Abstract
TAK1 kinase is an indispensable intermediate in several cytokine signaling pathways including tumor necrosis factor, interleukin-1, and transforming growth factor-β signaling pathways. TAK1 also participates in stress-activated intracellular signaling pathways such as osmotic stress signaling pathway. TAK1-binding protein 1 (TAB1) is constitutively associated with TAK1 through its C-terminal region. Although TAB1 is known to augment TAK1 catalytic activity when it is overexpressed, the role of TAB1 under physiological conditions has not yet been identified. In this study, we determined the role of TAB1 in TAK1 signaling by analyzing TAB1-deficient mouse embryonic fibroblasts (MEFs). Tumor necrosis factor- and interleukin-1-induced activation of TAK1 was entirely normal in Tab1-deficient MEFs and could activate both mitogen-activated protein kinases and NF-κB. In contrast, we found that osmotic stress-induced activation of TAK1 was largely impaired in Tab1-deficient MEFs. Furthermore, we showed that the C-terminal 68 amino acids of TAB1 were sufficient to mediate osmotic stress-induced TAK1 activation. Finally, we attempted to determine the mechanism by which TAB1 activates TAK1. We found that TAK1 is spontaneously activated when the concentration is increased and that it is totally dependent on TAB1. Cell shrinkage under the osmotic stress condition increases the concentration of TAB1-TAK1 and may oligomerize and activate TAK1 in a TAB1-dependent manner. These results demonstrate that TAB1 mediates TAK1 activation only in a subset of TAK1 pathways that are mediated through spontaneous oligomerization of TAB1-TAK1.
TAK1 kinase is an indispensable intermediate of several innate immune signaling pathways including cytokines TNF2 and IL-1 as well as Toll-like receptors and intracellular bacterial sensor NOD-like receptor NOD1/2 pathways (1–5). In those pathways, TAK1 is recruited into the IκB kinase (IKK) complex through a polyubiquitin chain and activates transcription factor NF-κB (6). The innate stimuli-activated TAK1 also induces activation of transcription factor AP-1 through MAPKs such as c-Jun N-terminal kinase (JNK) and p38. NF-κB and AP-1 cooperatively modulate gene expression to induce inflammation and cell survival (7, 8). TAK1 is involved in several other signaling pathways, for example, in TGF-β signaling pathways, TAK1 participates in the non-Smad pathway by activating p38 and SnoN degradation (9, 10). TAK1 is critically involved in stress-activated cell signaling (11–13). Among the stress conditions, we found that osmotic stress-induced JNK activation requires TAK1 (12).
We have identified several TAK1-binding proteins including TAK1-binding protein 1 (TAB1) (14) and TAK1-binding protein 2/3 (TAB2/3) (15, 16). TAB1 and TAB2 are isolated by the yeast two-hybrid screening using TAK1 protein as a bait, and both endogenous TAB1 and TAB2 are coprecipitated with endogenous TAK1 in many types of cells. TAB2 and its homolog TAB3 are found to bind to ubiquitin and function as an adaptor tethering TAK1 to the IKK complex (17, 18). In contrast, the role of TAB1 in TAK1 signaling under the physiological setting has not yet been explored. In culture cells, TAB1 is found to be constitutively associated with TAK1 (19). Ectopic expression of TAB1 together with TAK1 induces TAK1 autophosphorylation and thereby activates TAK1 kinase in vitro (19). Only 68 amino acid residues of C-terminal TAB1 are essential and sufficient for binding to TAK1 and induction of autophosphorylation/activation of TAK1 (20).
Disruption of Tab1 causes embryonic lethality with several developmental dysregulations including failure of cardiovascular morphogenesis (21). Disruption of Tak1 also causes early embryonic lethality presumably because of its importance for regulating multiple cytokine signaling pathways (4, 22, 23). These facts raise the possibility that TAB1 may be involved in TAK1-mediated signaling pathways during development. However, Tab1-deficient embryos are largely normal by embryonic day 14.5 (21), which is different from Tak1-deficient embryos that are lethal by embryonic day 9.5 (4, 22, 23). This reveals that TAB1 is not essential for all of the TAK1-mediated signaling pathways, or functions of TAB1 can be compensated by other gene products. An earlier study using Tab1-deficient MEFs has reported that TNF-, and IL-1-induced activation of NF-κB, JNK and p38 is not affected by Tab1 deletion (4). However, Mendoza et al. (24) have recently reported that IL-1-induced TAK1 activation may be reduced in Tab1-deficient MEFs. Our goal is to define the essential roles of TAB1 under physiological conditions. We have recently generated a Tab1-floxed mouse line that allows us to investigate the role of TAB1 in several different tissues (25). In the current study, we started determining the TAB1-mediated signaling and investigated which types of TAK1 pathways require TAB1 by using TAB1-deficient MEFs prepared from our newly generated Tab1-floxed mice.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection—Heterozygous mice for the Cre-recombined allele of Tab1 were generated by crossing Sox2-Cre mice and Tab1-floxed (Tab1flox/flox) mice (25). The heterozygous mice were intercrossed, and Tab1 control and Tab1-deficient MEFs were isolated from wild type and homozygous mutant embryos at embryonic day 14.5 and spontaneously immortalized by the standard method. Preparation of TAK1+/+ and TAK1Δ/Δ MEFs were described previously (23). MEFs and 293 cells were cultured in Dulbecco's modified Eagle's medium with 10% bovine growth serum (Hyclone) and penicillin-streptomycin at 37 °C in 5% CO2. 293 cells were transfected with expression vectors for hemagglutinin (HA)-tagged TAK1 (pCMV-HA-TAK1) and TAB1 (pCMV-TAB1) as described previously (26).
Reagents—The reagents used were IL-1β (mouse recombinant; Roche Applied Science), TNFα, TGF-β (human recombinant, Roche), calyculin A, and okadaic acid (Calbiochem). The following polyclonal antibodies were used: TAK1 and Tab1 described previously (3), Thr(P)187 TAK1 (27) (Cell Signaling), JNK1 (FL), p38 (N-20), IκBα, p65 (Santa Cruz), phospho-IκB, phospho-p38 (Thr180/Tyr182), and AMPK (Cell Signaling). Rabbit monoclonal antibody phospho-AMPK (Cell Signaling) and mouse monoclonal antibodies phospho-JNK (Thr183/Tyr185) (Cell Signaling) and FLAG-M2 (Sigma) were also used.
Electrophoretic Mobility Shift Assay—The binding reactions contained radiolabeled 32P-NF-κB oligonucleotide probe (Promega), cell extracts, 4% glycerol, 1 mm MgCl2, 0.5 mm EDTA, 0.5 mm dithiothreitol, 50 mm NaCl, 10 mm Tris-HCl (pH 7.5), 500 ng of poly(dI-dC) (GE Healthcare), and 10 μg of bovine serum albumin to a final volume of 10 μl. The reaction mixtures were incubated at 25 °C for 15 min, separated by 5% (w/v) polyacrylamide gel, and visualized by autoradiography.
Immunoblotting—The cells were washed once with ice-cold phosphate-buffered saline, and whole cell extracts were prepared using lysis buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 12.5 mm β-glycerophosphate, 1.5 mm MgCl2, 2 mm EGTA, 10 mm NaF, 2 mm dithiothreitol, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 20 μm aprotinin, 0.5% Triton X-100). The cell extracts were resolved on SDS-PAGE and transferred to Hybond-P membranes (GE Healthcare). The membranes were immunoblotted with various antibodies, and the bound antibodies were visualized with horseradish peroxidase-conjugated antibodies against rabbit or mouse IgG using the ECL Western blotting system (GE Healthcare).
Retroviral Infection—Retroviral vectors for Tab1 (pMXs-neo-Tab1) was generated by inserting human TAB1 cDNAs into the retroviral vector pMXs-neo (28). To generate retroviral vectors for FLAG-TAB1N (pMXs-puro-FLAG-TAB1N), the ClaI-HincII fragment of pCMV-FLAG-TAB1 (encodes FLAG-tagged TAB1 N terminus 1–418 amino acids) (29) was inserted into pMXs-puro. To generate retroviral vectors for FLAG-TAB1C (pMXs-puro-FLAG-TAB1C), the FLAG tag was fused to C terminus of TAB1 (437–504 amino acids) and inserted into pMXs-puro. EcoPack293 cells (BD Biosciences) were transiently transfected with pMX-neo-TAB1, pMX-puro-FLAG-TAB1N or pMX-puro-FLAG-TAB1C. After 48 h of culture, the growth medium containing retrovirus was collected and filtered with a 0.45-μm cellulose acetate membrane to remove packaging cells. MEFs were incubated with the collected virus-containing medium with 8 μg/ml polybrene for 24 h. The uninfected cells were removed by G418 or puromycin selection.
Concentration-dependent Activation of TAK1—Cell lysates from Tab1+/+ and –/– MEFs were incubated and immunoprecipitated with anti-TAK1 at a high protein concentration (10 mg proteins/ml) or at a low protein concentration (2 mg protein/ml) at room temperature for 3 h. Immunoprecipitates were incubated in a kinase buffer (20 mm HEPES, pH 7.4, 1 mm dithiothreitol, 10 mm MgCl, 1 mm ATP) at 37 °C for 20 min and subjected to an immunoblot analysis and kinase assay described previously (12). Loading amounts of immunoprecipitates were adjusted to yield the same loading of TAK1.
RESULTS AND DISCUSSION
Conventional Tab1 knock-out mice have demonstrated that TAB1 is essential for proper embryogenesis (21). However, the roles of TAB1 at the molecular level in cell signaling and morphogenesis are still elusive. To define the in vivo role of TAB1 in several different cell types and tissues, we have recently generated the floxed Tab1 mouse (25). In this mouse with floxed Tab1, Cre-dependent recombination results in deletion of C-terminal amino acid residues 308–504 of TAB1 protein. Because TAB1 C terminus contains the TAK1-binding domain (20), the C-terminal truncated TAB1, if produced from the Cre recombined allele, should be functionally inactive in TAK1-mediated signaling pathways. To begin characterizing the role of TAB1, Cre-dependent DNA recombination was introduced to generate heterozygous mice for Tab1. Intercross of the resulted heterozygous mice for Tab1 was set up, and subsequently Tab1-deficient MEFs were isolated from the homozygous embryos for Cre-recombined Tab1 allele at embryonic day 14.5. These cells along with MEFs from control littermates were utilized to determine the essential role of TAB1 in TAK1 signaling pathways.
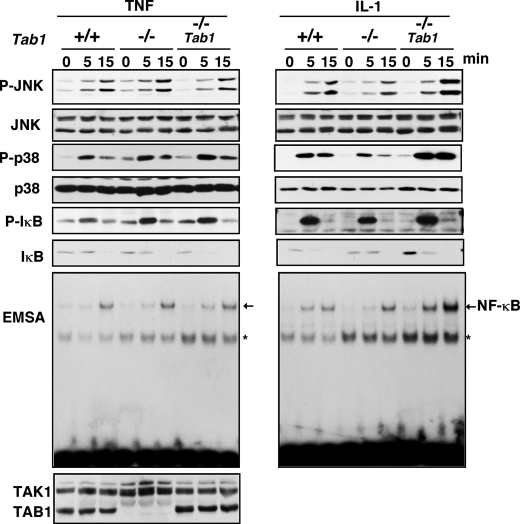
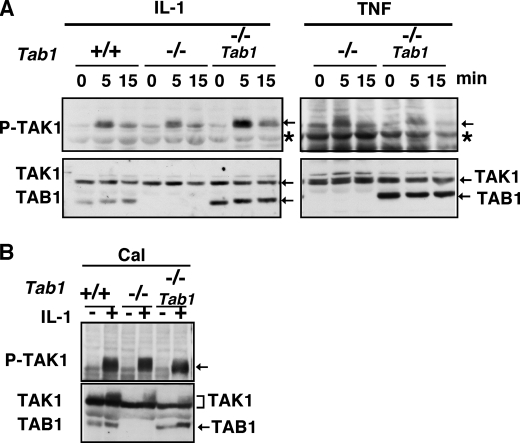
TAK1 is activated by IL-1 and TNF and plays a central role in inflammatory responses by activating JNK, p38, and NF-κB (30). We first examined whether TAB1 participates in IL-1- and TNF-induced TAK1 signaling pathway. Because it is a formal possibility that immortalization process may genetically and epigenetically alter cells, we generated Tab1 restored Tab1-deficient MEFs (Tab1-restored) to determine whether the alteration found is truly TAB1-dependent. To generate Tab1-restored cells, we infected Tab1-deficient MEFs with retrovirus expressing Tab1, and a pool of Tab1 expressing cells was used for the following experiments. We treated Tab1 control, -deficient, and -restored MEFs with IL-1 or TNF and determined the activation of JNK, p38, and NF-κB (Fig. 1). Inconsistent with the earlier study (4), we found that IL-1- and TNF-induced activation of JNK, p38, and NF-κB was not altered by TAB1 deficiency. We asked whether IL-1 and TNF could activate TAK1 in our Tab1-deficient MEFs. We detected an active form of TAK1 by using the Thr(P)187-specific TAK1 antibody (27). The levels of TAK1 activation in response to IL-1 or TNF were not significantly reduced by TAB1 deficiency (Fig. 2A). IL-1 and TNF activate TAK1 in MEFs not as strongly as in other types of cells such as 293 that we have previously shown (27, 31). Therefore, to further confirm the activation of TAK1, we pretreated MEFs with type 2A protein phosphatases inhibitor calyculin A, which inhibits down-regulation of TAK1 (27), and examined activation of TAK1 in response to IL-1. The activation of TAK1 in the presence of calyculin A was also not reduced by Tab1 deletion (Fig. 2B). Thus, TAB1 is dispensable for TNF- and IL-1-induced activation of TAK1.
FIGURE 1.
TAB1 is dispensable for TNF and IL-1 signaling pathways. Tab1 control (+/+), Tab1-deficient (–/–), and Tab1-restored (–/– Tab1) MEFs were stimulated with 20 ng/ml TNF or 5 ng/ml IL-1. Activation of JNK and p38 was detected with anti-phospho-JNK and anti-phospho-p38, and the total amounts of JNK and p38 were detected with anti-JNK and anti-p38. Activation of NF-κB was monitored by phosphorylation of IκB with anti-phospho-IκB, degradation of IκB with anti-IκB, and electrophoretic mobility shift assay (EMSA). TAK1 and TAB1 were detected with anti-TAK1 and anti-TAB1. The asterisks indicate nonspecific bands.
FIGURE 2.
TAB1 is dispensable for TNF- and IL-1-induced activation of TAK1. A, MEFs were stimulated with 5 ng/ml IL-1 (left panels) or 20 ng/ml TNF (right panels). Activation of TAK1 was monitored by immunoblotting with anti-phospho-TAK1. TAK1 and TAB1 were detected with anti-TAK1 and anti-TAB1. The asterisks indicate nonspecific bands. B, MEFs were pretreated with 10 nm calyculin A for 1 h and stimulated with 5 ng/ml IL-1 for 10 min. Activation of TAK1 was analyzed by immunoblots.
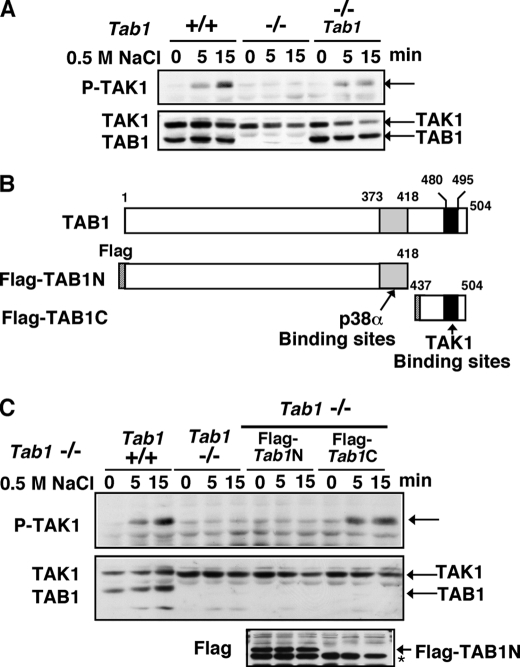
TAK1 is also activated by stress conditions such as osmotic stress (12) and arsenic treatment.3 Among the stress conditions, we have previously found that stringent osmotic stress using 0.5–0.7 m NaCl strongly activates TAK1 and that TAK1 is essential for NaCl-induced JNK activation (12). We examined whether osmotic stress-induced signaling events are altered by Tab1 deletion, and measured the osmotic stress-induced activation of TAK1 (Fig. 3A). TAK1 was activated at 5–15 min following 0.5 m NaCl treatment in Tab1 control MEFs, whereas osmotic stress-induced activation of TAK1 was greatly impaired in Tab1-deficient MEFs (Fig. 3A). Expression of exogenous TAB1 in the Tab1-deficient MEFs was able to restore the activation of TAK1. These results demonstrate that TAB1 is essential for osmotic stress-induced TAK1 activation. TAB1 consists of 504 amino acid residues in humans. TAB1 binds to TAK1 through its C-terminal amino acid residues 480–495 (20) (Fig. 3B). TAB1 also binds to and activates p38α through the amino acid residues 373–418 (32). Therefore, besides direct interaction between TAB1 and TAK1, TAB1 might indirectly mediate TAK1 activation through p38α. We next asked which possibility is more likely for activation of TAK1 in osmotic stress signaling. We infected Tab1-deficient MEFs with retrovirus expressing the N-terminal amino acid residues 1–418 of Tab1 (Tab1N), which include p38α but not the TAK1-binding region, or the C-terminal amino acid residues 437–504 of TAB1 (TAB1C), which only binds to TAK1. Subsequently, pools of the MEFs expressing Tab1Nor Tab1C were treated with 0.5 m NaCl (Fig. 3C). We found that TAB1C but not TAB1N was able to restore the activation of TAK1 in response to the osmotic stress. TAB1C is only 68 amino acids, and we could not detect the TAB1C protein by immunoblotting. To confirm whether TAB1C mediates TAK1 activation, we utilized other Tab1-deficient MEFs isolated from conventional Tab1 knock-out embryos (21), which are completely different source from the MEFs used in this study and generated Tab1C expressing Tab1-deficient. Those Tab1C expressing MEFs but not the Tab1-deficient MEFs activate TAK1 in response to the osmotic stress (data not shown). These results suggest that TAB1 association with TAK1 is important for osmotic stress-induced activation of TAK1. Moreover, this demonstrates that the C-terminal 68 amino acid residues of TAB1 are sufficient to mediate osmotic stress-induced TAK1 activation.
FIGURE 3.
TAB1 mediates osmotic stress-induced activation of TAK1 through its C terminus. A, MEFs were stimulated with 0.5 m NaCl. Activation of TAK1 was analyzed by immunoblots. B, schematic diagram of TAB1 and truncated mutants of TAB1. p38α- and TAK1-binding sites are shown. C, Tab1 control (+/+), Tab1-deficient (–/–) and Tab1-deficient but restored by expressing FLAG-tagged Tab1N (–/– FLAG-Tab1N) or FLAG-tagged Tab1C (–/– FLAG-Tab1C) MEFs were stimulated with 0.5 m NaCl. Activation of TAK1 was analyzed by immunoblots. The expression of FLAG-TAB1N was detected by the immunoblot with anti-FLAG. The asterisk indicates a nonspecific band. FLAG-TAB1C was too small to detect by immunoblot analysis.
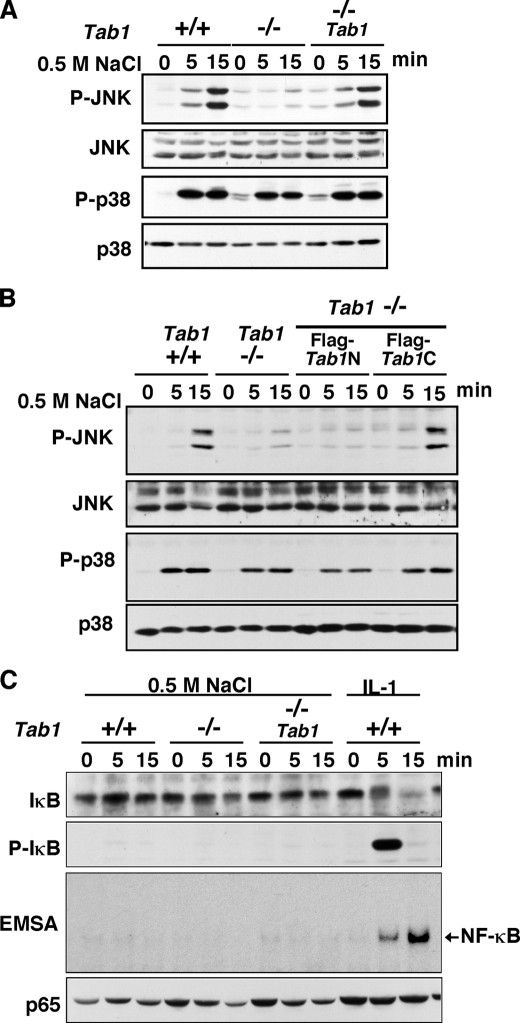
We next investigated the cellular responses involving osmotic stress induction that are mediated by the TAB1-TAK1 pathway. We examined activation of JNK and p38 in Tab1 control and Tab1-deficient MEFs (Fig. 4A). Activation of JNK but not of p38 was impaired in Tab1-deficient MEFs. We confirmed that TAB1C but not TAB1N was able to restore osmotic stress-induced JNK activation (Fig. 4B). We have previously reported that osmotic stress-induced activation of JNK but not the p38 is impaired in TAK1-deficient cells (12). Therefore, TAB1-TAK1 is predominantly function upstream of JNK but not of p38 in osmotic stress signaling pathway. It has been reported that another MAPK kinase, MEKK3, is important for osmotic stress-induced activation of p38 (33). Thus, it is likely that TAB1-TAK1 is the major mediator of JNK activation, whereas MEKK3 is the major mediator of p38 activation.
FIGURE 4.
TAB1 is essential for osmotic stress-induced activation of JNK. A and B, MEFs were stimulated with 0.5 m NaCl. Activation of JNK and p38 was analyzed by immunoblots. C, MEFs were stimulated with 0.5 m NaCl or 5 ng/ml IL-1. Activation of NF-κB was monitored by phosphorylation of IκB with anti-phospho-IκB, degradation of IκB with anti-IκB and electrophoretic mobility shift assay (EMSA). The amount of p65 is shown as a loading control.
We also examined activation of NF-κB (Fig. 4C). Although TAK1 is capable of activating NF-κB and is highly activated under osmotic stress conditions, NF-κB pathway was not activated at all even in wild type MEFs, which is consistent with our previous observation (12). We think that TAK1 is directed to the JNK pathway in response to osmotic stress by binding to TAO2 kinase as described in our previous report (12).
In addition to JNK and p38, many stress conditions activate the AMPK pathway that regulates energy metabolism (34). TAK1 is previously implicated in activation of AMPK (35). We would like to note that AMPK was activated upon 0.5 m NaCl treatment, and the level of activation was not markedly reduced either by Tab1 deletion or by Tak1 deletion (supplemental Fig. S1). These results demonstrate that osmotic stress activates TAK1 in a TAB1-dependent manner, which is essential for activation of JNK but not of p38 or AMPK.
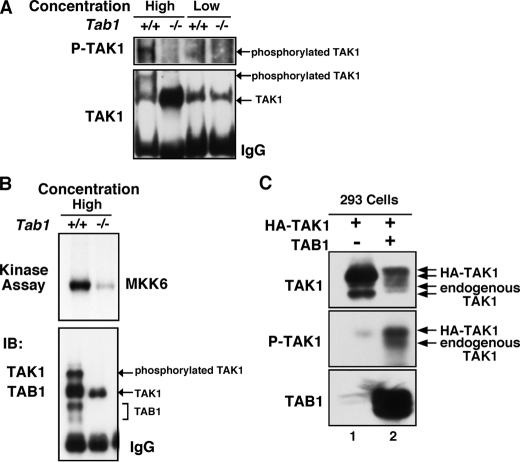
Our results demonstrated that among the TAK1 stimuli, only osmotic stress signaling utilizes TAB1 to activate TAK1. We then attempted to determine the mechanism by which TAB1 activates TAK1 in response to osmotic stress. In IL-1 and TNF pathways, it has been demonstrated that TAK1 is associated with a large signaling complex consisting of a number of proteins including TAB2/3 and TNF receptor-associated factors (36). TAK1 oligomerization in the signaling complexes is essential for activation of TAK1. In contrast to cytokine signaling, formation of such signaling complexes has not been identified in the osmotic stress pathway. Because oligomerization is one of common mechanisms of enzymatic activation of kinases, we speculated that TAK1 might be oligomerized and activated under an osmotic stress condition in a TAB1-dependent manner. How is TAK1 oligomerized by osmotic stress? There is no specific sensor molecule for osmotic stress in mammalian cells. It has been well known that osmotic stress shrinks cells and that the reduced cell volume is the trigger of cell signaling (37, 38). Thus, we speculated that the TAB1-TAK1 complex might be spontaneously oligomerized and activated when the concentration of TAB1-TAK1 is increased. To test this possibility, we examined whether TAB1-TAK1 can be spontaneously activated when the protein concentration is increased. We incubated cell lysates from Tab1 control or Tab1-deficient MEFs at a high (10 mg protein/ml) or low (2 mg protein/ml) concentration and measured TAK1 activity (Fig. 5, A and B). A slowly migrating TAK1 on SDS-PAGE, which was phosphorylated, was detected when incubated at the high concentration in the presence of TAB1. In contrast, TAK1 was not phosphorylated at any concentrations in the lysate from Tab1-deficient MEFs. We confirmed that catalytic activity of TAK1 was greatly increased after incubation at the high concentration in a TAB1-dependent manner (Fig. 5B), which suggests that when TAB1-TAK1 concentration is increased, TAK1 is oligomerized and thereby activated. further test this idea, we utilized 293 cells to overexpress HA-tagged TAK1 with TAB1 to increase the concentration of TAB1-TAK1. If an increase of TAB1-TAK1 concentration causes oligomerization and activation of TAK1, exogenously overexpressed HA-TAK1-TAB1 could activate not only HA-TAK1 but also endogenous TAK1. Because HA-tagged TAK1 is significantly bigger than endogenous TAK1, we were able to detect a slower migrating HA-TAK1 band on SDS-PAGE when HA-TAK1 alone was overexpressed (Fig. 5C, top panel, lane 1). We found that when cells were transfected with HA-TAK1 and TAB1, both HA-TAK1 and endogenous TAK1 migrated as slower smear bands on SDS-PAGE (Fig. 5C, top panel, lane 2). The slowly migrated HA-TAK1 and endogenous TAK1 were confirmed as phosphorylated forms (Fig. 5C, middle panel). This suggests that exogenously overexpressed TAB1-TAK1 can interact with and induced activation of endogenous TAK1. These results indicate that TAB1 is essential for concentration-dependent spontaneous activation of TAK1, which may be mediated by TAB1-TAK1 oligomerization. We next attempted to determine whether TAB1-TAK1 is oligomerized under osmotic stress conditions. However, the interaction of TAK1 with TAB1 was not altered by osmotic stress (data not shown), and TAB1 was always coprecipitated with TAK1 regardless of treatment of stimuli including osmotic stress and IL-1, which is consistent with our previous observation (19). We assume that TAB1-TAK1 complexes are preformed and that some of the complexes may be oligomerized upon stimuli challenges. The necessity of overexpression of both TAK1 and TAB1 for activation of TAK1 (Fig. 5C) supports this idea. We speculate that cell shrinkage under osmotic stress conditions increases the TAB1-TAK1 concentration, which may be sufficient to induce oligomerization of some TAB1-TAK1 in the cells.
FIGURE 5.
TAB1 is essential for concentration-dependent activation of TAK1. A and B, cell lysates from Tab1 control (+/+) and Tab1-deficient (–/–) MEFs were incubated and immunoprecipitated at a high or low protein concentration as described under “Experimental Procedures.” The immunoprecipitates were incubated in a kinase buffer and analyzed by immunoblots (IB) or by in vitro kinase assay using bacterially expressed MKK6 as an exogenous substrate. C, 293 cells were transfected with expression vectors for HA-TAK1 and TAB1. At 48 h post-transfection, ∼3 μg of cell lysates form HA-TAK1 alone (lane 1) or HA-TAK1 + TAB1 (lane 2) transfected cells were loaded onto SDS-PAGE and analyzed by immunoblots. The sizes of the upper bands of exogenous and endogenous TAK1 shown in the top panel correspond to the sizes of phosphorylated forms shown in the middle panel.
In summary, TAB1 is essential for osmotic stress-induced activation of TAK1, which may be mediated by concentration-dependent oligomerization of TAK1. In contrast, TAK1 is activated in a TAB1-independent manner in response to IL-1 and TNF, which is presumably because TAK1 is oligomerized through other adaptor molecules including TAB2.
In addition to these pathways, TAB1 is known to be critically involved in TGF-β family signaling pathways. TAB1 is associated with X-linked inhibitor of apoptosis protein and thereby recruiting TAK1 to the TGF-β receptor complex (29, 39). We examined TGF-β-induced activation of plasminogen activator inhibitor 1, which was previously identified as one of the TAK1-dependent events in TGF-β signaling pathways (10). We found that the induction of plasminogen activator inhibitor 1 mRNA was not impaired in Tab1-deficient MEFs (supplemental Fig. S2). This suggests that TAB1 is dispensable at least for TGF-β-induced plasminogen activator inhibitor 1 induction in MEFs. Because TGF-β family ligand-induced cellular responses are not effectively detected in MEFs, we have not yet determined whether other TGF-β signaling pathways are mediated by TAB1.
Mendoza et al. (24) have recently reported that IL-1-induced TAK1 activation is impaired in Tab1-deficient MEFs. In contrast, our results demonstrated that TAB1 is dispensable for IL-1-induced TAK1 activation. Mendoza et al. used MEFs from the conventional Tab1 knock-out embryos (21). This Tab1 knock-out embryo lacks exons 10 and 11 of Tab1, which presumably express a C-terminal truncated form of TAB1 that is almost identical to the truncated TAB1 generated in our Cre-mediated Tab1 deletion system. We would like to note that we conducted most of the experiments in this study not only using our Tab1-deficient MEFs but also MEFs from the conventional Tab1 knock-out embryos and that all of the results were the same in both Tab1-deficient MEFs. Therefore, we do not know the reason for the discrepancy between their and our results. It might be possible that MEFs respond differently to IL-1 because of genetic or epigenetic alterations generated during immortalization but not because of TAB1 deficiency. In our study, we utilized Tab1-restored MEFs to confirm the TAB1-dependent cellular responses and found that Tab1 deletion does not significantly affect TNF and IL-1 signaling pathways. It has been well documented that TAK1 deletion almost completely abolishes TNF- and IL-1-induced JNK, p38, and NF-κB activation in several types of cells (4, 5, 23, 40). This suggests that there is no compensatory mechanism that can activate JNK, p38, and NF-κB in the absence of TAK1 in IL-1 and TNF signaling pathways, which is consistent with our results that activation of TAK1, NF-κB, JNK, and p38 was all intact in Tab1-deficient MEFs. Thus, we conclude that TAB1 is dispensable for TAK1 activation, at least in TNF and IL-1 signaling pathways.
In this study, we identified that the C-terminal region of TAB1 is essential and sufficient for osmotic stress-induced activation of TAK1. The C-terminal region of TAB1 binding to TAK1 may be important for oligomerization of TAK1, which in turn activates TAK1. Further studies to define the molecular mechanism by which such a small region of TAB1 mediates TAK1 activation in response to osmotic stress will be important. We show here that TAB1 is totally dispensable for IL-1 and TNF signaling pathways but essential for osmotic stress-induced JNK activation. Based on these findings, we assume that TAB1-TAK1 signaling is involved in a subset of stress responses but not in IL-1 or TNF signaling in vivo. Utilizing our Tab1-floxed mice, we anticipate that we can explore the role of TAB1-TAK1 signaling in an in vivo setting.
Supplementary Material
Acknowledgments
We thank S. Akira for materials.
This work was supported, in whole or in part, by National Institutes of Health Grants ES071003-10 (Y. M.) and GM068812 (to J. N.-T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: TNF, tumor necrosis factor; AMPK, AMP-activated protein kinase; HA, hemagglutinin; MEF, mouse embryonic fibroblasts; TAB, TAK1-binding protein; IL, interleukin; TGF, transforming growth factor; MAPK, mitogen-activated protein kinase; MEKK, MAPK/extracellular signal-regulated kinase kinase kinase; JNK, c-Jun N-terminal kinase.
M. Inagaki, E. Omori, J.-Y. Kim, Y. Komatsu, G. Scott, M. K. Ray, G. Yamada, K. Matsumoto, Y. Mishina, and J. Ninomiya-Tsuji, unpublished results.
References
- 1.Hasegawa, M., Fujimoto, Y., Lucas, P. C., Nakano, H., Fukase, K., Nunez, G., and Inohara, N. (2008) EMBO J. 27 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim, J. Y., Omori, E., Matsumoto, K., Nunez, G., and Ninomiya-Tsuji, J. (2008) J. Biol. Chem. 283 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ninomiya-Tsuji, J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z., and Matsumoto, K. (1999) Nature 398 252–256 [DOI] [PubMed] [Google Scholar]
- 4.Shim, J. H., Xiao, C., Paschal, A. E., Bailey, S. T., Rao, P., Hayden, M. S., Lee, K. Y., Bussey, C., Steckel, M., Tanaka, N., Yamada, G., Akira, S., Matsumoto, K., and Ghosh, S. (2005) Genes Dev. 19 2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takaesu, G., Surabhi, R. M., Park, K. J., Ninomiya-Tsuji, J., Matsumoto, K., and Gaynor, R. B. (2003) J. Mol. Biol. 326 105–115 [DOI] [PubMed] [Google Scholar]
- 6.Adhikari, A., Xu, M., and Chen, Z. J. (2007) Oncogene 26 3214–3226 [DOI] [PubMed] [Google Scholar]
- 7.Karin, M. (2006) Nature 441 431–436 [DOI] [PubMed] [Google Scholar]
- 8.Weston, C. R., and Davis, R. J. (2007) Curr. Opin. Cell Biol. 19 142–149 [DOI] [PubMed] [Google Scholar]
- 9.Hanafusa, H., Ninomiya-Tsuji, J., Masuyama, N., Nishita, M., Fujisawa, J.-I., Shibuya, H., Matsumoto, K., and Nishida, E. (1999) J. Biol. Chem. 274 27161–27167 [DOI] [PubMed] [Google Scholar]
- 10.Kajino, T., Omori, E., Ishii, S., Matsumoto, K., and Ninomiya-Tsuji, J. (2007) J. Biol. Chem. 282 9475–9481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, W., White, M. A., and Cobb, M. H. (2002) J. Biol. Chem. 277 49105–49110 [DOI] [PubMed] [Google Scholar]
- 12.Huangfu, W.-C., Omori, E., Akira, S., Matsumoto, K., and Ninomiya-Tsuji, J. (2006) J. Biol. Chem. 281 28802–28810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singhirunnusorn, P., Suzuki, S., Kawasaki, N., Saiki, I., and Sakurai, H. (2005) J. Biol. Chem. 280 7359–7368 [DOI] [PubMed] [Google Scholar]
- 14.Shibuya, H., Yamaguchi, K., Shirakabe, K., Tonegawa, A., Gotoh, Y., Ueno, N., Irie, K., Nishida, E., and Matsumoto, K. (1996) Science 272 1179–1182 [DOI] [PubMed] [Google Scholar]
- 15.Ishitani, T., Takaesu, G., Ninomiya-Tsuji, J., Shibuya, H., Gaynor, R. B., and Matsumoto, K. (2003) EMBO J. 22 6277–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takaesu, G., Kishida, S., Hiyama, A., Yamaguchi, K., Shibuya, H., Irie, K., Ninomiya-Tsuji, J., and Matsumoto, K. (2000) Mol. Cell 5 649–658 [DOI] [PubMed] [Google Scholar]
- 17.Kanayama, A., Seth, R. B., Sun, L., Ea, C. K., Hong, M., Shaito, A., Chiu, Y. H., Deng, L., and Chen, Z. J. (2004) Mol. Cell 15 535–548 [DOI] [PubMed] [Google Scholar]
- 18.Kishida, S., Sanjo, H., Akira, S., Matsumoto, K., and Ninomiya-Tsuji, J. (2005) Genes Cells 10 447–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishimoto, K., Matsumoto, K., and Ninomiya-Tsuji, J. (2000) J. Biol. Chem. 275 7359–7364 [DOI] [PubMed] [Google Scholar]
- 20.Ono, K., Ohtomo, T., Sato, S., Sugamata, Y., Suzuki, M., Hisamoto, N., Ninomiya-Tsuji, J., Tsuchiya, M., and Matsumoto, K. (2001) J. Biol. Chem. 276 24396–24400 [DOI] [PubMed] [Google Scholar]
- 21.Komatsu, Y., Shibuya, H., Takeda, N., Ninomiya-Tsuji, J., Yasui, T., Miyado, K., Sekimoto, T., Ueno, N., Matsumoto, K., and Yamada, G. (2002) Mech. Dev. 119 239–249 [DOI] [PubMed] [Google Scholar]
- 22.Jadrich, J. L., O'Connor, M. B., and Coucouvanis, E. (2006) Development 133 1529–1541 [DOI] [PubMed] [Google Scholar]
- 23.Sato, S., Sanjo, H., Takeda, K., Ninomiya-Tsuji, J., Yamamoto, M., Kawai, T., Matsumoto, K., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 1087–1095 [DOI] [PubMed] [Google Scholar]
- 24.Mendoza, H., Campbell, D. G., Burness, K., Hastie, J., Ronkina, N., Shim, J. H., Arthur, J. S., Davis, R. J., Gaestel, M., Johnson, G. L., Ghosh, S., and Cohen, P. (2008) Biochem. J. 409 711–722 [DOI] [PubMed] [Google Scholar]
- 25.Inagaki, M., Komatsu, Y., Scott, G., Yamada, G., Ray, M., Ninomiya-Tsuji, J., and Mishina, Y. (2008) Genesis 46 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uemura, N., Kajino, T., Sanjo, H., Sato, S., Akira, S., Matsumoto, K., and Ninomiya-Tsuji, J. (2006) J. Biol. Chem. 281 7863–7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kajino, T., Ren, H., Iemura, S., Natsume, T., Stefansson, B., Brautigan, D. L., Matsumoto, K., and Ninomiya-Tsuji, J. (2006) J. Biol. Chem. 281 39891–39896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitamura, T. (1998) Int. J. Hematol. 67 351–359 [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi, K., Nagai, S., Ninomiya-Tsuji, J., Nishita, M., Tamai, K., Irie, K., Ueno, N., Nishida, E., Shibuya, H., and Matsumoto, K. (1999) EMBO J. 18 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayden, M. S., and Ghosh, S. (2008) Cell 132 344–362 [DOI] [PubMed] [Google Scholar]
- 31.Takaesu, G., Ninomiya-Tsuji, J., Kishida, S., Li, X., Stark, G. R., and Matsumoto, K. (2001) Mol. Cell Biol. 21 2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge, B., Gram, H., Di Padova, F., Huang, B., New, L., Ulevitch, R. J., Luo, Y., and Han, J. (2002) Science 295 1291–1294 [DOI] [PubMed] [Google Scholar]
- 33.Uhlik, M. T., Abell, A. N., Johnson, N. L., Sun, W., Cuevas, B. D., Lobel-Rice, K. E., Horne, E. A., Dell'Acqua, M. L., and Johnson, G. L. (2003) Nat. Cell Biol. 5 1104–1110 [DOI] [PubMed] [Google Scholar]
- 34.Hardie, D. G. (2007) Nat. Rev. Mol. Cell Biol. 8 774–785 [DOI] [PubMed] [Google Scholar]
- 35.Xie, M., Zhang, D., Dyck, J. R., Li, Y., Zhang, H., Morishima, M., Mann, D. L., Taffet, G. E., Baldini, A., Khoury, D. S., and Schneider, M. D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17378–17383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, Z. J., Bhoj, V., and Seth, R. B. (2006) Cell Death Differ. 13 687–692 [DOI] [PubMed] [Google Scholar]
- 37.Burg, M. B., Ferraris, J. D., and Dmitrieva, N. I. (2007) Physiol. Rev. 87 1441–1474 [DOI] [PubMed] [Google Scholar]
- 38.Roger, F., Martin, P.-Y., Rousselot, M., Favre, H., and Feraille, E. (1999) J. Biol. Chem. 274 34103–34110 [DOI] [PubMed] [Google Scholar]
- 39.Lu, M., Lin, S. C., Huang, Y., Kang, Y. J., Rich, R., Lo, Y. C., Myszka, D., Han, J., and Wu, H. (2007) Mol. Cell 26 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omori, E., Matsumoto, K., Sanjo, H., Sato, S., Akira, S., Smart, R. C., and Ninomiya-Tsuji, J. (2006) J. Biol. Chem. 281 19610–19617 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.