Abstract
The small heat shock protein Hsp20 protects cardiomyocytes against apoptosis, and phosphorylation at its Ser16 site enhances its cardioprotection. To determine whether genetic variants exist in human Hsp20, which may modify these beneficial effects, we sequenced the coding region of the Hsp20 gene in 1347 patients suffering from dilated cardiomyopathy and 744 subjects with no heart disease. We identified a C59T substitution in the human Hsp20 gene in one patient and three individuals without heart disease. All subjects were heterozygous for this mutation, which changes a fully conserved proline residue into leucine at position 20 (P20L), resulting in secondary structural alterations. To examine the potential functional significance of the P20L-Hsp20 human variant, adult rat cardiomyocytes were infected with Ad.GFP (where Ad is adenovirus and GFP is green fluorescent protein), Ad.WT-Hsp20 (where WT is wild-type), and Ad.P20L-Hsp20 and subjected to simulated ischemia/reperfusion injury. Expression of WT-Hsp20 resulted in significant attenuation of apoptosis compared with the GFP control. However, the P20L-Hsp20 mutant showed no protection against apoptosis, assessed by Hoechst staining and DNA fragmentation. The loss of cardioprotection by the mutant Hsp20 was associated with its diminished phosphorylation at Ser16 compared with WT-Hsp20. Furthermore, maximal stimulation of cardiomyocytes with isoproterenol or protein kinase A-mediated phosphorylation in vitro confirmed the impaired ability of the mutant Hsp20 to become phosphorylated at Ser16. In conclusion, we have identified a P20L substitution in human Hsp20, which is associated with diminished phosphorylation at Ser16 and complete abrogation of the Hsp20 cardioprotective effects which may adversely affect the ability of human carriers to cope with cellular stress.
Heat shock proteins (Hsp)2 are key mediators of cytoprotection upon a broad range of potentially deleterious stimuli (1). In fact, it has been shown that the expression of these molecular chaperones increases transiently under cellular conditions of stress, such as heat (2, 3), oxidative stress (2, 4, 5), and β-adrenergic stimulation (6, 7) as a protective mechanism. Hsps are a group of well conserved proteins, which are differentially classified by molecular mass into various families. Within this superfamily of cellular chaperones, the small heat shock proteins comprise a subgroup with 10 members, whose molecular mass ranges from 12 to 43 kDa (8, 9). These proteins contain a unique N terminus and a conserved α-crystallin domain at their C terminus, which facilitates their chaperone activity (9).
Of particular interest in this subfamily is a protein of ∼20 kDa, namely Hsp20. This is the only small heat shock protein that has the consensus motif (RRAS) for protein kinase A(PKA)/protein kinase G-dependent phosphorylation at Ser16 (10). In recent years, Hsp20 and its phosphorylation have emerged to be functionally significant in the heart. Interestingly, the levels of Hsp20 have been shown to increase under the physiological stress of exercise training (11), as well as in the pathological setting of chronic β-adrenergic stimulation (7) and in tachycardia-induced heart failure (12). Using adenovirus-infected cardiomyocytes and transgenesis, we and others have demonstrated that this increase is cardioprotective (7, 13–17). More specifically, overexpression of Hsp20 protected the heart against ischemia/reperfusion (I/R)-induced injury (13–16), β-agonist-mediated remodeling (17), and isoproterenol (Iso)-induced apoptosis (7, 17). Importantly, our data show that the phosphorylation of Hsp20 at Ser16 may be instrumental to its anti-apoptotic properties. In particular, it has been reported that the phosphorylation of Hsp20 at Ser16 increased after prolonged exposure to β-agonist stimulation (7), during I/R (16, 18), as well as in congestive heart failure (12). To assess the role of enhanced phosphorylation at this site, a constitutively phosphorylated form of Hsp20 (S16D) was expressed in cardiomyocytes (7). This mutant Hsp20 protected cardiomyocytes to a higher extent, after Iso-induced apoptosis, compared with WT-Hsp20. Conversely, the non-phosphorylatable mutant (S16A) showed no antiapoptotic effects. Overall, these data implicate Hsp20 and its phosphorylation at Ser16 as important mediators of cardioprotection.
Given the cytoprotective nature of Hsp20 in the heart, this study was designed to examine whether genetic variants in human Hsp20 may diminish its cardioprotective effects and render cells more susceptible to insults. Thus, heart failure patients and individuals with no heart disease were screened for variations in the human Hsp20 gene. We have identified a C59T substitution in exon 1 of Hsp20, which changes a proline residue to leucine at position 20 (P20L). We present evidence here that this substitution is associated with structural alterations, diminished phosphorylation at Ser16, and complete negation of the anti-apoptotic effects of Hsp20, suggesting that human carriers of the P20L-Hsp20 genetic variant may present with an impaired ability to cope with cellular stress in the heart.
EXPERIMENTAL PROCEDURES
Population Screening and Mutation Identification—Participating subjects provided written consent. The protocols used were approved by the Institutional Review Board of the Onassis Cardiac Surgery Center, the Georg August University of Goettingen, or the University of Cincinnati College of Medicine. Genomic DNA was isolated from whole blood from the participants. The entire Hsp20 gene (NCBI accession number NM_144617), consisting of three exons, was initially screened in 100 dilated cardiomyopathic (DCM) patients and in 100 individuals with no heart disease. Exon 1 of the human Hsp20 gene was amplified in a single PCR. Although exons 2 and 3 were amplified together in a separate PCR, using Accuprime GC-rich DNA polymerase (Invitrogen). The primers used for exon 1 were 5′-CAT CCA GCA GGC GCT TAA TA-3′ (sense) and 5′-ATC ACC AGC CTC CTT CAG AG-3′ (antisense). The primers used for exons 2 and 3 were 5′-GTA GGA GAG CTG ACT GC-3′ (sense) and 5′-AGA GGA CAG TCC TTG GC-3′ (antisense). The conditions were as follows: one cycle at 95 °C for 2 min, linked to 35 cycles at 95 °C for 30 s, 63 °C for 30 s, 72 °C for 30 s, followed by a final extension at 72 °C for 10 min. The generated PCR product was purified using a PCR-purification kit (Qiagen) and sequenced using automated dye-primer chemistry. The obtained sequences were compared against the known human Hsp20 gene using an NIH software program (Blast 2 sequences). In addition, the chromatograms were also inspected individually for heterozygosis. This initial screen led to the identification of a C59T transversion in exon 1. A total of 1347 DCM patients and 744 individuals with no heart disease were analyzed for this substitution. Due to legal restrictions, clinical data were available for 1111/1347 DCM patients and 744/744 non-cardiomyopathic individuals. The data indicated that the ages (mean ± S.D.) of DCM and non-cardiomyopathic individuals were 62.58 ± 11.46 and 55.91 ± 11.28, respectively. Males composed 58.3% and 50.3% of the DCM and non-cardiomyopathic populations, respectively.
Secondary Structure Predictions and Circular Dichroism— The program nnpredict, available through the University of California at San Francisco, was used to generate the predicted secondary structure content of the WT and mutant Hsp20. This was aligned with the primary sequence using the Polyview protein structure visualization server, available through the Cincinnati Children's Hospital Medical Center, to generate a schematic representation of the secondary structure. Circular dichroism measurements were performed on recombinant proteins at a concentration of 0.1 mg/ml in 50 mm Tris-HCl, pH 7.0, using a Jasco 715 spectropolarimeter. A 1-cm path length cell was used, and the measurements were recorded at 25 °C. The analysis was performed by PrimeSyn Lab Inc. Estimation of secondary structure parameters was performed using the SOMCD algorithm (19).
Generation of Hsp20 Recombinant Proteins and Adenoviruses— The human Hsp20 cDNA (Open Biosystems) was cloned into the pGEX-6P-3 plasmid. The P20L mutant was generated using the Quik-Change site-directed mutagenesis II kit (Stratagene). Subsequently, WT and mutant Hsp20 were expressed as glutathione-S-transferase-fusion proteins in BL21 CodonPlus (DE3)-RIPL competent cells as described previously (20). Briefly, the proteins were purified using the B-PER glutathione-S-transferase fusion protein purification kit (Pierce), and the glutathione-S-transferase tag was digested using the PreScission protease (GE Healthcare). Samples were analyzed by SDS-PAGE to estimate the extent of cleavage and protein yield after purification. The protein concentration was determined using the MicroBCA assay (Pierce). The adenoviruses were generated as previously described (20). Briefly, the Ad-Easy XL system (Stratagene) was used to generate adenoviruses expressing the WT-Hsp20 (Ad.WT-Hsp20), the Hsp20 mutant (Ad.P20L-Hsp20), or green fluorescent protein (Ad.GFP). The viruses were purified using the adenovirus mini purification kit (Virapur) and titered using the Adeno-X Rapid Titer kit (Clontech).
Isolation of Adult Rat Cardiomyocytes and Simulated I/R— Animals were handled according to the Institutional Animal Care and Use Committee at the University of Cincinnati. Myocytes from adult male Sprague-Dawley rats (∼300 g) were isolated by collagenase digestion as previously described (20). Myocytes were resuspended in modified culture medium (M199, Sigma), counted, and plated on laminin-coated plates or dishes for 2 h at 37°C in a humidified, 5% CO2 incubator and subsequently infected with the adenoviruses at a multiplicity of infection of 500 for 2 h. At 48 h post-infection, the cells were subjected to simulated ischemia/reperfusion. Specifically, the medium was replaced with ischemic buffer containing 1.13 mm CaCl2, 5 mm KCl, 0.3 mm KH2PO4, 0.5 mm MgCl2, 0.4 mm MgSO4, 128 mm NaCl, 4 mm NaHCO3, 10 mm HEPES, pH 6.8 (13), and placed in a chamber mimicking the hypoxic (1% O2) and hypercapnic conditions (20% CO2), observed during ischemia (21). The ischemic buffer was replaced with normal medium and the cells were placed back in the humidified chamber at atmospheric conditions to allow reperfusion for 3 h. The cells were either collected for the DNA fragmentation studies and Western blotting or fixed in plates for Hoechst staining.
Detection of Apoptosis through Hoechst Staining and DNA Fragmentation—For Hoechst staining, the cells were fixed using 4% paraformaldehyde for 25 min at 4 °C. The cells were then washed with phosphate-buffered saline and stored at -20 °C in 70% ethanol. Prior to staining, the cells were washed with phosphate-buffered saline. Finally, the cells were stained with Hoechst and visualized under a fluorescent microscope. DNA fragmentation was assessed using a commercially available ELISA kit (Roche Applied Science), which measures cytosolic mono- and oligonucleosomes (16). 100 μg of cytosolic protein were incubated with 80 μl of immunoreagent, consisting of anti-histone biotin and anti-DNA-peroxidase antibodies, in streptavidin-coated plates for 2 h at room temperature. Color development was initiated with the colorimetric substrate ABTS. Optical density values were recorded in an ELISA plate reader at 405 nm.
Generation of a pSer16-Hsp20 Antibody—The pSer16-Hp20 antibody was generated by Affinity Bioreagents (Golden, CO) using the BioSpecific™ plan. Specifically, specific-pathogen-free-rabbits were immunized with the synthetic phosphopeptide SWLLRRA-S-(PO3)-APLPG conjugated to keyhole limpet hemocyanin using glutaraldehyde as a cross-linker. The adjuvant used during the immunizations was Freund's complete adjuvant for the primary injection and Freund's incomplete adjuvant for the following boosts. The phosphorylation state-specific antibody was then antigen affinity-purified from the antisera through a two-step purification process utilizing the non-phosphorylated peptide followed by the phosphorylated peptide.
Immunoblot Analysis—Proteins were separated by 15% SDS-PAGE and transferred to nitrocellulose membranes. Nonspecific binding was blocked for 1 h using 5% dried milk in Tris-buffered saline, pH 7.4, containing 0.1% Tween 20. Membranes were probed overnight at 4 °C with primary antibodies. A secondary peroxidase-labeled antibody (Amersham Biosciences) was used in combination with an enhanced chemiluminescent detection system (Supersignal West Pico chemiluminescent, Pierce) to visualize the primary antibodies. The optical density of the bands was analyzed using ImageQuant 5.2 software. For the peptide competition experiments, the primary antibody was preincubated with the peptide in excess (5-fold) for 1 h and then added to the membranes. The phosphopeptide was SWLLRRA-S-(PO3)-APLPG, and the dephosphopeptide was SWLLRRASAPLPG.
Protein Kinase A Phosphorylation in Vitro—Recombinant proteins (2–25 μm) were phosphorylated in vitro with 1.5 μg of the catalytic subunit of protein kinase A (Promega) in 50 mm Tris-HCl, pH 7.0, 5 mm MgCl2, 5 mm NaF, and 1 mm EGTA. The reaction was initiated with the addition of 250 μm ATP at 37 °C, and it was terminated after 30 min by the addition of SDS sample buffer. The extent of phosphorylation was determined by Western blotting using the pSer16-Hsp20 antibody. The same membrane was stripped and probed with an antibody against total Hsp20 (RDI). Densitometric analysis was performed with ImageQuant 5.2, and the extent of phosphorylation was expressed as the ratio of pSer16-Hsp20/Hsp20 levels. Michaelis-Menten parameters were determined using GraphPad Prism v.4.
Statistical Analysis—All the values are expressed as mean ± S.E. for n experiments. Comparisons between two groups were evaluated by Student's t test for unpaired data. Statistical analysis of multiple groups was carried out by one-way ANOVA with a Tukey test for post hoc analysis. Results were considered statistically significant at p < 0.05.
RESULTS
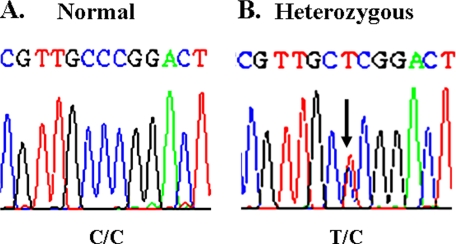
Identification of a Human Hsp20 Mutant (C59T)—To identify variants in the human Hsp20 gene that could potentially alter its cardioprotective properties, the entire Hsp20 gene, which consists of three exons, was screened in 100 DCM patients and in 100 subjects with no heart disease. We identified a genetic variant entailing substitution of cytosine with thymine at position 59 (C59T) in exon 1 of the Hsp20 gene (Fig. 1). Interestingly, this Hsp20 genetic variant has been previously described in the single nucleotide polymorphism data base (ref-SNP ID rs11549029), but it is not currently known whether it is a common polymorphism and whether it may affect the function of Hsp20. Thus, 1347 DCM patients and 744 individuals with no heart disease were subsequently examined for this Hsp20 variant. This analysis identified one DCM patient and three individuals without heart disease who were heterozygous carriers of the C59T substitution. These data show that C59T is a rare mutation found in both the cardiomyopathic and non-cardiomyopathic populations.
FIGURE 1.
Identification of a C59T substitution in the human Hsp20 gene. A partial nucleotide sequence of the Hsp20 coding region in an individual with normal Hsp20 (A) and in an individual who was heterozygous for the C59T substitution (B) is shown.
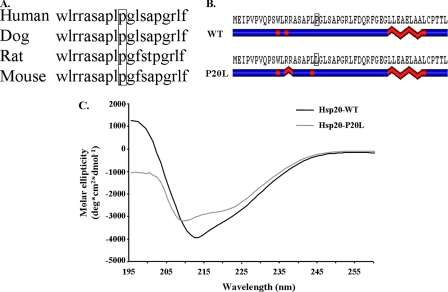
The C59T Genetic Variant Results in Alterations of Hsp20 Secondary Structure—The C59T mutation changes a proline residue at position 20 to leucine (P20L). As shown in Fig. 2A, comparison of this site in human, dog, rat, and mouse revealed that it is fully conserved among these species, suggesting a critical role in Hsp20 function. Considering the fact that proline residues are structural disruptors, it was important to examine the functional significance of this mutation to the secondary structure of Hsp20. As such, protein structure analysis software was initially used to predict any potential alterations. Interestingly, residue 20 is found near the PKA/protein kinase G phosphorylation site (Ser16) in a region predicted to be characterized by short helical segments in the WT protein (Fig. 2B). Similar analysis for the mutant revealed a potentially extended helical region around the RRAS phosphorylation motif (Fig. 2B). Importantly, the secondary structure was also determined experimentally. Specifically, recombinant proteins encoding WT-Hsp20 (WT) and mutant-Hsp20 were generated and analyzed using circular dichroism (CD) spectroscopy, a commonly used methodology that yields information on the relative structural constitution of a protein. The far UV CD measurements showed that the WT protein yielded a profile with negative maxima at 212 nm (Fig. 2C). This spectrum is typical of proteins predominated by β-sheets and is consistent with previously reported CD data from Hsp20 and other small Hsps (22–24). Interestingly, the P20L substitution shifted the negative maxima to 209 nm and decreased the CD spectrum amplitude in the far-UV wavelength range (Fig. 2C). Secondary structure estimates obtained using the SOMCD algorithm (19) showed that there was an increase in helical content (from 7.4% to 9.1%) and in unordered conformation (from 36.9% to 40.1%), whereas there was a decrease in β-sheet content (from 46.5% to 41.7%) with no change in β-turns (9.2%) in the mutant Hsp20. Overall, these results indicated that the P20L substitution in human Hsp20 is associated with structural perturbations. On the basis of these secondary structure alterations, we postulated that this mutation may prevent efficient phosphorylation of Hsp20 at Ser16, which may result in loss or attenuation of its cardioprotective effects.
FIGURE 2.
The C59T substitution changes a fully conserved proline at position 20 to a leucine (P20L), which results in secondary structural alterations. A, partial amino acid sequence alignment (amino acids 11–29) in human, dog, rat, and mouse shows that proline 20 is fully conserved. B, secondary structure predictions revealed alterations in the helical content around the Ser16 phosphorylation site as well as around the site of the mutation. Blue, linear coil; red, helical content. C, far-UV CD data obtained from analysis of recombinant proteins indicated that the P20L substitution shifted the negative maxima from 212 nm to 209 nm and decreased the CD amplitude in the far-UV wavelength range. Secondary structure estimates obtained using the SOMCD algorithm (19) showed that the secondary structure constitution was altered (see “Results”).
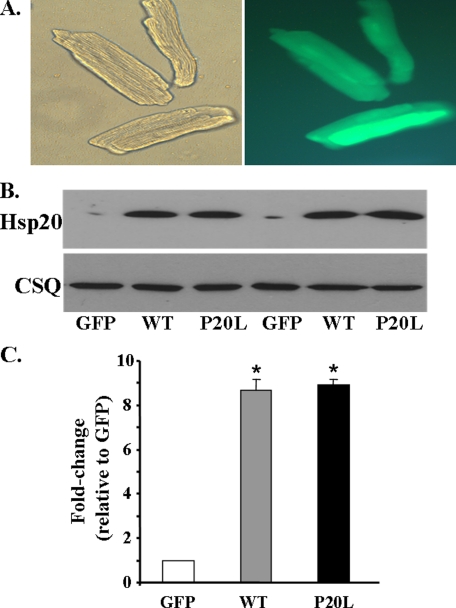
Human P20L-Hsp20 Is Associated with Loss of Cardioprotection—To delineate the functional significance of the human P20L-Hsp20 variant, adult rat cardiomyocytes were infected with Ad.GFP as a control, Ad.WT-Hsp20, and Ad.P20L-Hsp20. Adenoviral infection efficiency, assessed at 48 h post-infection by green fluorescence (Fig. 3A, right panel) indicated that ∼100% of the cells were infected. As shown in Fig. 3B, this resulted in comparable levels of overexpression between the WT-Hsp20 and P20L-Hsp20 groups (∼9-fold compared with the GFP control). Because the anti-apoptotic properties of Hsp20 are now well recognized (7, 13–17), it was important to examine any potential alterations by the mutant Hsp20 protein on programmed cell death. Thus, we determined the extent of apoptosis in the control GFP, WT-Hsp20, and P20L-Hsp20 groups. First, the effect of the P20L substitution on basal levels of apoptosis was examined using Hoechst staining to visualize nuclei. Cells undergoing apoptosis have pyknotic nuclei, which appear round, although longitudinal nuclei are indicative of non-apoptotic cells. As shown in Fig. 4 (A and B), no differences were detected between the three groups at basal levels. Further evaluation of the effect of the P20L mutant, using a more quantitative method to measure the extent of DNA fragmentation, indicated no differences in the basal levels of apoptosis between the three groups (Fig. 4C), similar to the Hoechst staining.
FIGURE 3.
Adenovirus-mediated expression of WT-Hsp20 and P20L-Hsp20 in adult rat cardiomyocytes. A, cardiomyocytes were infected with Ad.WT-Hsp20 and Ad.P20L-Hsp20. The Ad.GFP vector was used as a control. Bright-field image of adult rat cardiomyocytes 48 h post-infection (left panel) and fluorescent image (right panel) that show GFP expression indicates ∼100% infectivity. B, a representative Western blot of Hsp20 expression 48 h post-infection. C, quantification of the relative overexpression of WT- and P20L-Hsp20, normalized to calsequestrin, revealed a 9-fold increase in both groups compared with the GFP control. Values represent mean ± S.E. obtained from five independent myocyte lysates.*, p < 0.0001 versus GFP.
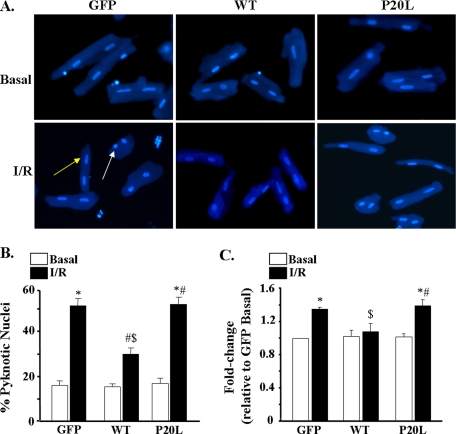
FIGURE 4.
The P20L substitution abrogates the cardioprotective effects of Hsp20. A, representative images of cardiomyocytes stained with Hoechst 33342 to detect apoptotic nuclei. Apoptotic nuclei are pyknotic (white arrow), whereas non-apoptotic nuclei are longitudinal (yellow arrow). B, pyknotic nuclei were counted and expressed as the percentage of total nuclei. Bars represent mean ± S.E. from three independent experiments with >200 nuclei counted per heart per group. C, the extent of DNA fragmentation was quantified in an ELISA-based method, which measures cytoplasmic histone-associated DNA fragments. Optical density values from the GFP basal group were used as a reference, and data were expressed as –fold change relative to this group. Values represent mean ± S.E. from six independent myocyte lysates. *, p < 0.05 versus GFP basal; #, p < 0.05 basal versus I/R; $, p < 0.05 versus GFP I/R.
Because Hsp20 has been previously shown to be cardioprotective against I/R-induced injury (13–16), infected myocytes were subjected to simulated I/R (1 h of ischemia/3 h of reperfusion) to assess any potential alterations elicited by the P20L mutation on the beneficial effects of Hsp20. We found that the percentage of pyknotic nuclei increased significantly from 16% to 52% in the control Ad.GFP-infected cells following simulated I/R (Fig. 4, A and B). However, the percentage of pyknotic nuclei in the WT-Hsp20 group was significantly lower (29%) than in the control group after I/R (Fig. 4, A and B), consistent with the cardioprotective effects of Hsp20 (7, 13–17). Interestingly, the mutant P20L-Hsp20 showed no cardioprotection (53% of nuclei were pyknotic after I/R). In further experiments, these findings were confirmed using the DNA fragmentation assay, described above. The extent of DNA fragmentation was reduced by 23% in the WT-Hsp20 group compared with the control following I/R (Fig. 4C). On the contrary, the P20L-Hsp20 group exhibited similar values to the control group (Fig. 4C). Overall, these data show that the P20L-Hsp20 mutant completely abrogates the anti-apoptotic effects of Hsp20.
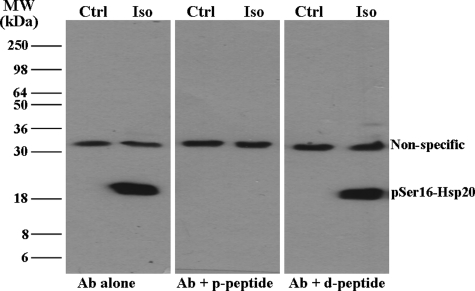
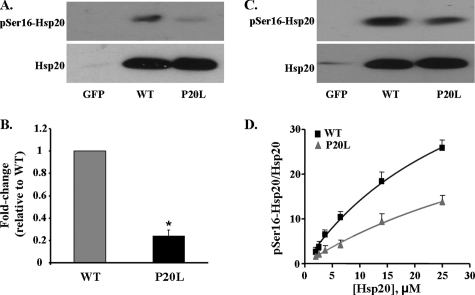
Effect of the P20L Substitution on Phosphorylation at Ser16 of Hsp20—The observed secondary structure alterations (Fig. 2, B and C) suggested that the ability of the mutant Hsp20 to become phosphorylated at Ser16 may be compromised. To assess this experimentally, an antibody against pSer16-Hsp20 was generated in rabbits. Our initial experiments focused on characterizing this antibody (Fig. 5). Myocytes were infected with Ad.WT-Hsp20 and either collected under control, non-stimulated conditions or after stimulation with 100 nm isoproterenol (Iso) to activate PKA. Upon Western blotting, two bands were observed in the cells treated with Iso, one of which migrated at ∼20 kDa, which is consistent with the molecular mass of Hsp20 (Fig. 5, left panel). At basal levels, no phosphorylated Hsp20 could be detected, consistent with the low activity of PKA in isolated myocytes. To test the specificity of this antibody, peptide competition was performed with the antigenic peptide used to generate the antibody. The band at ∼20 kDa was the only band out-competed by the phosphopeptide (Fig. 5, middle panel), but it was still detectable in the presence of the dephosphopeptide (Fig. 5, right panel). These findings suggest that the band at ∼20 kDa is specific for Hsp20 phosphorylated at Ser16. Therefore, the phosphorylation status of the adenovirus-infected myocytes was subsequently tested (Fig. 6, A and B). Our results showed that the P20L mutant was phosphorylated to a significantly lower extent than the WT protein (23% of WT) after simulated I/R (Fig. 6, A and B). Taken together, these data suggest that the conformational changes in the mutant Hsp20 (Fig. 2, B and C) may make it a poor substrate for phosphorylation. To further test this hypothesis in a more direct manner, cardiomyocytes infected with Ad.GFP, Ad.WT-Hsp20, or Ad.P20L-Hsp20 were subjected to maximal β-adrenergic stimulation (100 nm isoproterenol) to activate PKA and phosphorylate Hsp20. As shown in Fig. 6C, phosphorylation of the P20L mutant protein was decreased by 39% compared with the WT protein. The differences in the extent of the decrease upon I/R and after Iso stimulation are probably related to the extent of the kinase/phosphatase activities under the two different conditions. To better understand the mechanism by which the P20L substitution in Hsp20 reduces its ability to become phosphorylated efficiently at its Ser16 site, enzyme kinetic parameters were determined from PKA-dependent phosphorylation of WT- and P20L-Hsp20 recombinant proteins in vitro. The P20L substitution did not alter the apparent Km of the reaction but significantly decreased the Vmax value (by 33%), suggesting that the affinity of the mutant Hsp20 for PKA was unaltered but the turnover number and catalytic efficiency was significantly decreased by the P20L substitution (Fig. 6D). Collectively, these results show that the human P20L-Hsp20 mutant is associated with diminished phosphorylation at Ser16, which may be associated with the observed loss of cardioprotection.
FIGURE 5.
Generation and characterization of a pSer16-Hsp20 antibody. A newly raised antibody (Ab) against pSer16-Hsp20 recognized a band at ∼20 kDa in myocytes infected with Ad.WT-Hsp20 only after Iso stimulation (left panel). Competition with the phosphopeptide (p-peptide) and dephosphopeptide (d-peptide) (middle and right panels, respectively) showed that this band is specific for Hsp20 phosphorylated at Ser16.
FIGURE 6.
The P20L mutation is associated with diminished phosphorylation at Ser16. A, Western blot analysis from lysates of cardiomyocytes in the GFP, WT-Hsp20, or P20L-Hsp20 groups after simulated I/R revealed that the mutant Hsp20 failed to become phosphorylated to the same extent as the WT protein. B, quantification of the relative pSer16-Hsp20 levels, normalized to total Hsp20 levels, revealed that the P20L-Hsp20 mutant was phosphorylated only to 23% of WT-Hsp20. Bars show means ± S.E. from five independent experiments. *, p < 0.0005 versus WT I/R. C, Western blot analysis of cell lysates in the GFP, WT-Hsp20, and P20L-Hsp20 groups treated with maximal isoproterenol (100 nm) showed that phosphorylation at Ser16 was decreased in the P20L mutant protein compared with the WT protein. D, the plot represents Michaelis-Menten analysis of protein kinase A-dependent phosphorylation of WT- and P20L-Hsp20 recombinant proteins in vitro. The extent of phosphorylation was expressed as the pSer16-Hsp20 levels normalized to total Hsp20 levels in that sample determined by Western blotting and using substrate concentrations ranging from 2 to 25 μm. Values represent mean ± S.E. from four independent experiments.
DISCUSSION
This study is the first to show that a human genetic variant in the Hsp20 gene, which results in the substitution of a highly conserved proline for leucine at amino acid position 20 (P20L), is associated with structural alterations, diminished phosphorylation at Ser16, and loss of the cardioprotective effects of Hsp20 under stress conditions. Although this variant has been previously described in the single nucleotide polymorphism data base, we have shown that the C59T transversion is a rare human mutation found in individuals with no heart disease as well as dilated cardiomyopathic patients.
Similar to other small heat shock proteins, the α-crystallin domain of Hsp20 is dominated by β-sheets, whereas its N terminus is characterized by short helices (25, 26). Indeed, the spectra from the circular dichroism analysis in the present study also revealed a pattern typical of proteins rich in β-sheets and low α-helical content. Notably, the amino acid sequence in the Hsp20 N-terminal region contains several proline residues, which probably facilitate proper helical structure by serving as secondary structure breakers in this region. Interestingly, proline 20 is located just four amino acids downstream of the Ser16 phosphorylation site. Our results suggest that the P20L substitution alters Hsp20 structure by creating an extended helical region, which is predicted to be located around the Ser16 phosphorylation site, and by decreasing the protein's β-sheet content. Future studies may be designed to further investigate the effects of the P20L substitution on Hsp20 secondary structure under different conditions, including increased temperature, to allow for partial unfolding of the protein.
Importantly, these alterations make Hsp20 a poorer substrate for phosphorylation. Our study is the first to show that the P20L substitution in Hsp20 disrupts its structure and alters its ability to become phosphorylated at Ser16. This diminished phosphorylation results in complete loss of the cytoprotective properties of Hsp20, pointing to an instrumental role of phosphorylated Hsp20 in cardioprotection. Indeed, our previous studies have shown that the constitutively phosphorylated mutant S16D-Hsp20 conferred cardioprotection against Iso-induced apoptosis in cultured myocytes (7). Conversely, the constitutively dephosphorylated mutant, namely S16A-Hsp20, displayed no anti-apoptotic properties (7), similar to our findings with the P20L mutant. Increased phosphorylation at Ser16 may provide protection at multiple levels. First, we have previously shown that Hsp20 confers protection via inhibition of the Bax/caspase-3 pro-apoptotic pathway (16), which may be subject to regulation by its phosphorylation state. Second, pSer16-Hsp20 may help maintain cardiomyocyte integrity via stabilization of the cytoskeleton (16, 27). Finally, Hsp20 may form both homologous and heterologous macromolecular structures (3, 27, 28), which may be affected by its phosphorylation status. Overall, loss of the Hsp20 ability to become phosphorylated may have detrimental effects on its cellular functions. Future studies expressing either S16A- or S16D-Hsp20 in vivo may further elucidate the physiological role of phosphorylation at Ser16.
Because we have identified this mutation at low frequencies in the cardiomyopathic and non-cardiomyopathic populations, its relevant contribution to cardiac pathogenesis is not clear at this point. However, we did observe loss of Hsp20 anti-apoptotic effects by the P20L substitution. This may be of relevance to both experimental (29–31) and human (32, 33) heart failure, where apoptosis has been shown to be increased. Furthermore, a recent study in transgenic mice expressing a conditionally active form of caspase-8 demonstrated that induction of apoptosis as low as 4–10-fold lower than the degree observed in human heart failure is deleterious as it causes lethal, dilated cardiomyopathy (34). Thus, the need for increased Hsp20 expression becomes more important in the pathophysiological setting of heart failure. Indeed, previous studies have shown that the levels of Hsp20 as well as its phosphorylation levels at Ser16 increase in the diseased myocardium (7, 10, 12, 16, 18) as a protective mechanism (7, 13, 14–17). Consistent with these findings, we observed that the extent of apoptosis was reduced significantly in cells infected with Ad.WT-Hsp20 after simulated I/R compared with the control group. Because we have shown that the mutant Hsp20 loses these beneficial effects, it is intriguing to speculate that this may render the heart more susceptible to insults in a scenario where Hsp20 levels would increase, but would not confer protection.
In summary, we have identified a human Hsp20 mutation that is associated with diminished phosphorylation at Ser16 and negation of the protein's anti-apoptotic effects. Future studies may be designed to elucidate the functional role of this human variant in vivo.
Acknowledgments
We thank Dr. D. T. Kremastinos and Dr. F. Kolokathis for obtaining the samples from the Greek subjects. We also thank S. Figueira for excellent technical assistance and Dr. L. Conforti for providing the hypoxic chamber.
This work was supported, in whole or part, by National Institutes of Health Grants HL-26507, HL-64018, HL-77101 (to E. G. K.), and HL-087861 (to G.-C. F.). This work was also supported by the Leducq Foundation (to E. G. K.) and American Heart Association Predoctoral Fellowship 0715500B (to P. N.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Hsp, heat shock protein; PKA, protein kinase A; Ad, adenovirus; WT, wild-type; GFP, green fluorescent protein; DCM, dilated cardiomyopathy; I/R, ischemia/reperfusion; Iso, isoproterenol; GST, glutathione S-transferase; ELISA, enzyme-linked immunosorbent assay.
References
- 1.Young, J. C., Agashe, V. R., Siegers, K., and Hartl, F. U. (2004) Nat. Rev. Mol. Cell Biol. 5 781-791 [DOI] [PubMed] [Google Scholar]
- 2.Currie, R. W. (1987) J. Mol. Cell. Cardiol. 19 795-808 [DOI] [PubMed] [Google Scholar]
- 3.Kato, K., Goto, S., Inaguma, Y., Hasegawa, K., Morishita, R., and Asano, T. (1994) J. Biol. Chem. 269 15302-15309 [PubMed] [Google Scholar]
- 4.Dillmann, W. H., Mehta, H. B., Barrieux, A., Guth, B. D., Neeley, W. E., and Ross, J., Jr. (1986) Circ. Res. 59 110-114 [DOI] [PubMed] [Google Scholar]
- 5.Donnelly, T. J., Sievers, R. E., Vissern, F. L., Welch, W. J., and Wolfe, C. L. (1992) Circulation 85 769-778 [DOI] [PubMed] [Google Scholar]
- 6.White, F. P., and White, S. R. (1986) Cardiovasc. Res. 20 512-515 [DOI] [PubMed] [Google Scholar]
- 7.Fan, G. C., Chu, G., Mitton, B., Song, Q., Yuan, Q., and Kranias, E. G. (2004) Circ. Res. 94 1474-1482 [DOI] [PubMed] [Google Scholar]
- 8.Kappé, G., Franck, E., Verschuure, P., Boelens, W. C., Leunissen, J. A., and de Jong, W. W. (2003) Cell Stress Chaperones 8 53-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narberhaus, F. (2002) Microbiol. Mol. Biol. Rev. 66 64-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu, G., Egnaczyk, G. F., Zhao, W., Jo, S. H., Fan, G. C., Maggio, J. E., Xiao, R. P., and Kranias, E. G. (2004) Circ. Res. 94 184-193 [DOI] [PubMed] [Google Scholar]
- 11.Boluyt, M. O., Brevick, J. L., Rogers, D. S., Randall, M. J., Scalia, A. F., and Li, Z. B. (2006) Proteomics 6 3154-3169 [DOI] [PubMed] [Google Scholar]
- 12.Dohke, T., Wada, A., Isono, T., Fujii, M., Yamamoto, T., Tsutamoto, T., and Horie, M. (2006) J. Card. Fail. 12 77-84 [DOI] [PubMed] [Google Scholar]
- 13.Islamovic, E., Duncan, A., Bers, D. M., Gerthoffer, W. T., and Mestril, R. (2007) J. Mol. Cell. Cardiol. 42 862-869 [DOI] [PubMed] [Google Scholar]
- 14.Zhu, Y. H., Ma, T. M., and Wang, X. (2005) Acta Pharmacol. Toxicol. 26 1193-1200 [DOI] [PubMed] [Google Scholar]
- 15.Zhu, Y. H., and Wang, X. (2005) Acta Pharm. Sin. 26 1076-1080 [DOI] [PubMed] [Google Scholar]
- 16.Fan, G. C., Ren, X., Qian, J., Yuan, Q., Nicolaou, P., Wang, Y., Jones, W. K., Chu, G., and Kranias, E. G. (2005) Circulation 111 1792-1799 [DOI] [PubMed] [Google Scholar]
- 17.Fan, G. C., Yuan, Q., Song, G., Wang, Y., Chen, G., Qian, J., Zhou, X., Lee, Y. J., Ashraf, M., and Kranias, E. G. (2006) Circ. Res. 99 1233-1242 [DOI] [PubMed] [Google Scholar]
- 18.De Celle, T., Vanrobaeys, F., Lijnen, P., Blankesteijn, W. M., Heeneman, S., Van Beeumen, J., Devreese, B., Smits, J. F., and Janssen, B. J. (2005) Exp. Physiol. 90 593-606 [DOI] [PubMed] [Google Scholar]
- 19.Unneberg, P., Merelo, J. J., Chacón, P., and Morán, F. (2001) Proteins 42 460-470 [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez, P., Mitton, B., Waggoner, J. R., and Kranias, E. G. (2006) J. Biol. Chem. 281 38599-38608 [DOI] [PubMed] [Google Scholar]
- 21.Qin, Y., Vanden Hoek, T. L., Wojcik, K., Anderson, T., Li, C., Shao, Z., Becker, L. B., and Hamann, K. J. (2004) Am. J. Physiol. 286 H2280-H2286 [DOI] [PubMed] [Google Scholar]
- 22.van de Klundert, F. A., Smulders, R. H., Gijsen, M. L., Lindner, R. A., Jaenicke, R., Carver, J. A., and de Jong, W. W. (1998) Eur. J. Biochem. 258 1014-1021 [DOI] [PubMed] [Google Scholar]
- 23.Merck, K. B., Groenen, P. J., Voorter, C. E., de Haard-Hoekman, W. A., Horwitz, J., Bloemendal, H., and de Jong, W. W. (1993) J. Biol. Chem. 268 1046-1052 [PubMed] [Google Scholar]
- 24.Kim, M. V., Seit-Nebi, A. S., Marston, S. B., and Gusev, N. B. (2004) Biochem. Biophys. Res. Commun. 315 796-801 [DOI] [PubMed] [Google Scholar]
- 25.Kim, K. K., Kim, R., and Kim, S. H. (1998) Nature 394 595-599 [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Ranea, J. A., Mirey, G., Camonis, J., and Valencia, A. (2002) FEBS Lett. 529 162-167 [DOI] [PubMed] [Google Scholar]
- 27.Pipkin, W., Johnson, J. A., Creazzo, T. L., Burch, J., Komalavilas, P., and Brophy, C. (2003) Circulation 107 469-476 [DOI] [PubMed] [Google Scholar]
- 28.Fontaine, J. M., Sun, X., Benndorf, R., and Welsh, M. J. (2005) Biochem. Biophys. Res. Commun. 337 1006-1011 [DOI] [PubMed] [Google Scholar]
- 29.Sharov, V. G., Sabbah, H. N., Shimoyama, H., Goussev, A. V., Lesch, M., and Goldstein, S. (1996) Am. J. Pathol. 148 141-149 [PMC free article] [PubMed] [Google Scholar]
- 30.Teiger, E., Than, V. D., Richard, L., Wisnewsky, C., Tea, B. S., Gaboury, L., Tremblay, J., Schwartz, K., and Hamet, P. (1996) J. Clin. Investig. 97 2891-2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Z., Bing, O. H., Long, X., Robinson, K. G., and Lakatta, E. G. (1997) Am. J. Physiol. 272 H2313-H2319 [DOI] [PubMed] [Google Scholar]
- 32.Olivetti, G., Abbi, R., Quaini, F., Kajstura, J., Cheng, W., Nitahara, J. A., Quaini, E., Di Loreto, C., Beltrami, C. A., Krajewski, S., Reed, J. C., and Anversa, P. (1997) N. Engl. J. Med. 336 1131-1141 [DOI] [PubMed] [Google Scholar]
- 33.Guerra, S., Leri, A., Wang, X., Finato, N., Di Loreto, C., Beltrami, C. A., Kajstura, J., and Anversa, P. (1999) Circ. Res. 85 856-866 [DOI] [PubMed] [Google Scholar]
- 34.Wencker, D., Chandra, M., Nguyen, K., Miao, W., Garantziotis, S., Factor, S. M., Shirani, J., Armstrong, R. C., and Kitsis, R. N. (2003) J. Clin. Investig. 111 1497-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]