ROC1 was incubated with GST-CAND1 at room temperature for 20 min and rocked at 4 °C for 30 min. SCCRO, SCCRO-D241N and/or testis lysate from SCCRO+/+ or SCCRO–/– mice was added onto the beads and rotated at 4 °C for 1 h followed by three 20× bead volume washes with EBC buffer. 6× Laemmli buffer was added to the beads, and bound proteins were analyzed by Western blot for Cul1.
RESULTS
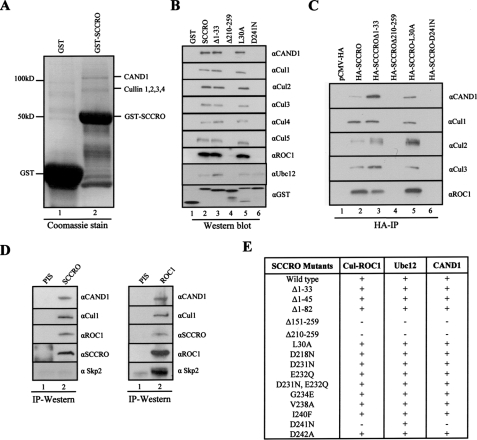
SCCRO Interacts with Components of the Neddylation Pathway—We identified SCCRO by fluorescent in situ hybridization-based fine resolution mapping of a recurrent amplification at 3q26.3 that is present in multiple human cancers (18, 20–22). As a step toward understanding the molecular function of SCCRO, we sought to identify interacting proteins. GST-SCCRO pulldown from HeLa lysates identified two unique bands relative to GST alone (Fig. 1A). These bands were excised and subjected to mass spectrometric analysis (MALDI-ReTOF) identifying CAND1 and members of the cullin family of proteins (Cul1, Cul2, Cul3, and Cul4) as putative binding partners (Fig. 1A). We confirmed these interactions by GST-SCCRO pulldown from HeLa lysates followed by Western blot for each of the identified proteins. In addition, we found that SCCRO also binds to Cul5 and ROC1 (Fig. 1B, lane 2). GST pulldown assays using purified proteins showed that SCCRO directly binds to ROC1, consistent with findings in yeast (data not shown) (23). Because they are soluble only as a complex with ROC1, we could not assess if SCCRO can also bind to cullins directly.
FIGURE 1.
SCCRO interacts with Cullins, CAND1, and Ubc12. A, Coomassie-stained SDS-PAGE gel showing GST (lane 1) and GST-SCCRO (lane 2) pulldown products from HeLa extracts. Unique bands from GST-SCCRO pulldown assays were excised and subjected to tandem mass spectroscopy analysis (MALDI-ReTOF) revealing the 120-kDa band as CAND1 and the 80–90-kDa bands as cullins 1–4. B, Western blot on the pulldown products of GST-SCCRO and GST-SCCRO mutants from HeLa extracts probed with indicated antibodies showing SCCRO binds to CAND1, cullins 1–5, ROC1, and Ubc12 (lane 2). The C-terminal (SCCROΔ210–259; lane 4) but not the N-terminal deletion of SCCRO (SCCROΔ1–33; lane 3) lost interaction with the Cullin-ROC1, CAND1, and Ubc12 (lane 4). SCCRO-D241N lost interaction with the Cullin-ROC1 and CAND1 while retaining binding to Ubc12 (lane 6). The level of the various GST-tagged proteins used in the pulldown experiment was confirmed by probing a Western blot with anti-GST antibody (bottom panel). C, Western blot showing products from an HA immunoprecipitation (IP) of lysates from HeLa cells transfected pCMV-HA-SCCRO or selected SCCRO deletions/mutants probed with indicated antibodies showing SCCRO maintains binding to neddylation components in vivo (lane 2). SCCROΔ210–259 (lane 4) and SCCRO-D241N (lane 6) lose binding to cullins, ROC1, and CAND1. D, Western blot on immunoprecipitates from HeLa extract prepared with anti-SCCRO (left panel; lane 2) anti-ROC1 (right panel; lane 2) or preimmune serum (PIS; lane 1) probed with indicated antibodies showing in vivo interaction between native SCCRO and CAND1, Cul1, and ROC1 but not SKP2 (left panel), whereas ROC1 binds to CAND1, Cul1, SCCRO, and SKP2 (right panel). E, summary of results from pulldown assays using GST-SCCRO or SCCRO deletions/mutations showing binding of CAND1, Cul-ROC1, and Ubc12 requires C-terminal 50 amino acids. SCCRO-D241N loses binding to CAND1 and Cul1-ROC1 but retains binding to Ubc12.
Given the established role of CAND1 and the cullin family of proteins in neddylation, we assessed for binding to other proteins in the neddylation pathway, including NEDD8, APPBP1/Uba3, and Ubc12 and found that SCCRO interacts with Ubc12 (Fig. 1B, lane 2). The co-immunoprecipitation of SCCRO with each of its putative binding partners in HA-SCCRO-transfected HeLa cells confirmed that the observed interactions occur in cells (Fig. 1C, lane 2). SCCRO does not bind to Ubc9 (an E2 for sumoylation), SKP1, or SKP2, suggesting that its interactions are specific to the components of the neddylation pathway (data not shown).
To confirm these interactions occur in vivo, we performed immunoprecipitation experiments using a SCCRO monoclonal antibody that is highly sensitive and specific for SCCRO. Western blots on the immunoprecipitates showed that SCCRO binds to Cul1, ROC1, and CAND1 (Fig. 1D, left panel). Similarly, reciprocal immunoprecipitation for ROC1 followed by Western blotting confirmed the interaction with SCCRO as well as CAND1, Cul1 and SKP2 (Fig. 1D, right panel). While SCCRO did not interact with SKP2, ROC1 bound, as expected.
To map binding domains, we created a series of SCCRO deletions and point mutants as GST fusions, including those involving the N-terminal UBA (amino acids 8–45) and C-terminal DCUN1 domain (amino acids 60–259) (Fig. 1, B, C, and E, and supplemental Fig. 2). Whereas the N-terminal deletions (SCCROΔ1–33, SCCROΔ1–45, and SCCROΔ1–82) retained binding, C-terminal deletions (SCCROΔ151–259 and SCCROΔ210–259) lost detectable binding to CAND1, Cul-ROC1, and Ubc12, suggesting that these proteins interact with the C-terminal 49 amino acids within the DCUN1 domain of SCCRO (Fig. 1, B and C, lane 3 and 4, and E). Point mutations were generated in highly conserved residues within the C-terminal region of SCCRO and tested for binding to cullins, CAND1, and Ubc12. SCCRO-D241N lost detectable binding to CAND1 and Cul-ROC1 complex but not to Ubc12, suggesting that Ubc12 and CAND1/Cul-ROC1 bind to different regions within the C terminus of SCCRO (Fig. 1B, lane 6). Our findings using a biochemical approach are consistent with results from structure-based analyses of Caenorhabditis elegans and Saccharomyces cerevisiae DCN-1/Dcn1p where mutations in a similar residue in the SCCRO paralogue DCN1 (Asp-259) also resulted in loss of binding to cullin-ROC1 (23–25).
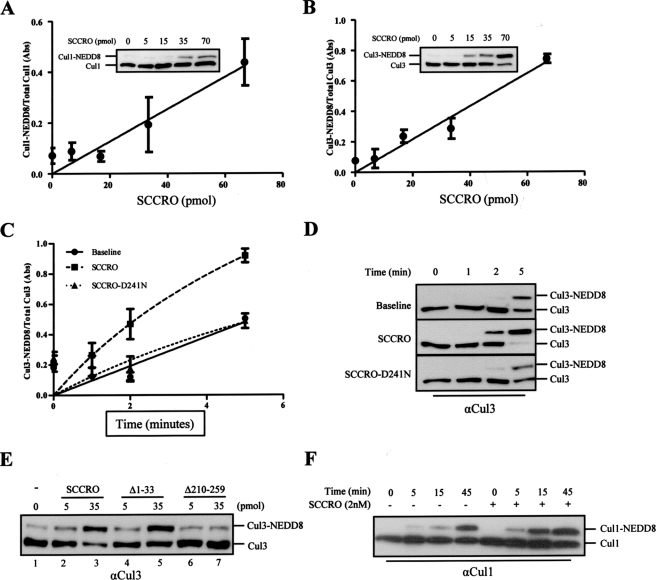
SCCRO Augments Cullin Neddylation—Recently it was reported that DCN-1/Dcn1p, the C. elegans and S. cerevisiae orthologs of SCCRO, facilitate cullin neddylation (23–25). To test if the human ortholog also promotes cullin neddylation, we performed in vitro neddylation assays in the presence of varying concentrations of SCCRO. Reactions containing NEDD8, recombinant APPBP1/Uba3 (E1), Ubc12 (E2), ATP, and whole cell lysate from HeLa cells (as a source of cullin-ROC1 substrate) showed a dose-dependent increase in cullin neddylation with SCCRO (Fig. 2, A and B). A time course reaction showed that SCCRO also enhances the rate of cullin neddylation (Fig. 2, C and D).
FIGURE 2.
SCCRO augments Cullin neddylation in vitro. A and B, plot showing levels of Cul1 (A) and Cul3 (B) (mean ± S.E.) neddylation quantified by densitometry of Western blots from three independent in vitro neddylation reactions containing HeLa extract (as a source of cullin substrate), E1, E2, ATP, NEDD8, and a concentration gradient of SCCRO. The fraction of neddylated Cul1 (A) and Cul3 (B) increased with increasing SCCRO concentration. Representative Western blots from in vitro neddylation reactions are shown as insets. C, a plot showing the fraction of neddylated Cul3 (means ± S.E.) against time in minutes from three independent neddylation assays. SCCRO but not SCCRO-D241N enhances neddylation efficiency. D, representative Western blot from C showing Cul3 neddylation in presence of SCCRO or SCCRO-D241N. E, results from in vitro neddylation reaction supplemented with SCCRO (lanes 2 and 3) or SCCRO deletions showing SCCROΔ1–33 (lanes 4 and 5) but not the C-terminal deletion SCCROΔ210–259 (lanes 6 and 7) retained neddylation activity. F, Western blot for Cul1 from a time course in vitro reaction containing purified, bacterially expressed Cul1-ROC1 showing SCCRO enhances efficiency but is not required for neddylation.
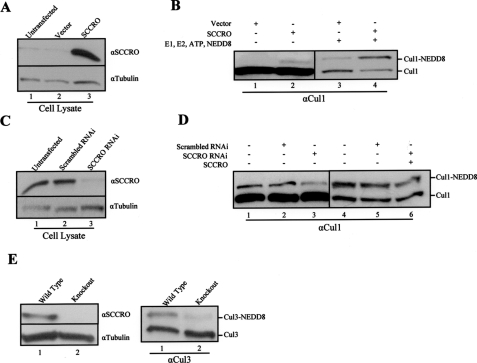
To determine whether binding is required for the observed functional effects, we supplemented neddylation reactions with SCCRO or selected SCCRO deletions. Whereas the N-terminal deletion (SCCROΔ1–33) enhanced the rate of Cul3 neddylation to levels similar to SCCRO, the C-terminal deletion (SCCROΔ210–259) that loses binding to Ubc12 and Cul-ROC1 failed to augment cullin neddylation beyond basal levels (Fig. 2E). Consistent with these findings, we found that SCCRO, but not the loss of cullin binding mutant SCCRO-D241N, increased neddylation efficiency beyond basal levels (Fig. 2, C and D). These findings suggest that the effects of SCCRO on cullin neddylation require its interaction with Cul-ROC1 and/or Ubc12 and implicate it as a component of the neddylation E3. Furthermore, cullin neddylation increased with transient expression of HA-SCCRO and decreased with knockdown of SCCRO by RNAi (Fig. 3B). The decrease in cullin neddylation as a result of SCCRO protein knockdown by RNAi was rescued by the addition of recombinant SCCRO to the lysate (Fig. 3D).
FIGURE 3.
SCCRO augments cullin neddylation in vivo. A, Western blot on HeLa lysates showing elevated SCCRO protein levels in SCCRO transfected (lane 3) relative to untransfected (lane 1) or vector-transfected (lane 2) cells. B, Western blot showing a higher level of neddylated cullins in lysates from SCCRO transfected (lane 2) relative to vector-transfected cells (lane 1). Western blot on HeLa lysates from B after addition of neddylation components (E1, E2, NEDD8, and ATP) showing increased neddylated Cul1 levels in SCCRO-transfected (lane 4) relative to empty vector (lane 3)-transfected cells. C, Western blot on lysates from SCC15 cells showing a decrease in SCCRO protein levels in cells transfected with specific RNAi against SCCRO (lane 3) relative to untransfected (lane 1) or scrambled RNAi-transfected cells (lane 2). D, in vitro neddylation reaction of the same lysates showing decreased Cul1 neddylation in SCCRO-RNAi transfected (lane 3) compared with untransfected (lane 1) or scrambled RNAi (lane 2)-transfected cells. The addition of recombinant SCCRO to the lysate from SCCRO RNAi-transfected SCC15 cells (lane 6) recovers Cul1 neddylation to levels observed in controls (lanes 4 and 5). E, Western blot showing the absence of detectable SCCRO protein in testis lysates from SCCRO–/– mice (left panel, lane 2) in contrast to a SCCRO+/+ (left panel, lane 1) litter mate control. The same lysates were subjected to neddylation assays (right panel) showing a significant decrease in neddylated Cul3 levels in SCCRO–/– mice (lane 2).
SCCRO Is Not Required for Neddylation in Vitro—Several in vitro studies suggest that ROC1 functions as an E3 ligase and is sufficient to promote neddylation by itself (26–29) To determine the effect of SCCRO on cullin neddylation, we performed a time course reaction using recombinant purified components (E1, E2, ATP, NEDD8, and Cul1-ROC1). Although Cul1 neddylation occurred in its absence, the addition of SCCRO enhanced the rate of cullin neddylation (Fig. 2F). This is consistent with published reports showing cullin neddylation occurs in the absence of SCCRO in vitro (26, 28). Neddylation is reduced in testis lysates from SCCRO–/– mice (Fig. 3E, right panel) as well as in C. elegans and S. cerevisiae where DCN-1/Dcn1p knockouts lose neddylation activity (24) Combined, these findings suggest that although SCCRO is not required for neddylation in vitro, it may be required in vivo.
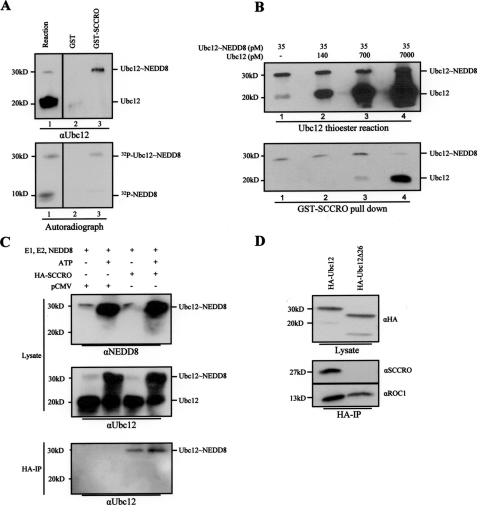
SCCRO Preferentially Interacts with Ubc12 Thioester Intermediate—The mechanisms and reaction processivity involved in the conjugation of Ub and Ublp are highly conserved. A mutually exclusive interaction between E1 and E3 with E2 is a key aspect in maintaining reaction processivity. Accordingly, an E2 conjugating enzyme must dissociate from its cognate E1 before binding to E3 (30). One way this is achieved is the differential affinity of E1 and E3 for free and conjugated E2. Several studies show that the affinity of E2 for E1 (and E3 to E2) is higher when the latter is loaded with Ub or Ublp (13, 31, 32). If SCCRO functions as a component of the neddylation E3, it should have greater affinity for Ubc12∼NEDD8 thioester over free Ubc12. GST-SCCRO pulldown from a Ubc12 thioester reaction followed by Western blot for Ubc12 showed that SCCRO preferentially binds to Ubc12∼NEDD8 thioester even in the presence of a large excess of free Ubc12 (Fig. 4A, lane 1 and 3). To quantify the differences in binding affinity of SCCRO for Ubc12 and Ubc12∼NEDD8, competition experiments were performed. GST-SCCRO pulldown assays from a mixture containing Ubc12∼NEDD8 (generated and purified as discussed under “Experimental Procedures”) and a gradient of free Ubc12 followed by Western blot showed that SCCRO preferentially binds to Ubc12∼NEDD8 even in the presence of a ≥20-fold excess of free Ubc12 (Fig. 4B, lane 3). Similarly, when lysates from HA-SCCRO-transfected HeLa cells that were subjected to in vitro thioester reaction (Fig. 4C, top and middle panel) were immunoprecipitated with anti-HA antibody and probed for Ubc12, preferential binding of SCCRO to Ubc12∼NEDD8 was detected (Fig. 4C, bottom panel). The preferential interaction with Ubc12∼NEDD8 thioester over free Ubc12 suggests that SCCRO conforms to the conserved processivity paradigm of Ub and Ublp pathway and supports its role as a component of the E3 complex for neddylation.
FIGURE 4.
SCCRO interacts with the unique N terminus of Ubc12∼NEDD8. A, Western blot for Ubc12 after a thioester reaction showing generation of Ubc12∼NEDD8 thioester (top panel, lane 1, upper band). GST (lane 2) and GST-SCCRO (lane 3) pulldown on the same reaction products showing preferential interaction of SCCRO with Ubc12∼NEDD8 despite a large excess of free Ubc12 (top panel, lane 3). Autoradiograph of products from a thioester reaction containing E1, E2, ATP, and 32P-labeled PK-NEDD8 showing Ubc12∼NEDD8 (bottom panel, lane 1, upper band) and free NEDD8 (bottom panel, lane 1, lower band). GST (lane 2) or GST-SCCRO (lane 3) pulldown assays on thioester reaction components showing SCCRO binds to Ubc12∼NEDD8 but not to free NEDD8 (bottom panel, lane 3) B, Western blot showing levels of Ubc12∼NEDD8 and Ubc12 from a thioester reaction supplemented with varying concentrations of free Ubc12 after quenching with EDTA (see “Experimental Procedures”; top panel). The same reactions were subjected to pulldown assays using limiting amounts of GST-SCCRO (70 pm) showing preferential binding to Ubc12∼NEDD8 even in the presence of 20-fold excess of free Ubc12 (bottom panel). C, Western blots for NEDD8 (top panel) and Ubc12 (middle panel) on thioester reactions with and without ATP in lysates from HeLa cells transfected with pCMV or pCMV-SCCRO showing lower levels of Ubc12∼NEDD8 in reactions not containing ATP (compare first and third lanes with the second and fourth lanes). HA immunoprecipitation of the same reaction mix showing preferential binding of SCCRO (in limiting concentrations) to Ubc12∼NEDD8 (bottom panel, third and fourth lanes), even in the presence of a large excess of free Ubc12 (bottom panel, third lane). D, Western blot for HA after thioester reactions on HeLa cell lysates transfected with HA-Ubc12 or HA-Ubc12Δ26 showing equal amounts of NEDD8 thioester formation after 10 and 30 min of incubation, respectively. HA immunoprecipitation on the same reaction products followed by Western blot with the indicated antibody showing SCCRO interacts with full-length Ubc12∼NEDD8 but not with Ubc12Δ26∼NEDD8, whereas ROC1 interacts with both (bottom right).
SCCRO Interacts with the Unique N Terminus of Ubc12—A second mechanism that ensures processivity in Ub and Ublp conjugation results from the presence of overlapping binding sites on E2 for E1 and E3, making their interactions mutually exclusive (30). Like all E2s, Ubc12 contains a ∼150 residue catalytic core domain. However, it is distinguished from other E2s by the presence of a unique N-terminal extension (33). Studies have shown that both the N-terminal extension and the catalytic core domain of Ubc12 are involved in its interaction with the NEDD8∼E1 (33). ROC1 binds to the catalytic core domain of Ubc12, with no known role for the N-terminal extension in the Ubc12-ROC1 interaction (11, 34) To determine whether SCCRO binds to the unique N-terminal region of Ubc12 or to the catalytic core domain, we performed HA immunoprecipitation on lysates from HeLa cells transfected with either HA-Ubc12 or HA-Ubc12Δ1–26 (N-terminal deleted Ubc12) and probed for SCCRO on a Western blot. Because SCCRO preferentially interacts with the Ubc12∼NEDD8 thioester, we performed a thioester reaction on the lysate to generate NEDD8 loaded HA-Ubc12 and HA-Ubc12Δ1–26 before immunoprecipitation (Fig. 4D, top). Because the efficiency of thioester formation is lower for Ubc12Δ1–26 deletion, we allowed the reaction adequate time to run to completion so as to generate an equivalent amount of the NEDD8 thioester. Even though longer incubation time was required to generate HA-Ubc12Δ1–26∼NEDD8, this interaction was reducible with DTT, suggesting the presence of a thioester bond (data not shown). Despite equal loading of NEDD8 to Ubc12 and Ubc12Δ1–26 (Fig. 4D, top), only HA-Ubc12 pulled down SCCRO (Fig. 4D, middle). However, ROC1, which is known to interact with the conserved core domain of Ubc12, bound to both HA-Ubc12 and HA-Ubc12Δ1–26 (Fig. 4D, bottom). These observations suggest that the binding site of SCCRO on Ubc12 (E2) overlaps with the NEDD8∼E1. Moreover, unlike ROC1 which interacts with the conserved core domain of Ubc12 (E2), SCCRO interacts with its unique N-terminal extension, raising the possibility that SCCRO-Ubc12 interaction is specific to the neddylation pathway. This is further strengthened by lack of interaction between SCCRO and other E2s (Ubc9) in GST pulldown assays (data not shown). Taken together, these observations suggest that SCCRO conforms to the conserved reaction processivity paradigms, further supporting its candidacy as a component of the E3 complex.
SCCRO Binds to CAND1 Only When It Is in Complex with Cul1-ROC1—Reflecting the increasing complexity of protein regulation, the neddylation pathway is also more complex in higher organisms. For example, in contrast to S. cerevisiae, where CAND1 is absent, in higher organisms unneddylated cullin-ROC1 exists almost exclusively in complex with CAND1 (11, 35, 36). Binding to CAND1 inhibits cullin neddylation and subsequent ubiquitination E3 complex assembly. Once neddylated, cullin-ROC1 is released from CAND1 and forms active E3 ubiquitination complexes. Analysis of the crystal structure shows that CAND1 binds to Cul1-ROC1 in a head to tail arrangement, burying the otherwise solvent-exposed lysine residue on Cul1 and making it inaccessible to the neddylation machinery. In contrast, a β-hairpin motif from CAND1 directly binds to two helices in Cul1 that are involved in its interaction with SKP1. In this arrangement it is possible for neddylation of cullins to proceed if modifications expose the target lysine, whereas binding to SKP1 would require dissociation of CAND1.
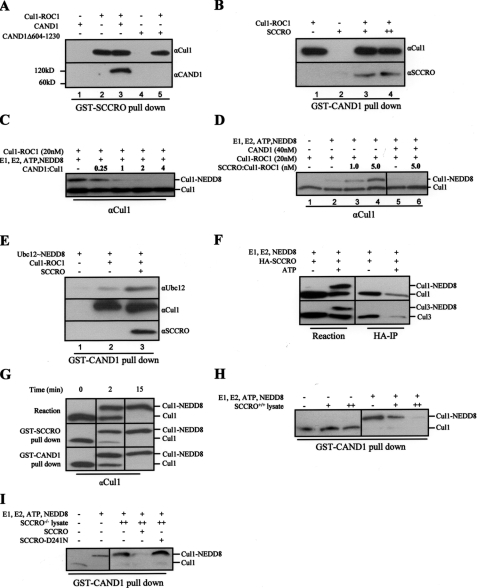
Given that SCCRO interacts with Cul-ROC1 and CAND1, we aimed to determine whether SCCRO binds to CAND1 directly or indirectly through its interaction with Cul-ROC1. To begin to define the steric interactions, we looked for the presence of SCCRO in exclusive complexes with either Cul1-ROC1 or CAND1 in cellular systems using gel filtration analyses. SCCRO was not found in fractions containing significant amounts of CAND1, Cul1, or ROC1, suggesting that the majority of endogenous cellular SCCRO is not stably associated with CAND1 or cullin-containing complexes, thereby limiting our ability to draw conclusions about the individual protein interactions (data not shown). GST-SCCRO pulldown assays using purified proteins showed that CAND1 only interacts with SCCRO when in complex with Cul1-ROC1 (Fig. 5A, compare lanes 1 and 3). A deletion of CAND1 (CAND1Δ604–1230), which loses binding to Cul1-ROC1, does not interact with SCCRO (Fig. 5A, lane 4 and 5). In a reciprocal pulldown assay, GST-CAND1 only pulls down SCCRO when the latter is in complex with Cul1-ROC1 (Fig. 5B, lane 3 and 4). These findings suggest that the interaction of SCCRO with CAND1 is mediated by Cul1-ROC1. Moreover, the binding stoichiometry between SCCRO and Cul1-ROC1 was not altered by the presence of CAND1 (data not shown). Given that in cells unneddylated cullins exist exclusively as ternary complexes with CAND1 and ROC1 and that SCCRO preferentially interacts with unneddylated cullins (see below), it is likely that SCCRO binds to the cullin-ROC1 complexes before they are released by CAND1. These observations raised the possibility that SCCRO may be involved in overcoming CAND1 inhibition of cullin neddylation.
FIGURE 5.
SCCRO does not overcome the inhibitory effects of CAND1 on neddylation. A, Western blot on GST-SCCRO pulldown products after incubation with purified, bacterially expressed Cul1-ROC1 and CAND1 showing CAND1 is pulled down by SCCRO only when it is complexed with Cul1-ROC1 (lanes 1 and 3). A CAND1 deletion (CAND1Δ604–1230) that loses binding to cullins is not pulled down by SCCRO (lane 4 and 5). B, Western blot from reciprocal GST-CAND1 pulldown assay showing interaction of CAND1 with SCCRO requires Cul1-ROC1 (lanes 2–4). C, Western blot for Cul1 following in vitro neddylation reaction using purified recombinant components showing complete inhibition of neddylation in the presence of 2-fold or higher molar excess of CAND1. D, Western blot on products from an in vitro neddylation reaction confirming increasing Cul1 neddylation with increasing amounts of SCCRO (lanes 2–4). The addition of CAND1 inhibits Cul1 neddylation (lane 5), which is not rescued even by 5-fold molar excess of SCCRO (lane 6; same blot, excess lanes removed). E, Western blot showing GST-CAND1 pulldown products after incubation with purified, bacterially expressed Cul1-ROC1, Ubc12∼NEDD8, and SCCRO. CAND1 interacts with Ubc12∼NEDD8 only when Cul1-ROC1 is present (lanes 1 and 2), and this interaction is enhanced by the presence of SCCRO (lane 3). F, Western blot on lysates from HA-SCCRO-transfected HeLa cells subjected to neddylation reaction showing an increase in cullin neddylation with the addition of ATP (lanes 1 and 2). HA immunoprecipitation (IP) of the same reaction products showed SCCRO preferentially interacts with unneddylated cullins (lanes 3 and 4). G, Western blot on products from an in vitro neddylation reaction using purified recombinant components showing an increase in the fraction of neddylated Cul1 with increasing reaction time (top panel). Western blot on the same reaction after GST-SCCRO (middle panel) and GST-CAND1 (bottom panel) pulldown assays showing that SCCRO and CAND1 interact with both free and neddylated cullins (same blot, excess lanes removed). H, Western blot on products from pulldown assays after GST-CAND1 complexed with either NEDD8-Cul1-ROC1 or free Cul1-ROC1 was incubated with increasing amounts of testis lysate from SCCRO+/+ mice showing release of NEDD8-Cul1-ROC1 but not unneddylated Cul1-ROC1. I, Western blot on products from pulldown assays after GST-CAND1 complexed with either NEDD8-Cul1-ROC1 or free Cul1-ROC1 was incubated with increasing amounts of testis lysate from SCCRO–/– mice showing release of Cul1-ROC1-NEDD8 only with addition of SCCRO and not SCCRO-D241N (same blot, excess lanes removed).
SCCRO Does Not Overcome Inhibition of Cullin Neddylation by CAND1—Given the observed binding interactions, we wanted to determine whether SCCRO could overcome the inhibitory effects of CAND1 on cullin neddylation. We confirmed previous findings that CAND1 inhibits cullin neddylation in an in vitro purified recombinant system (Fig. 5C) (12, 35). To assess the ability of SCCRO to overcome CAND1 inhibition, we preincubated Cul1-ROC1 with the minimal concentration of CAND1 required to completely inhibit cullin neddylation. The addition of up to a 5-fold molar excess of SCCRO failed to overcome the inhibitory effects of CAND1 on cullin neddylation (Fig. 5D). To determine whether the effects of CAND1 on cullin neddylation were related to recruitment of Ubc12∼NEDD8, we performed pulldown assays. We found CAND1-Cul1-ROC1 is able to bind to Ubc12∼NEDD8. This binding is enhanced by the addition of SCCRO (Fig. 5E, lane 3). However, assembly of the protein complex was not sufficient to overcome CAND1 inhibition. As expected, this complex was not detected in HeLa lysates where its assembly should lead to efficient transfer of the NEDD8 to cullins and dissociation of CAND1 (data not shown). These observations suggest that SCCRO is not sufficient to overcome the inhibitory effects of CAND1 and raise the possibility of additional factors involved in CAND1 release.
Neddylated Cul1 Can Bind to CAND1 and SCCRO in Vitro—To assess if SCCRO binding to Cul1-ROC1 is affected by neddylation, lysates from HeLa cells transfected with HA-SCCRO were subjected to in vitro neddylation reaction with and without ATP (Fig. 5F, left panel). Western blot analysis showed increased cullin neddylation in reactions containing ATP. When HA immunoprecipitation was performed on these lysates, only unneddylated cullins were pulled down (Fig. 5F, right panel). In contrast, binding assays using purified proteins showed that both neddylated and unneddylated Cul1-ROC1 were pulled down by GST-SCCRO and GST-CAND1 (Fig. 5G). These results suggest that neddylation of cullins may not be sufficient by itself to dissociate CAND1. Because we could not detect any binding between neddylated cullins and CAND1 in lysates, a factor in the lysate may be required to release neddylated cullins. To address this issue, GST-CAND1 complexed with either neddylated or unneddylated Cul1-ROC1 was incubated with lysate. Incubation with HeLa lysate or SCCRO+/+ mice testis lysate resulted in the dissociation of neddylated but not unneddylated cullins from CAND1 (Fig. 5H). This dissociation was not due to competition with native cullin complexes, as these complexes were not detected on the GST-CAND1 pulldown assays (data not shown). In contrast, incubation with testis lysate from SCCRO–/– mice failed to dissociate GST-CAND1 from neddylated Cul1-ROC1. The addition of recombinant SCCRO, but not SCCRO-D241N, to the SCCRO–/– lysate allowed dissociation of neddylated Cul1-ROC1 from CAND1 (Fig. 5I) These data suggest that neddylated cullins are dissociated from CAND1 by a factor in lysate and that SCCRO may be required for this dissociation.
DISCUSSION
Hershko and Ciechanover (1) define ubiquitin ligases as enzymes that bind directly or indirectly to specific protein substrates and promote the transfer of Ub directly or indirectly from a thioester intermediate to amide linkages with proteins or polyubiquitin chains. Our data suggest that SCCRO is part of a complex that functions as the E3 for neddylation. The neddylation E3 ligase complex (Cul-ROC1-SCCRO) shares a modular architecture similar to CRL-containing ubiquitin ligases. CRL complexes have a constant catalytic module (ROC1 bound to E2 thioester) that is preserved in the neddylation E3 complex. SCCRO has a unique regulatory role by helping to recruit the Ubc12∼NEDD8 thioester to the neddylation E3 complex. Because SCCRO promotes cullin neddylation, which serves as a signal for assembly of ubiquitination E3 complexes, factors controlling SCCRO likely regulate ubiquitination by CRL-containing E3 complexes. Given the diversity of proteins regulated by CRL-containing E3s, it is not surprising that dysregulation of Dcn1p/DCN-1 results in severe developmental defects in S. cerevisiae, C. elegans, and mice5 and that SCCRO appears to be a key target activated by amplification in a vast array of human cancers (18).
Assembly of E3 complexes is a key regulatory mechanism for ubiquitination. CAND1 affects the assembly of both the neddylation E3 complex as well as the CRL complexes. Several studies as well as our own findings suggest that unneddylated cullins exist almost exclusively in complexes with CAND1, whereas neddylated cullins are predominantly in active ubiquitination E3 complexes (10–12). Although it is accepted that neddylation serves as a signal for CAND1 dissociation from Cul1-ROC1 and subsequent assembly of ubiquitination E3 complexes, the precise mechanisms leading to the release remain ill defined. The finding that only endogenous Cul1-ROC1 (immunoprecipitated from cells) and not the recombinant protein bound to CAND1 can undergo neddylation leads to speculation that a factor(s) in lysate allows release of CAND1 inhibition (10, 12). Several experimental observations from our studies may begin to help explain the mechanisms involved in CAND1 release. First, we found that the entire E3 neddylation complex (including Ubc12∼NEDD8 and SCCRO) can assemble onto purified Cul1-ROC1, when it is in complex with CAND1. However, this is neither sufficient to promote cullin neddylation nor to induce CAND1 dissociation, suggesting that the assembly of the neddylation E3 complex by itself is not sufficient to overcome the inhibitory effects of CAND1 and the subsequent CRL assembly. However, the CAND1-Cul1-ROC1-Ubc12∼NEDD8 complex is not detectable in lysates, consistent with the fact that the assembly of SCCRO and Ubc12 onto the CAND1-Cul1-ROC1 results in rapid neddylation and dissociation of the complex. This is supported by the increase in neddylation of endogenous cullins that are thought to be bound to CAND1 consequent to the addition of Ubc12 and/or SCCRO to lysates. Given that SCCRO is not required for neddylation in purified systems but is important in SCCRO–/– mice5 and DCN-1/Dcn1p knockouts in C. elegans and S. cerevisiae raises the possibility that SCCRO may be required to overcome CAND1 associated inhibition of cullin neddylation in vivo where unneddylated cullins are in complex with CAND1. Consistent with this hypothesis, the addition of recombinant SCCRO, but not Ubc12∼NEDD8, can rescue neddylation in lysates from both DCN-1, Dcn1p, and SCCRO knockouts in C. elegans, S. cerevisiae, and mice,5 respectively.
The prevailing theory has been that neddylation makes the binding between CAND1 and cullins unfavorable. This led to speculation that either neddylation serves as a signal for the release or occurs after dissociation from CAND1. We found that, in contrast to cellular lysates where they exist in exclusive complex with unneddylated cullins, CAND1 and SCCRO can bind to both neddylated and unneddylated cullins in assays using recombinant proteins. This raises the possibility that neddylation by itself, although required, may not be sufficient for CAND1 dissociation. When incubated with HeLa or lysate from SCCRO+/+ mouse testis, CAND1 is dissociated from neddylated Cul1-ROC1 but not unneddylated Cul1-ROC1, suggesting that cellular factors are involved in the release of neddylated cullins from CAND1. In contrast, neddylated Cul1 was not released by CAND1 when incubated in lysates from SCCRO–/– mice unless supplemented with recombinant SCCRO (but not SCCRO-D241N). Factors that compete for binding of neddylated cullins (i.e. SKP1/SKP2) dissociate CAND1 from Cul1-ROC1 only in lysates (which contains wild type SCCRO) but not in purified protein systems, suggesting that SCCRO is required for release. The combined results from these experiments suggest that SCCRO plays a critical role in both cullin neddylation and CAND1 release. Our findings indicate that SCCRO by itself is not sufficient for cullin neddylation and CAND1 release, and a factor(s) in lysate is required in these processes. Goldenberg et al. (12) suggest that this factor may be a small molecule that binds the CAND1-Cul1-ROC1 complex to modify its structure, exposing the neddylation site on Cul1. Because CAND1 binds to both neddylated and unneddylated cullins with equal affinity, neddylation in itself, although required, may not be sufficient for CAND1 release.
Based on these findings, we propose a two-step model leading to cullin neddylation and CAND1 release. We propose that neddylation occurs through the assembly of the competent neddylation E3 complex onto cullin-ROC1-CAND1, including SCCRO, Ubc12∼NEDD8, and an unidentified CAND1 opening factor present in lysate. In this model neddylation serves as a signal for recruitment of release factors required to dissociate CAND1. The release factor remains to be defined but can include a novel binding protein or posttranslational modifications in reaction components. Although provocative, further studies are required to validate this model. The other critical question that remains is what is downstream of SCCRO. Katanin is a key target in C. elegans and yeast, as its levels are altered in DCN1/DCN1p knockouts. The identification of CRL targets downstream of SCCRO is more complex given the presence of multiple paralogues that retain neddylation activity and the complex phenotype in SCCRO–/– mice.
Supplementary Material
Acknowledgments
We thank Christopher Lima and Andrew Koff for many helpful discussions and critical reading of the manuscript, Zhen-Qiang Pan (The Mount Sinai School of Medicine) for PK-NEDD8 protein, Nikola Pavletich (Memorial Sloan-Kettering Cancer Center) for Cul1-ROC1 and Ubc12 constructs, Paul Tempst and Hediye Erdjument-Bromage for assistance with mass spectroscopy sequencing, Chris Sander and Boris Reva for assistance with sequence analysis, Aarti Ravikumar, Christina Cordeiro, Anne Conway, Peter Meisel, and Sarina Bains for technical assistance, Nancy Bennett and Chan-Bene Lin for editorial assistance, and Drs. Valerie Rusch and Jatin Shah for support and encouragement.
This work was supported in part a George H. A. Clowes Memorial Research Career Development Award from the American College of Surgeons (to B. S.), a Clinical Innovator Award from Flight Attendant Medical Research Institute (to B. S. and Y. R.), the T. J. Martel Fund (to B. S. and V. Rusch), the Falcone Fund (to B. S. and J. Shah), an National Institutes of Health training grant (to L. C.), and a Matheson Fellowship (to Y. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: Ub, ubiquitin; CRL, Cullin RING ligases; Ublp, ubiquitin-like protein; SCCRO, squamous cell carcinoma-related oncogene; E1, ubiquitin (or Nedd8)-activating enzyme; E2, ubiquitin (or Nedd8) carrier protein; E3, ubiquitin (or Nedd8)-protein isopeptide ligase; RNAi, RNA-mediated interference; GST, glutathione S-transferase; MALDI-reTOF, matrix-assisted laser desorption/ionization reflectron time-of-flight; HA, hemagglutinin; DTT, dithiothreitol.
A. Kaufman, L. Hyurn, A. Conway, A. Kim, D. Pham, S. Talbot, S. Broderick, G. Huang, P. Morris, K. Manova, G. Hannicut, Y. Ramanathan, and B. Singh, manuscript in preparation.
References
- 1.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Hicke, L. (2001) Nat. Rev. Mol. Cell Biol. 2 195–201 [DOI] [PubMed] [Google Scholar]
- 3.Kerscher, O., Felberbaum, R., and Hochstrasser, M. (2006) Annu. Rev. Cell Dev. Biol. 22 159–180 [DOI] [PubMed] [Google Scholar]
- 4.Mukhopadhyay, D., and Riezman, H. (2007) Science 315 201–205 [DOI] [PubMed] [Google Scholar]
- 5.Aguilar, R. C., and Wendland, B. (2003) Curr. Opin. Cell. Biol. 15 184–190 [DOI] [PubMed] [Google Scholar]
- 6.Pickart, C. M., and Eddins, M. J. (2004) Biochim. Biophys. Acta 1695 55–72 [DOI] [PubMed] [Google Scholar]
- 7.Petroski, M. D., and Deshaies, R. J. (2005) Nat. Rev. Mol. Cell Biol. 6 9–20 [DOI] [PubMed] [Google Scholar]
- 8.Hotton, S. K., and Callis, J. (2008) Annu. Rev. Plant Biol. 59 467–489 [DOI] [PubMed] [Google Scholar]
- 9.Wu, K., Chen, A., and Pan, Z. Q. (2000) J. Biol. Chem. 275 32317–32324 [DOI] [PubMed] [Google Scholar]
- 10.Liu, J., Furukawa, M., Matsumoto, T., and Xiong, Y. (2002) Mol. Cell 10 1511–1518 [DOI] [PubMed] [Google Scholar]
- 11.Zheng, J., Yang, X., Harrell, J. M., Ryzhikov, S., Shim, E. H., Lykke-Andersen, K., Wei, N., Sun, H., Kobayashi, R., and Zhang, H. (2002) Mol. Cell 10 1519–1526 [DOI] [PubMed] [Google Scholar]
- 12.Goldenberg, S. J., Cascio, T. C., Shumway, S. D., Garbutt, K. C., Liu, J., Xiong, Y., and Zheng, N. (2004) Cell 119 517–528 [DOI] [PubMed] [Google Scholar]
- 13.Kawakami, T., Chiba, T., Suzuki, T., Iwai, K., Yamanaka, K., Minato, N., Suzuki, H., Shimbara, N., Hidaka, Y., Osaka, F., Omata, M., and Tanaka, K. (2001) EMBO J. 20 4003–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakata, E., Yamaguchi, Y., Miyauchi, Y., Iwai, K., Chiba, T., Saeki, Y., Matsuda, N., Tanaka, K., and Kato, K. (2007) Nat. Struct. Mol. Biol. 14 167–168 [DOI] [PubMed] [Google Scholar]
- 15.Wimuttisuk, W., and Singer, J. D. (2007) Mol. Biol. Cell 18 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D. A., Wei, N., and Deshaies, R. J. (2001) Science 292 1382–1385 [DOI] [PubMed] [Google Scholar]
- 17.Wei, N., and Deng, X. W. (2003) Annu. Rev. Cell Dev. Biol. 19 261–286 [DOI] [PubMed] [Google Scholar]
- 18.Sarkaria, I., P, O. c., Talbot, S. G., Reddy, P. G., Ngai, I., Maghami, E., Patel, K. N., Lee, B., Yonekawa, Y., Dudas, M., Kaufman, A., Ryan, R., Ghossein, R., Rao, P. H., Stoffel, A., Ramanathan, Y., and Singh, B. (2006) Cancer Res. 66 9437–9444 [DOI] [PubMed] [Google Scholar]
- 19.Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., Conaway, R. C., Conaway, J. W., Harper, J. W., and Pavletich, N. P. (2002) Nature 416 703–709 [DOI] [PubMed] [Google Scholar]
- 20.Singh, B., Gogineni, S. K., Sacks, P. G., Shaha, A. R., Shah, J. P., Stoffel, A., and Rao, P. H. (2001) Cancer Res. 61 4506–4513 [PubMed] [Google Scholar]
- 21.Singh, B., Reddy, P. G., Goberdhan, A., Walsh, C., Dao, S., Ngai, I., Chou, T. C., P, O. C., Levine, A. J., Rao, P. H., and Stoffel, A. (2002) Genes Dev. 16 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh, B., Stoffel, A., Gogineni, S., Poluri, A., Pfister, D. G., Shaha, A. R., Pathak, A., Bosl, G., Cordon-Cardo, C., Shah, J. P., and Rao, P. H. (2002) Am. J. Pathol. 161 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang, X., Zhou, J., Sun, L., Wei, Z., Gao, J., Gong, W., Xu, R. M., Rao, Z., and Liu, Y. (2007) J. Biol. Chem. 282 24490–24494 [DOI] [PubMed] [Google Scholar]
- 24.Kurz, T., Ozlu, N., Rudolf, F., O'Rourke, S. M., Luke, B., Hofmann, K., Hyman, A. A., Bowerman, B., and Peter, M. (2005) Nature 435 1257–1261 [DOI] [PubMed] [Google Scholar]
- 25.Kurz, T., Chou, Y. C., Willems, A. R., Meyer-Schaller, N., Hecht, M. L., Tyers, M., Peter, M., and Sicheri, F. (2008) Mol. Cell 29 23–35 [DOI] [PubMed] [Google Scholar]
- 26.Kamura, T., Conrad, M. N., Yan, Q., Conaway, R. C., and Conaway, J. W. (1999) Genes Dev. 13 2928–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dharmasiri, S., Dharmasiri, N., Hellmann, H., and Estelle, M. (2003) EMBO J. 22 1762–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morimoto, M., Nishida, T., Nagayama, Y., and Yasuda, H. (2003) Biochem. Biophys. Res. Commun. 301 392–398 [DOI] [PubMed] [Google Scholar]
- 29.Sufan, R. I., and Ohh, M. (2006) Neoplasia 8 956–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eletr, Z. M., Huang, D. T., Duda, D. M., Schulman, B. A., and Kuhlman, B. (2005) Nat. Struct. Mol. Biol. 12 933–934 [DOI] [PubMed] [Google Scholar]
- 31.Huang, D. T., Paydar, A., Zhuang, M., Waddell, M. B., Holton, J. M., and Schulman, B. A. (2005) Mol. Cell 17 341–350 [DOI] [PubMed] [Google Scholar]
- 32.Siepmann, T. J., Bohnsack, R. N., Tokgoz, Z., Baboshina, O. V., and Haas, A. L. (2003) J. Biol. Chem. 278 9448–9457 [DOI] [PubMed] [Google Scholar]
- 33.Huang, D. T., Miller, D. W., Mathew, R., Cassell, R., Holton, J. M., Roussel, M. F., and Schulman, B. A. (2004) Nat. Struct. Mol. Biol. 11 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng, N., Wang, P., Jeffrey, P. D., and Pavletich, N. P. (2000) Cell 102 533–539 [DOI] [PubMed] [Google Scholar]
- 35.Hwang, J. W., Min, K. W., Tamura, T. A., and Yoon, J. B. (2003) FEBS Lett. 541 102–108 [DOI] [PubMed] [Google Scholar]
- 36.Oshikawa, K., Matsumoto, M., Yada, M., Kamura, T., Hatakeyama, S., and Nakayama, K. I. (2003) Biochem. Biophys. Res. Commun. 303 1209–1216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.