Abstract
Toll-like receptors (TLRs) recognize pathogen-associated molecules and play a vital role in promoting an immune response against invading microbes. TLR2, one of the key members of the TLR family, recognizes a wide variety of microbial products, including lipoproteins and lipopeptides, from a number of pathogens. Recent studies from our laboratory indicate that glycopeptidolipids (GPLs), a major surface component of Mycobacterium avium and other non-tuberculosis mycobacteria, are ligands for TLR2. However, the molecular requirements necessary for the GPL-TLR2 interaction were not defined in this report. In the present study we isolated different GPL species from M. avium, and using mass spectrometry and NMR analyses, characterized the molecular requirements of the GPL-TLR2 interaction. Interestingly, the extent of the respective acetylation and methylation of the 6-deoxytalose and rhamnose contained within the core GPL structure dictated whether the GPL signaled through TLR2. These experiments illustrate how subtle changes in a complex TLR2 ligand can alter its affinity for this important receptor, and suggest that M. avium could potentially modify its GPL structure to limit its interaction with TLR2.
Toll-like receptors (TLRs)3 are a class of pattern-recognition receptors expressed by cells of the innate immune system. These receptors are known for their ability to recognize a wide variety of conserved molecules expressed by pathogens. In mice there are 11 members of the TLR family that recognize an array of structures, including unmethylated CpG DNA, flagellin, glycolipids, peptidoglycan, heat-shock proteins, taxol, among others (1). Upon stimulation, TLRs dimerize and couple to myeloid differentiation primary response protein 88 (MyD88), a common cytoplasmic adapter molecule required for TLR signaling, leading to the activation and nuclear translocation of nuclear factor-κB (NFκB) and the production of pro-inflammatory mediators, such as TNF-α. TLRs are therefore important in activating the innate immune system and instructing the development of the adaptive response upon recognition of the various pathogen-associated molecular patterns (2). The pro-inflammatory responses elicited by TLR2 and TLR4 have been widely examined with respect to ligand recognition and signaling. TLR4 was originally characterized as the receptor that plays a pivotal role in enterobacterial lipopolysaccharide (LPS)-induced septic shock (3). TLR2 is also noteworthy as it is able to recognize an extremely diverse array of microbial products, such as lipoproteins and lipopeptides from different pathogens, including Staphylococcus aureus (4), Borrelia burgdorferi (5), and Mycoplasma fermentans (6). Moreover, TLR2 has also been shown to respond to non-enterobacterial LPS (7). TLR2 is able to form heterodimers with TLR1 or TLR6, further expanding the repertoire of bacterial products that it can recognize (8–10).
The importance of TLR-mediated responses during M. tuberculosis infections has been extensively examined. Fenton and colleagues (11) performed in vitro studies and observed that M. tuberculosis activated both TLR2 and TLR4, whereas heat-killed M. tuberculosis activated only TLR2. Similarly, macrophages expressing dominant negative TLR2 resulted in the inhibition of TNF-α production in response to both virulent and avirulent M. tuberculosis strains (12). Another study revealed that mice lacking TLR2 had increased bacterial load, developed chronic pneumonia, and succumbed to infection more readily, as compared with wild-type mice, when challenged with M. tuberculosis (13). A role for TLRs has also been evaluated for M. avium, a major opportunistic pathogen in immunocompromised patients and a significant cause of increased morbidity and mortality in patients with an advanced stage of AIDS (14, 15). In particular, in vitro and in vivo studies have implicated TLR2 in promoting an immune response following M. avium challenge. Lien et al. (6) observed that TNF-α release from peripheral blood mononuclear cells in response to M. avium was blocked when cells were pre-treated with a TLR2-specific antibody. Additionally, TLR2-deficient mice had increased bacterial loads compared with wild-type mice when challenged with a virulent strain of M. avium (16). Another study evaluated the response to M. avium infection in mice that were deficient in TLR2, TLR4, and MyD88. It was observed that MyD88- and TLR2-deficient mice had an increased bacterial load and increased susceptibility to M. avium infection, and failed to recruit neutrophils to the site of infection, as compared with TLR4-deficient and wild-type mice (17). These studies, together, highlight the importance of TLRs, particularly TLR2, in combating M. tuberculosis and M. avium infections.
TLR2-, TLR4-, and TLR9-specific ligands produced by M. tuberculosis have been identified and include the 19-kDa lipoprotein, heat-shock protein 65, and mycobacterial DNA, respectively (18–20). However, the TLR ligands produced by other mycobacteria are less well defined. Recently, we showed that the surface-exposed glycopeptidolipids (GPLs) produced by M. avium can serve as TLR2 but not TLR4 ligands (21). GPLs consist of a fatty acyl chain N-linked to a tripeptide-aminoalcohol core containing d-phenylalanine, d-allo-threonine, d-alanine, and l-alaninol. The allo-threonine is glycosidically linked to a 6-deoxy-α-l-talopyranosyl residue (6-deoxytalose), and the alaninol is glycosidically linked to a methylated α-l-rhamnopyranosyl residue (rhamnose). The structure of GPLs makes them unique TLR2 agonists since they essentially constitute a “mixture” of glycolipids and lipopeptides. Moreover, they contain only one N-linked fatty acid chain, thereby differing from other TLR2 (and TLR4) ligands, which usually contain multiple fatty acids of varying length that can be either O-linked or N-linked (7, 22–24). In the present study, we show that specific acetylation and methylation patterns of the 6-deoxytalose and rhamnose, respectively, which comprise the diglycosylated lipopeptide core of GPL, are required for GPL signaling through the macrophage TLR2. These results further illustrate the complexity and specificity of TLR2-ligand interactions and suggest that M. avium could potentially modify its GPL structure to limit its interaction with TLR2.
EXPERIMENTAL PROCEDURES
Extraction and Purification of GPLs—M. avium strains 104 and 2151 were grown on Middlebrook 7H11 agar (Difco), and total lipids were extracted with a solution of chloroform:methanol (C:M, 2:1 v/v) as described in previous reports (21, 25). GPLs were purified by preparative TLC, using 10 cm × 20 cm Silica gel 60 plates (EMD Chemicals) that were loaded with 3–5 mg of total lipid and developed lengthwise. Nonspecific GPLs were purified using a C:M:H2O (90:10:1, v/v)-developing solvent, and serovar 2 GPLs were purified using a C:M:H2O (50:10:1, v/v)-developing solvent. Side strips were cut from the TLC plate and visualized with a solution of 1% α-naphthol (w/v)/5% sulfuric acid (v/v) in ethanol, and heating to 100 °C. Respective bands were scraped from the TLC plate, re-extracted from the scraped silica in C:M (2:1) overnight at 37 °C, and Folch-washed with endotoxin-free water. Purity of GPLs was checked by TLC using a developing solvent composed of C:M:H2O (50:10:1).
Isolation and Cultivation of Bone Marrow-derived Macrophages—Bone marrow-derived macrophages (BMMΦs) were harvested from the bone marrow of 6- to 8-week-old C57BL/6 mice as described previously (26). Mature macrophages were harvested and stored at –140 °C. Thawed macrophages were cultured on non-tissue culture plates for 3–7 days, and after one passage, were re-plated on tissue-culture plastic at 3 × 105 cells/35-mm plate.
GPL Deacetylation—Native total lipid and purified GPLs were deacetylated by a standard mild alkali treatment (27). Briefly, dried lipids were dissolved in one part C:M (2:1) and one part 0.2 n NaOH in methanol at a total volume of ∼6 ml per 50 mg or less of lipid, and incubated at 37 °C for 40 min. The solution was then neutralized with 2–3 drops of glacial acetic acid, dried down, Folch-washed as described above, dried down, and re-suspended at known concentrations.
Sugar Analyses (GC/MS)—Purified native and deacetylated GPLs were subjected to a standard alditol acetate derivatization procedure where the GPL sugar moieties were hydrolyzed, reduced, and per-acetylated as described (21). Alditol acetates were dissolved in ∼20 μl of high-performance liquid chromatography-grade chloroform (Fisher), and then 1 μl of sample was injected into a Fisons Instruments 8035 gas chromatograph connected to a Fisons Instruments MD 800 quadrupole mass spectrometer. Derivatized sugars were separated on a 60 m × 0.25 mm ID column with a film thickness of 0.25 μm composed of 5% phenyl and 95% dimethylpolysiloxane (Zebron ZB5), using a temperature program beginning at 50 °C for 5 min and increasing to 250 °C at a rate of 10 °C/min, and then holding the temperature at 250 °C for 15 min. Electron ionization mass spectra were acquired in the positive ionization mode in a scan range of 40–340 atomic mass units at a rate of 1 scan/s for the entire run time totaling at 40 min/run.
GPL Exposure to BMMΦs—Native and deacetylated total lipid and purified GPLs were dissolved in high-performance liquid chromatography-grade methanol and directly applied to tissue-culture grade plastic at the indicated concentrations. The methanol was allowed to completely evaporate prior to adding BMMΦs, after which the cells were incubated at 37 °C in 5% CO2 for defined time periods.
TNF-α Enzyme-linked Immunosorbent Assay and Western Blots—TNF-α was measured by enzyme-linked immunosorbent assay as described (21). For Western blots, 10 μg of native and deacetylated total lipid and purified GPLs were exposed to 3 × 105 BMMΦs for 4 h. BMMΦs were then washed with ice-cold phosphate-buffered saline and treated for 10 min with ice-cold lysis buffer as previously described (26). Equal amounts of protein as determined by the MicroBCA protein assay (Pierce) were analyzed by Western blot for phosphorylated and total p38 and phosphorylated and total p65 NFκB (21).
ESI/MS and ESI/MS/MS Analyses—Purified native and deacetylated GPLs were dissolved in an high-performance liquid chromatography-grade solution of methanol:methylene chloride (2:1, v/v) and analyzed using a Micromass (Beverly, MA) Quattro-LC triple quadrupole (Q1q-Q3) instrument employing MassLynx 4.0 software. The Quattro-LC Z-spray interface was operated in the ESI mode with the following parameters: capillary voltage = 2.4–3.0 kV, cone voltage = 25–40 V, source temperature = 120–125 °C, desolvation temperature = 150 °C, and desolvation gas (N2) flow rate = 271–414 liters/h. For direct infusion experiments, a Harvard apparatus (Holliston, MA) syringe pump delivered samples at a flow rate of 10 μl/min. For MS/MS experiments, the mass spectrometer tune was optimized by directly infusing a solution of GPL dissolved in methanol:methylene chloride (2:1, v/v). Collisionally activated dissociation (CAD) was employed for all MS/MS experiments by admitting argon into the collision cell (q). For product ion scans, argon was admitted into q until the precursor signal intensity was reduced by ∼50%, then the collision energy (voltage offset between Q1 and Q3) was increased to 20–40 eV. MS/MS spectra were acquired in the multichannel acquisition mode of data acquisition at a rate of 400 atomic mass units/s.
NMR Spectroscopy—NMR spectra were obtained at 22 °C on a Varian UNITYPlus FT-NMR spectrometer operating at 599.887 MHz for 1H. Samples were analyzed in 5-mm NMR tubes (Wilmad) at a concentration of ∼10 mm in CDCl3 solvent. Chemical shifts were referenced to the internal residual CHCl3 signal (7.270 ppm). 1H spectra were collected with a 7000-Hz spectral window and ∼5.0-s recycle time. Free induction decays were zero-filled to give final digital resolutions of <0.05 Hz/pt. The J-couplings are reported in hertz and are accurate to ±0.1 Hz unless otherwise indicated. The two-dimensional 1H-1H COSY spectra were obtained with a 6200-Hz spectral window and ∼2.0-s recycle time. 256 t1 increments of 16 scans each were sampled. Free induction decays were zero-filled to give 4K × 4K complex data sets that were Fourier-transformed with line broadening, sinebell, and/or Gaussian weighting functions in both domains. Two-dimensional rotating frame nuclear overhauser effect spectra were obtained with a 6200-Hz spectral window using a spin-lock mixing time of 200 ms, spin-lock field strength of 3731 Hz, and a relaxation delay of 2 s. 256 t1 increments of 64 scans each were sampled, and free induction decays were zero-filled to give 4K × 4K complex data sets. Data sets were Fourier-transformed with line broadening and Gaussian weighting functions in both domains, and a baseline correction was applied in the F2 domain.
RESULTS
Isolation and Characterization of the Serovar 2 GPLs—Previously, we observed that some GPLs could activate BMMΦsina MyD88- and TLR2-dependent manner while other GPLs failed to induce activation (21). We found that the serovar 2 GPL from M. avium 2151 activated p38 and IκBα, and stimulated TNF-α production from BMMΦs in a MyD88- and TLR2-dependent manner (21). However, in this previous study no attempt was made to separate the potential variants of the serovar 2 GPL to define the specific structural requirements necessary for activity. In the present study we used a TLC solvent system that allowed for a more refined separation of the different serovar 2 GPL species. Using this system, which consisted of chloroform: methanol:water (50:10:1, v/v), we were able to isolate two variants of serovar 2 GPL, which we designated S2A and S2B. The difference in migration on the TLC plate was lost if the GPLs were treated with mild base to remove acetyl groups, suggesting that the difference between the two species was due to the number of acetyl groups (Fig. 1A).
FIGURE 1.
Purified serovar 2 structural variants analyzed by TLC and GC/MS. A, TLC of native (N) and deacetylated (DA) serovar 2 structural variants. The more apolar serovar 2 GPL was named S2A, and the more polar species was named S2B. Upon deacetylation with a mild base treatment, both S2A and S2B increased in polarity, suggesting both species are acetylated in their native conformations. B, GC/MS analyses confirmed the presence of the sugars that constitute the serovar 2 GPL. Results were identical for both S2A and S2B (only S2B shown). Rha, rhamnose; Fuc, fucose; 6dTal, 6-deoxytalose.
To confirm that S2A and S2B were both serovar 2 GPLs, the carbohydrate composition was analyzed by alditol acetate derivatization and GC/MS. GC/MS analyses confirmed the presence of the sugars expected in the serovar 2 GPL, including the 3,4-di-O-methylrhamnose (m/z 131, 190, and 234), 2,3-di-O-methylfucose (m/z 118, 143, and 203), 6-deoxytalose (m/z 129, 171, and 231), and the rhamnose (m/z 129, 171, and 231). Results were nearly identical for both S2A and S2B (only GC trace for S2B shown, Fig. 1B). These data together indicate that there are two species of the serovar 2 GPL produced by M. avium 2151, where the glycosylation and methylation patterns are identical as revealed by GC/MS analyses, but that both species are acetylated to different extents as indicated by the TLC data.
Native but Not Deacetylated S2B GPL Stimulates Macrophage Activation—M. avium 2151 lipids were exposed to BMMΦs, as described under “Experimental Procedures,” where the BMMΦs were isolated from wild-type, MyD88–/–, and TLR2–/– C57BL/6 mice. The macrophages were exposed to native and deacetylated total lipid, S2A, S2B, methanol only (solvent control), or were left untreated. Culture supernatants were collected 24 h post-treatment and assayed for TNF-α production, and in separate experiments, cells were lysed 4 h post-treatment and assayed for p38 and NFκB phosphorylation. The results demonstrate that native total lipid and the S2B GPL induced TNF-α production in a dose-dependent manner, and that production was dependent on MyD88 and TLR2. Additionally, the ability of S2B to stimulate TNF-α production was abrogated upon deacetylation (Fig. 2A). In contrast, BMMΦs treated with native S2A produced minimal TNF-α (Fig. 2A). Furthermore, native S2B stimulated p38 and NFκB phosphorylation in a MyD88- and TLR2-dependent fashion, while native S2A failed to induce protein phosphorylation above what was observed in untreated cells (Fig. 2B). As predicted, deacetylation of the S2B GPL completely abrogated its ability to activate p38 and NFκB (Fig. 2B).
FIGURE 2.
A native acetylated serovar 2 GPL activates macrophages in a TLR2-dependent manner. Native (N) and deacetylated (DA) total lipid, S2A and S2B were exposed to wild-type (WT/BL/6), MyD88–/– (M–/–), and TLR2–/– (T–/–) macrophages from C57BL/6 mice and compared with methanol only (MeOH) and non-treated controls (NT). A, TNF-α production was analyzed by enzyme-linked immunosorbent assay after 24 h of exposure; n = 3 separate experiments ± S.E. B, p38 and NFκB phosphorylation was measured after 4 h of macrophage exposure to GPLs. pp38, phosphorylated p38; pNFκB; phosphorylated NFκB.
The O-Acetyl Groups of the Serovar 2 GPLs Are Located on the 6-Deoxytalose—ESI mass spectra were obtained in the positive ion mode for both native and deacetylated S2A and S2B. The ESI mass spectrum of native S2A showed high abundance peaks at m/z 1561 and 1595, as well as minor peaks representing possible differences in acyl chain length (14-atomic mass unit intervals) or those with sodium adducts (Table 1 and supplemental Fig. S1A). At m/z 1477 and 1511, there was a downward shift of ∼84 atomic mass units compared with the respective native S2A peaks at m/z 1561 and 1595, suggesting a difference of two acetyl groups between the native and deacetylated S2A species (Table 1 and supplemental Fig. S1A). The positive ion mode ESI mass spectrum for native S2B showed major peaks at m/z 1519 and 1553, as well as other peaks representing sodiation at m/z 1541 [1519-H+Na]+ and m/z 1575 [1553-H+Na]+ (Table 1 and Fig. 3A). The ESI mass spectrum of deacetylated S2B had prominent peaks at m/z 1477 and 1511, representing a downward shift of 42 atomic mass units compared with peaks in the ESI mass spectrum of native S2B, suggesting the native species peaks each contained one acetyl group (Table 1 and Fig. 3A).
TABLE 1.
Major intact GPL ions and product ions of S2A and S2B native (N) and de-acetylated (DA) species from ESI mass spectra and ESI production mass spectra, respectively
The product ion m/z values were obtained from CAD at a collision energy of 20 eV. The masses of the sugar moieties lost were determined by the difference between m/z values of product ions.
| Intact GPL ion m/z values | Product ion m/z values | Sugar moiety lost | |
|---|---|---|---|
| atomic mass units | |||
| S2A (N) | 1561 | 1386, 1240, 1065, 835 | 175, 146, 175, 230 |
| 1595 | 1420, 1274, 1099, 869 | 175, 146, 175, 230 | |
| S2A (DA) | 1477 | 1302, 1156, 1010, 835 | 175, 146, 146, 175 |
| 1511 | 1336, 1190, 1044, 869 | 175, 146, 146, 175 | |
| S2B (N) | 1519 | 1344, 1198, 1023, 835 | 175, 146, 175, 188 |
| 1553 | 1378, 1232, 1057, 869 | 175, 146, 175, 188 | |
| S2B (DA) | 1477 | 1302, 1156, 1010, 835 | 175, 146, 146, 175 |
| 1511 | 1336, 1190, 1044, 869 | 175, 146, 146, 175 |
FIGURE 3.
S2B is acetylated on the 6-deoxytalose. A, positive ion mode ESI mass spectra of native (N) and deacetylated (DA) biologically active S2B were compared and demonstrated a shift of approximately one acetyl group (42 atomic mass units) when comparing relevant peaks (i.e. m/z 1519 versus m/z 1477, and m/z 1553 versus m/z 1511). B, selected ions were subjected to undergo CAD. At a collision energy of 20 eV, product ions represented the protonated GPL ion with the sequential loss of sugar moieties.
To identify the location of the putative acetyl groups, product ion tandem mass spectrometry (MS/MS) experiments were performed on each of the major GPL peaks by subjecting individually selected ions to undergo CAD at collision energies of 20, 30, and 40 eV. Product ion mass spectra from native and deacetylated species were then compared. At a collision energy of 20 eV, product ion mass spectra consistently showed a fragmentation pattern signifying the sequential loss of individual sugar moieties from the GPL ions. Five major peaks were observed in each product ion mass spectrum, where the peak with the highest m/z value represented the intact GPL ion, and the subsequent peaks denoted the ions with the loss of an individual sugar moiety. As shown in Table 1, the m/z values of intact ions are listed with the respective product ions resulting from CAD at 20 eV. The “sugar moieties lost” column represents the numerical difference between each of the m/z values of the prominent adjacent peaks and indicates the mass (atomic mass units) of each of the sugars released from the intact GPL ion upon CAD (Table 1). For example, when analyzing the biologically active major native S2B intact GPL ion at m/z 1553, there was a sequential loss of a dimethylated 6-deoxyhexose (175 atomic mass units; product ion m/z 1378), an unmodified 6-deoxyhexose (146 atomic mass units; product ion m/z 1232), another dimethylated 6-deoxyhexose (175 atomic mass units; product ion m/z 1057), and lastly, a diacetylated 6-deoxyhexose (188 atomic mass units; product ion m/z 869) (Table 1 and Fig. 3, A and B). When comparing these m/z values to those of deacetylated S2B, the masses of the lost sugar moieties were identical, except for one moiety loss of 146 atomic mass units, which represented another unmodified 6-deoxyhexose (Table 1 and Fig. 3B). These results were consistent for each of the intact GPL ions analyzed (data not shown). Because the acetylated sugar was repeatedly the last carbohydrate to fragment from the intact GPL protonated ion, we hypothesized that the acetyl group was located on the 6-deoxytalose as this is the only non-methylated sugar attached to the lipopeptide core. However, the exact position of the acetyl group could not be determined by MS/MS experiments alone (Fig. 3B).
The same tandem MS experiments were performed to compare native non-active S2A with deacetylated S2A. Product ion mass spectra of intact GPL ions of native S2A consistently indicated the loss of two dimethylated 6-deoxyhexoses, representing the 3,4-di-O-methylrhamnose and 2,3-di-O-methylfucose (175 atomic mass units each), an unmodified 6-deoxyhexose (146 atomic mass units), and a diacetylated 6-deoxyhexose (230 atomic mass units) (Table 1 and supplemental Fig. S1B). The diacetylated sugar moiety was always the last component to fragment from the intact GPL protonated ions (Table 1 and supplemental Fig. S1B), supporting the assertion that the acetyl groups were likely located on the C3 and C4 carbons of the 6-deoxytalose, because according to published data these are the only positions available for modification, as the 6-deoxytalose is glycosidically linked to a rhamnose at the C2 carbon (28). Overall, these data suggest that the biologically active serovar 2 GPL (i.e. native S2B) has one O-acetyl group on the 6-deoxytalose, and that when this substitution is selectively removed by mild alkali treatment, the GPL loses its activity. Moreover, the addition of a second O-acetyl group on the 6-deoxytalose also rendered the serovar 2 GPL non-active (i.e. native S2A). Together, these data indicate that a minimal structural variation, namely the acetylation pattern of the 6-deoxytalose, can dictate whether the serovar 2 GPL molecule signals through the TLR2.
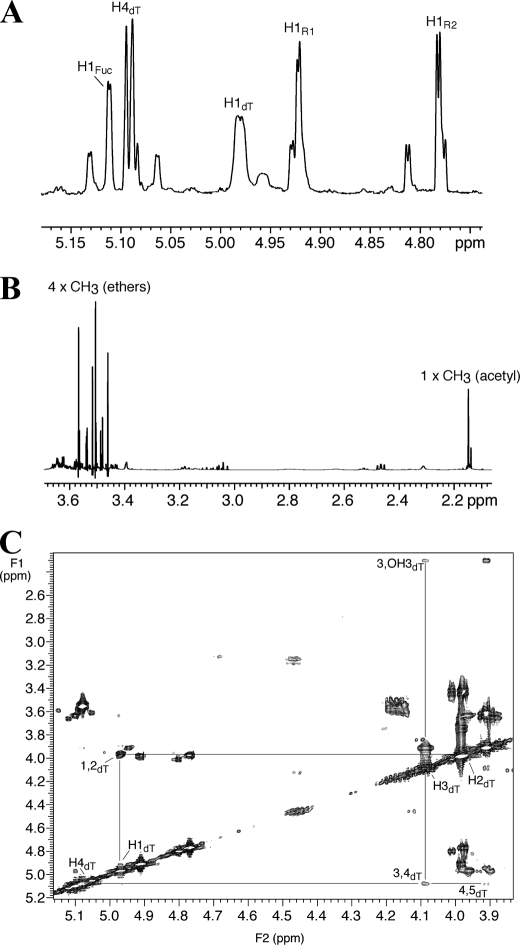
The Positioning of the O-Acetyl Group on the Biologically Active Serovar 2 GPL by NMR Spectroscopy—To establish the location and position of the O-acetyl groups on the serovar 2 GPL molecules, NMR spectroscopy was employed. The one-dimensional 1H NMR spectrum of biologically active native S2B contained four dominant anomeric proton signals at 5.112, 4.981, 4.922, and 4.782 ppm, and tentative assignments were made to fucose (Fuc), 6-deoxytalose (dT), and two rhamnose (R) residues, respectively (Fig. 4A). Assignment of the H1dT signal was based on a partial analysis of the two-dimensional 1H-1H COSY spectrum (Fig. 4C), which revealed a relatively deshielded H4dT signal (∼5.09 ppm; Fig. 4A) consistent with the presence of acetylation at O4 of this residue. Observation of an H3-OH3 cross-peak is consistent with a lack of substitution at O3 in the 6-deoxytalose. The one-dimensional 1H spectrum showed the presence of four dominant CH3 signals arising from -OCH3 groups, and a single dominant CH3 signal arising from a single acetyl group (Fig. 4B). On the other hand, the one-dimensional 1H NMR spectrum for S2A showed the presence of four dominant CH3 signals arising from -OCH3 groups, and two dominant signals arising from two acetyl groups (supplemental Fig. S2), corroborating GC/MS and ESI/MS/MS results.
FIGURE 4.
NMR analyses of S2B indicate the 6-deoxytalose is acetylated on the 4′ carbon. A, partial one-dimensional 1H NMR spectrum of native S2B in CDCl3 showing tentative assignments of the four predominant anomeric 1H signals, and assignment of the H4dT signal. B, partial one-dimensional 1H NMR spectrum of native S2B showing the presence of four predominant -OCH3 signals and one dominant CH3 signal arising from an acetyl group. C, partial two-dimensional 1H-1H COSY spectrum of native S2B in CDCl3 showing only correlations of the 6-deoxytalose residue. These data suggest the presence of an O-acetyl group at C4 of this residue based on the observation of the relatively deshielded H4dT signal. dT, 6-deoxytalose; R, rhamnose; Fuc, fucose.
Isolation and Characterization of nsGPLs—Many studies to date have focused on the activities of ssGPLs, however, none of these have demonstrated whether nsGPLs similarly possess biological activity (29–31). Therefore, we analyzed nsGPLs for the ability to stimulate macrophages in a TLR2-dependent manner since they could also hypothetically be acetylated on the 6-deoxytalose and thus resemble the active serovar 2 structural variant. Two nsGPL bands were isolated by preparative TLC (designated NS2 and NS3), with NS2 being the more apolar species. Purified native and deacetylated NS2 and NS3 were resolved on TLC plates to gain preliminary information about the methylation and acetylation patterns of these GPLs. The change in the migration pattern of NS2 upon deacetylation suggested that this species was acetylated in its native form (Fig. 5A). In contrast, NS3 appeared to be a mixture of two GPL species, because alkali treatment of NS3 resulted in the resolution of two distinct bands (Fig. 5A). These TLC data suggested that the original NS3 band consisted of two species that differed in both their methylation and acetylation patterns, with one GPL being more methylated and not acetylated, and the other species being acetylated but with fewer methyl groups. To test this prediction, deacetylated bands comprising NS2 and NS3 were purified, derivatized, and analyzed using GC/MS analyses. Results confirmed that NS2 consisted of one GPL containing a 3,4-di-O-methylrhamnose (m/z 131, 190, and 234) and a 3-O-methyl-6-deoxytalose (m/z 130, 143, 190, and 203) (Fig. 5B). NS3 comprised two GPLs, where the more apolar species contained a 3,4-di-O-methylrhamnose and a 3-O-methyl-6-deoxytalose, whereas the more polar GPL consisted of a 3-O-methylrhamnose and a 3-O-methyl-6-deoxytalose (both with m/z 130, 143, 190, and 203) (Fig. 5B).
FIGURE 5.
Isolated nonspecific GPLs (nsGPLs) contain both active and non-active species that differ in their acetylation and methylation patterns. A, TLC of native (N) and deacetylated (DA) nsGPL structural variants. The more apolar nsGPL was named NS2 and the more polar species was named NS3. Upon deacetylation, NS2 increased in polarity, suggesting that it is acetylated in its native conformation. NS3 appeared to be a mixture of two nsGPLs that differ in both their acetylation and methylation patterns. B, GC/MS analyses identified the number of methyl groups located on the sugars of the respective purified deacetylated nsGPLs. Lower panel, TNF-α production was measured upon exposure to total lipid (TL) and native and deacetylated (nsGPLs). NT, not-treated. n = 3 separate experiments ± S.E.
Native but Not Deacetylated NS3 Stimulates Macrophage Activation—M. avium 104 total lipid and purified nsGPLs, in their native and deacetylated forms, were exposed to BMMΦs. Culture supernatants and cell lysates were collected from treated macrophages as described above for the serovar 2 GPLs. Native NS3, but not NS2, activated BMMΦs to release TNF-α in a MyD88- and TLR2-dependent manner (Fig. 5, lower panel). As was observed with the serovar 2 GPL, the activity of native NS3 was abolished upon deacetylation (Fig. 5, lower panel). Moreover, only the native NS3 stimulated p38 and NFκB phosphorylation in a MyD88- and TLR2-dependent manner, which was again abolished upon deacetylation (data not shown).
The Biologically Active nsGPL Has One O-Acetyl Group—The biological assays implicate a role for O-acetyl groups in stimulating BMMΦs by native NS3. Moreover, TLC and GC/MS data suggest that the active NS3 GPL contains a 3-O-methylrhamnose, a 3-O-methyl-6-deoxytalose, and is acetylated. To determine the location of the acetyl group(s) on this nsGPL we obtained both ESI mass spectra and product ion mass spectra for the nsGPLs.
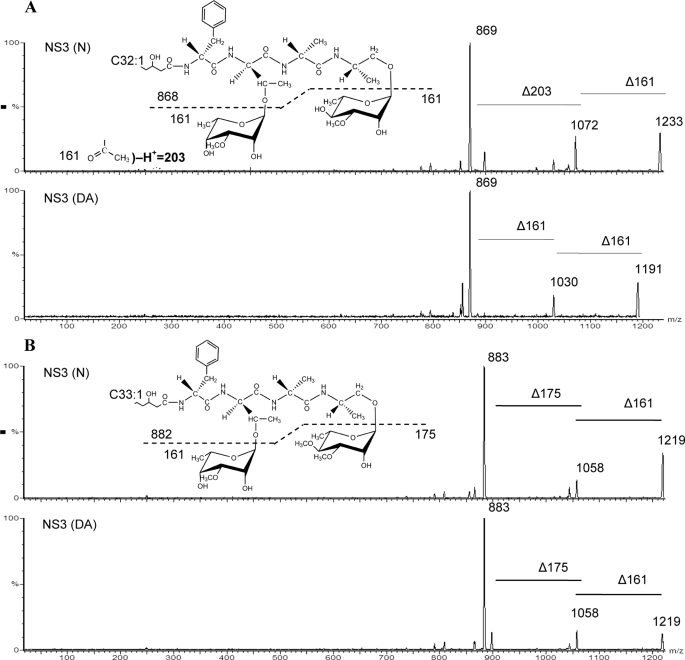
The positive mode ESI mass spectrum for native NS3 GPL showed major peaks with m/z 1219, 1233, and 1247 (Table 2 and supplemental Fig. S3). The positive ESI mass spectrum for deacetylated NS3 showed protonated species with m/z 1191, 1205, and 1219, plus these ions with sodium adducts (indicated by [M-H+Na]+) (Table 2 and supplemental Fig. S3). When comparing the ESI mass spectra for native and deacetylated NS3, there was a shift of 42 atomic mass units for some of the m/z values (Table 2 and supplemental Fig. S3). However, the peak at m/z 1219 appeared in both the native and deacetylated ESI mass spectra, and we hypothesized that this ion represented the non-acetylated species (Table 2 (Footnote a) and supplemental Fig. S3). To determine the structures of the acetylated and non-acetylated NS3 GPLs, product ion mass spectra were obtained. The GPL ions represented by peaks at m/z 1233 and 1247 both generated product ions indicating the presence of a monomethylated 6-deoxyhexose (161 atomic mass units) and a monomethylated, monoacetylated 6-deoxyhexose (203 atomic mass units) (Table 2 and Fig. 6A). Upon CAD, the product ions from peaks at m/z 1191 and 1205 signified the loss of two monomethylated 6-deoxyhexoses (161 atomic mass units each) (Table 2 and Fig. 6A). These results indicated the presence of one O-acetyl group located on either the 3-O-methylrhamnose or on the 3-O-methyl-6-deoxytalose of the native, active nsGPL (Fig. 6A). MS/MS experiments could not define the exact location of this O-acetyl substitution.
TABLE 2.
Major intact GPL ions and product ions of NS2 and NS3 native (N) and deacetylated (DA) species from ESI mass spectra and ESI product ion mass spectra, respectively
The product m/z values were obtained from CAD at a collision energy of 20 eV. The masses of the sugar moieties lost were determined by the difference between m/z values of product ions.
| Intact GPL ion m/z values | Product ion m/z values | Sugar moiety lost | |
|---|---|---|---|
| atomic mass units | |||
| NS2 (native) | 1247 | 1072, 869 | 175, 203 |
| 1261 | 1086, 883 | 175, 203 | |
| NS2 (DA) | 1205 | 1044, 869 | 161, 175 |
| 1219 | 1058, 883 | 161, 175 | |
| NS3 (native) | 1219a | 1058, 883 | 161, 175 |
| 1233 | 1072, 869 | 161, 203 | |
| 1247 | 1086, 883 | 161, 203 | |
| NS3 (DA) | 1191 | 1030, 869 | 161, 161 |
| 1205 | 1044, 883 | 161, 161 | |
| 1219a | 1058, 883 | 161, 175 |
The non-acetylated GPL ion found in both N and DA samples.
FIGURE 6.
Product ion MS/MS spectra from native NS3. A, ions for active native NS3 (N) and deacetylated (DA), m/z 1233 was subjected to CAD, which indicated the presence of one acetyl group on one of the 3-O-methyl-6-deoxyhexoses. B, product ion MS/MS results confirm that the NS3 with m/z 1219 is a non-acetylated nsGPL.
As mentioned above, ESI mass spectra showed a peak at m/z 1219 in both the native and deacetylated NS3 samples. Product ion mass spectra of this peak resulted in ions signifying the loss of sugars consistent with a dimethylated 6-deoxyhexose (175 atomic mass units; i.e. 3,4-di-O-methylrhamnose) and a monomethylated 6-deoxyhexose (161 atomic mass units; 3-O-methyl-6-deoxytalose) (Fig. 6B). The fragmentation pattern for this ion was identical in both the native and deacetylated samples, and as predicted, represented the second non-acetylated GPL structure in the native NS3 mixture, namely the non-active nsGPL containing a 3,4-di-O-methylrhamnose and a 3-O-methyl-6-deoxytalose, as detected by the TLC and GC/MS analyses. Similar analyses were performed on native and deacetylated NS2 and supported the presence of one O-acetyl group on the 6-deoxytalose (data not shown).
The O-Acetyl Group Is Located on C4 of the 6-Deoxytalose for Both NS2 and NS3: Assignment of 1H Chemical Shifts, Anomeric Configuration, and Substitution Pattern for Native NS3—The one-dimensional 1H NMR spectrum of NS3 contained four anomeric proton signals at 5.066, 5.036, 4.848, and 4.799 ppm (Fig. 7A). These signals gave nearly identical integrations and were split by a small 3JH1,H2 coupling (1.6–1.7 Hz). Analysis of the two-dimensional 1H-1H COSY spectrum (Fig. 8) provided chemical shift assignments of all non-anomeric protons in each residue. Based on the analysis of data obtained on the simpler NS2 structure (see supplemental material), the signal assignments were consistent with an approximately equimolar mixture of two species, denoted AdT/AR (active 6-deoxytalose/active rhamnose) and NdT/NR (non-active 6-deoxytalose/non-active rhamnose), respectively. NdT contains an -OCH3 group at C3, whereas AdT contains an -OCH3 group at C3 and an O-acetyl group at C4. NR contains two -OCH3 groups at C3 and C4, whereas AR contains only one -OCH3 group at C3. In all four residues, the hydroxyl group at C2 was unprotected. The structural assignments follow from the observation of the one-dimensional spectrum where there were five -OCH3 signals and one acetyl methyl signal (Fig. 7B) and six hydroxyl proton signals (Fig. 7C). The latter OH signals could be specifically assigned to each residue using the COSY data (Fig. 8B), thereby confirming the substitution patterns, and COSY correlations through each residue were extensive enough to allow assignment of the four H6 COSY signals (Fig. 8C). Similar analysis were performed on native NS2, which confirmed the presence of a GPL-containing a 3,4-di-O-methylated rhamnose and a 3-O-methyl-, 4-O-acetyl-6-deoxytalose (supplemental information and Figs. 4 and 5).
FIGURE 7.
One-dimensional 1H NMR spectrum of NS3. A, partial spectrum showing the four anomeric H1 signals and that of H4dT. B, partial spectrum showing the five methyl ether signals and the signal arising from the CH3 of an acetyl group. C, partial spectrum showing assignments of the six OH protons. AdT, active 6-deoxytalose; AR, active rhamnose; NdT, non-active 6-deoxytalose; NR, non-active rhamnose.
FIGURE 8.
Two-dimensional 1H-1H NMR spectrum of NS3 indicates that the active species is acetylated on the 6-deoxytalose at the 4′ carbon. A, partial COSY spectrum showing the four H1–H2 cross-peaks, and cross-peaks for H3–H4 and H4–H5 of the 6-deoxytalose of the active nsGPL comprising NS3 (AdT). B, partial COSY spectrum showing assignments of cross-peaks for the four residues: 6-deoxytalose of the non-active (NdT) and active species (AdT) species, and rhamnose of the non-active (NR) and active (AR) species. C, partial COSY spectrum showing the H5–H6 cross-peaks for the four residues.
DISCUSSION
The ability of the innate immune system to recognize pathogen-associated molecular patterns is critical for a protective immune response. The primary mediators of this recognition are the pattern recognition receptors, which include the TLRs, dectin-1, mannose receptor, among others (29). A significant body of research has identified the ligands recognized by the various pattern-recognition receptors, with much of the focus centered on TLRs. What is clear from these studies is that, although specific TLRs can bind structurally diverse ligands, minor modifications can lead to a loss in binding or a change in receptor specificity. This has been clearly demonstrated with LPS produced by Gram-negative bacteria whose structural variants have been shown to bind TLR4, TLR2, or neither (7, 30, 31). One study demonstrated that the number of phosphates and the number and length of the acyl chains appended to LPS molecules dictate whether they will bind TLR2 or TLR4 (7). Another study observed that a mutant Escherichia coli strain produced a penta-acylated LPS molecule that was considerably less stimulatory when compared with the well known hexaacylated structure (30), and yet others have demonstrated that the presence of a secondary acyl chain length of 10 carbons was optimal for hexa-acylated LPS to induce a TLR4-dependent pro-inflammatory response (24).
In the context of mycobacteria, non-pathogenic mycobacteria produce lipoarabinomannan (LAM) with arabinose caps (AraLAM) or phospho-myo-inositol caps (PILAM), while pathogenic mycobacteria, such as M. tuberculosis, produce mannosylated LAM (ManLAM) (32, 33). AraLAM and PILAM are known to elicit a signaling response through TLR2 (11, 33, 34). In contrast, ManLAM does not interact with TLR2, but rather with the mannose receptor (35). In addition to altering the glycosylation patterns, mycobacteria are also able to alter the acylation of glycolipids. For example, mycobacteria can modify the structure of lipomannan, a biosynthetic precursor for LAM. Lipomannan is produced in a variety of “acyl” forms, ranging from monoacylated to tetra-acylated (36). It was shown that lipomannans with three or four acyl chains stimulate TLR2 to elicit TNF-α production from murine and human macrophages, whereas monoacylated and diacylated forms failed to induce cytokine production (36). Together, these studies highlight how the structures of various pathogen-associated molecular patterns can influence their recognition by certain TLRs and emphasize the importance of the glycosylation and acylation states for many of the ligands.
In the context of GPLs, studies have implicated that the presence of O-acetyl groups on M. smegmatis GPLs are important in eliciting a macrophage response. In one study, GPLs of M. smegmatis were shown to inhibit phagocytosis by macrophages. However, when the inhibitory GPLs were deacetylated using mild alkali treatment, this activity was lost, supporting a role for O-acetyl groups in directing the macrophage response (37). Recht and Kolter examined the role of O-acetyl groups on GPLs by utilizing a M. smegmatis acetyltransferase (atf1)-deficient mutant, which lacked the two O-acetyl groups normally located on C3 and C4 carbons of the 6-deoxytalose (38). The authors demonstrated that the M. smegmatis atf1–/– mutant was defective in biofilm formation and sliding motility, properties associated with water system colonization and epithelial cell invasion (38–40).
Additional studies examining serovar-specific GPLs have focused on the role of oligosaccharides in modulating the immune response. Serovar 4 GPL was shown to promote phagocytosis more readily when coated on heat-killed Staphylococcus aureus as compared with the serovar 9 GPL (41). A study by Rastogi and colleagues (42) showed that human peripheral blood mononuclear cells produced significantly higher levels of pro-inflammatory cytokines such as interleukin-1β, interleukin-6, and TNF-α, in response to the serovar 8 GPL when compared with the serovar 4 GPL. The disparity in the response by the peripheral blood mononuclear cells to the different ssGPLs was attributed to the difference in their oligosaccharide composition. In the same study, it was also demonstrated that serovar 4 and serovar 8 could inhibit a T-cell response (42), an interesting observation considering some studies have observed a higher incidence of M. avium serovar 4 and serovar 8 strains in AIDS patients (43), although this conclusion has been questioned (44).
The above results initially led us to investigate whether different GPLs could differentially activate macrophages via TLR2. We observed that the serovar 1 and serovar 2 GPLs induced mitogen-activated protein kinase and NFκB activation as well as TNF-α production in a MyD88- and TLR2-dependent manner, however, nsGPLs and the serovar 4 GPL failed to induce activation in the same manner (21). From these studies we postulated that the oligosaccharides linked to the 6-deoxytalose were the components responsible for the differential macrophage response. In the present study, however, we observed that mild alkali treatment abrogated the ability of total lipid and active GPLs to signal through TLR2. Structural analyses indicated that the serovar 2 GPL with one O-acetyl group at C4 of 6-deoxytalose could induce a MyD88- and TLR2-dependent signaling response in macrophages. When this O-acetyl group was removed by mild alkali treatment, the GPL activity was lost. In addition, the serovar 2 GPL with two O-acetyl groups on 6-deoxytalose was inactive. Similarly, an active nsGPL was identified, which also stimulated macrophage activation in a MyD88- and TLR2-dependent manner. Again, the presence of an O-acetyl group at C4 of 6-deoxytalose was required for activity. However, the active species also required that the rhamnose be methylated only at the third carbon position. These observations indicate that the methylation patterns may also be important in determining whether an acetylated nsGPL can signal via TLR2.
When comparing the active serovar 2 GPL and the active nsGPL, we observed that 1) the rhamnose of the serovar 2 GPL was 3,4-di-O-methylated, and the rhamnose of the nsGPL is only methylated at C3 and 2) the 4-O-acetyl-6-deoxytalose of the serovar 2 GPL was linked to a rhamnose at C2, and the 4-O-acetyl-6-deoxytalose of the nsGPL was methylated at C3. Together, these observations suggest that the 4-O-acetyl group on the 6-deoxytalose is critical for TLR2 signaling induced by GPL. Moreover, when comparing the active serovar 2 GPL to the active nsGPL, there is one free hydroxyl group at C2 of the 6-deoxytalose of the nsGPL, and at C3 of the active serovar 2 GPL. Whether the free hydroxyl group on this sugar is an important determinant for GPL-TLR2 interaction remains to be determined.
Previous studies from our laboratory have implicated a role for GPLs in M. avium virulence. One study examined a serovar 1 M. avium methyltransferase (mtfD) mutant that was unable to methylate the rhamnose at C3, and as a result, produced only atypical nsGPLs that possessed an uncharacteristic non-methylated rhamnose attached to the alaninol (25). This mtfD mutant induced a more pro-inflammatory response in vitro and was more attenuated in vivo when compared with the wild-type and reconstituted strains (25). These observations are significant as they suggest that the GPL profile can influence the immune response during an M. avium infection.
In the present study, we have demonstrated that the presence of O-acetyl groups on the 6-deoxytalose of GPLs is essential for eliciting a TLR2-dependent response in vitro, but how this modification influences the virulence of M. avium in vivo has yet to be investigated. Moreover, whether the structure of GPLs can be modified during the course of an infection, so as to modulate or avoid immune surveillance, is presently unknown. This concept is not unreasonable as studies have shown that the LPS structure of various Gram-negative organisms is modified according to the environmental conditions (45, 46). The penta- and hexa-acylated LPS variants have been shown to be distinguished by human TLR4, with the hexa-acylated, but not the penta-acylated form, being stimulatory (47). Whether M. avium alters its GPL profile in response to environmental stimuli is currently unknown, although studies utilizing M. smegmatis have shown the production of an unusual polar GPL in response to carbon starvation (48, 49). Future studies should more closely examine the role of GPL constituents in modulating the immune response, using specific M. avium GPL mutants and by examining the GPL profiles during the course of infection.
Supplementary Material
Acknowledgments
We thank Dr. Michelle Joyce and Dennis Birdsell for their technical help in performing the mass spectrometry experiments, and for helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants AI056979 and AI052439 from NIAID. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS or NIH. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S5.
Footnotes
The abbreviations used are: TLR, Toll-like receptor; MyD88, myeloid differentiation primary response protein 88; TNF, tumor necrosis factor; LPS, lipopolysaccharide; GPL, glycopeptidolipid; GC, gas chromatography; MS, mass spectrometry; MS/MS, tandem MS; BMMΦ, bone marrow-derived macrophage; ESI, electrospray ionization; CAD, collisionally activated dissociation; nsGPL, non-serotype-specific GPL; ssGPL, serotype-specific GPL.
References
- 1.Gay, N. J., and Gangloff, M. (2007) Annu. Rev. Biochem. 76 141–165 [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., and Takeda, K. (2004) Nat. Rev. Immunol. 4 499–511 [DOI] [PubMed] [Google Scholar]
- 3.Poltorak, A., He, X., Smirnova, I., Liu, M. Y., Van Huffel, C., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., Freudenberg, M., Ricciardi-Castagnoli, P., Layton, B., and Beutler, B. (1998) Science 282 2085–2088 [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto, M., Tawaratsumida, K., Kariya, H., Aoyama, K., Tamura, T., and Suda, Y. (2006) Int. Immunol. 18 355–362 [DOI] [PubMed] [Google Scholar]
- 5.Hirschfeld, M., Kirschning, C. J., Schwandner, R., Wesche, H., Weis, J. H., Wooten, R. M., and Weis, J. J. (1999) J. Immunol. 163 2382–2386 [PubMed] [Google Scholar]
- 6.Lien, E., Sellati, T. J., Yoshimura, A., Flo, T. H., Rawadi, G., Finberg, R. W., Carroll, J. D., Espevik, T., Ingalls, R. R., Radolf, J. D., and Golenbock, D. T. (1999) J. Biol. Chem. 274 33419–33425 [DOI] [PubMed] [Google Scholar]
- 7.Erridge, C., Pridmore, A., Eley, A., Stewart, J., and Poxton, I. R. (2004) J. Med. Microbiol. 53 735–740 [DOI] [PubMed] [Google Scholar]
- 8.Farhat, K., Riekenberg, S., Heine, H., Debarry, J., Lang, R., Mages, J., Buwitt-Beckmann, U., Roschmann, K., Jung, G., Wiesmuller, K. H., and Ulmer, A. J. (2008) J. Leukoc. Biol. 83 692–701 [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi, O., Kawai, T., Muhlradt, P. F., Morr, M., Radolf, J. D., Zychlinsky, A., Takeda, K., and Akira, S. (2001) Int. Immunol. 13 933–940 [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi, O., Sato, S., Horiuchi, T., Hoshino, K., Takeda, K., Dong, Z., Modlin, R. L., and Akira, S. (2002) J. Immunol. 169 10–14 [DOI] [PubMed] [Google Scholar]
- 11.Means, T. K., Wang, S., Lien, E., Yoshimura, A., Golenbock, D. T., and Fenton, M. J. (1999) J. Immunol. 163 3920–3927 [PubMed] [Google Scholar]
- 12.Underhill, D. M., Ozinsky, A., Smith, K. D., and Aderem, A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14459–14463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drennan, M. B., Nicolle, D., Quesniaux, V. J., Jacobs, M., Allie, N., Mpagi, J., Fremond, C., Wagner, H., Kirschning, C., and Ryffel, B. (2004) Am. J. Pathol. 164 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsburgh, C. R., Jr. (1999) J. Infect. Dis. 179 Suppl. 3, S461–S465 [DOI] [PubMed] [Google Scholar]
- 15.Horsburgh, C. R., Jr., Havlik, J. A., Ellis, D. A., Kennedy, E., Fann, S. A., Dubois, R. E., and Thompson, S. E. (1991) Am. Rev. Respir. Dis. 144 557–559 [DOI] [PubMed] [Google Scholar]
- 16.Gomes, M. S., Florido, M., Cordeiro, J. V., Teixeira, C. M., Takeuchi, O., Akira, S., and Appelberg, R. (2004) Immunology 111 179–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, C. G., Scanga, C. A., Collazo-Custodio, C. M., Cheever, A. W., Hieny, S., Caspar, P., and Sher, A. (2003) J. Immunol. 171 4758–4764 [DOI] [PubMed] [Google Scholar]
- 18.Brightbill, H. D., Libraty, D. H., Krutzik, S. R., Yang, R. B., Belisle, J. T., Bleharski, J. R., Maitland, M., Norgard, M. V., Plevy, S. E., Smale, S. T., Brennan, P. J., Bloom, B. R., Godowski, P. J., and Modlin, R. L. (1999) Science 285 732–736 [DOI] [PubMed] [Google Scholar]
- 19.Quesniaux, V., Fremond, C., Jacobs, M., Parida, S., Nicolle, D., Yeremeev, V., Bihl, F., Erard, F., Botha, T., Drennan, M., Soler, M. N., Le Bert, M., Schnyder, B., and Ryffel, B. (2004) Microbes Infect. 6 946–959 [DOI] [PubMed] [Google Scholar]
- 20.Bulut, Y., Michelsen, K. S., Hayrapetian, L., Naiki, Y., Spallek, R., Singh, M., and Arditi, M. (2005) J. Biol. Chem. 280 20961–20967 [DOI] [PubMed] [Google Scholar]
- 21.Sweet, L., and Schorey, J. S. (2006) J. Leukoc. Biol. 80 415–423 [DOI] [PubMed] [Google Scholar]
- 22.Hirschfeld, M., Weis, J. J., Toshchakov, V., Salkowski, C. A., Cody, M. J., Ward, D. C., Qureshi, N., Michalek, S. M., and Vogel, S. N. (2001) Infect. Immun. 69 1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buwitt-Beckmann, U., Heine, H., Wiesmuller, K. H., Jung, G., Brock, R., and Ulmer, A. J. (2005) FEBS J. 272 6354–6364 [DOI] [PubMed] [Google Scholar]
- 24.Stover, A. G., Da Silva Correia, J., Evans, J. T., Cluff, C. W., Elliott, M. W., Jeffery, E. W., Johnson, D. A., Lacy, M. J., Baldridge, J. R., Probst, P., Ulevitch, R. J., Persing, D. H., and Hershberg, R. M. (2004) J. Biol. Chem. 279 4440–4449 [DOI] [PubMed] [Google Scholar]
- 25.Krzywinska, E., Bhatnagar, S., Sweet, L., Chatterjee, D., and Schorey, J. S. (2005) Mol. Microbiol. 56 1262–1273 [DOI] [PubMed] [Google Scholar]
- 26.Yadav, M., Roach, S. K., and Schorey, J. S. (2004) J. Immunol. 172 5588–5597 [DOI] [PubMed] [Google Scholar]
- 27.Brennan, P. J., Heifets, M., and Ullom, B. P. (1982) J. Clin. Microbiol. 15 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckstein, T. M., Belisle, J. T., and Inamine, J. M. (2003) Microbiology 149 2797–2807 [DOI] [PubMed] [Google Scholar]
- 29.Gordon, S. (2002) Cell 111 927–930 [DOI] [PubMed] [Google Scholar]
- 30.Somerville, J. E., Jr., Cassiano, L., Bainbridge, B., Cunningham, M. D., and Darveau, R. P. (1996) J. Clin. Invest. 97 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poltorak, A., Ricciardi-Castagnoli, P., Citterio, S., and Beutler, B. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 2163–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernardo, J., Billingslea, A. M., Blumenthal, R. L., Seetoo, K. F., Simons, E. R., and Fenton, M. J. (1998) Infect. Immun. 66 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briken, V., Porcelli, S. A., Besra, G. S., and Kremer, L. (2004) Mol. Microbiol. 53 391–403 [DOI] [PubMed] [Google Scholar]
- 34.Wieland, C. W., Knapp, S., Florquin, S., de Vos, A. F., Takeda, K., Akira, S., Golenbock, D. T., Verbon, A., and van der Poll, T. (2004) Am. J. Respir. Crit. Care Med. 170 1367–1374 [DOI] [PubMed] [Google Scholar]
- 35.Kang, B. K., and Schlesinger, L. S. (1998) Infect. Immun. 66 2769–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilleron, M., Nigou, J., Nicolle, D., Quesniaux, V., and Puzo, G. (2006) Chem. Biol. 13 39–47 [DOI] [PubMed] [Google Scholar]
- 37.Villeneuve, C., Etienne, G., Abadie, V., Montrozier, H., Bordier, C., Laval, F., Daffe, M., Maridonneau-Parini, I., and Astarie-Dequeker, C. (2003) J. Biol. Chem. 278 51291–51300 [DOI] [PubMed] [Google Scholar]
- 38.Recht, J., and Kolter, R. (2001) J. Bacteriol. 183 5718–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hilborn, E. D., Covert, T. C., Yakrus, M. A., Harris, S. I., Donnelly, S. F., Rice, E. W., Toney, S., Bailey, S. A., and Stelma, G. N., Jr. (2006) Appl. Environ. Microbiol. 72 5864–5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki, Y., Danelishvili, L., Wu, M., Hidaka, E., Katsuyama, T., Stang, B., Petrofsky, M., Bildfell, R., and Bermudez, L. E. (2006) Cell Microbiol. 8 806–814 [DOI] [PubMed] [Google Scholar]
- 41.Kano, H., Doi, T., Fujita, Y., Takimoto, H., Yano, I., and Kumazawa, Y. (2005) Biol. Pharm. Bull. 28 335–339 [DOI] [PubMed] [Google Scholar]
- 42.Horgen, L., Barrow, E. L., Barrow, W. W., and Rastogi, N. (2000) Microb. Pathog. 29 9–16 [DOI] [PubMed] [Google Scholar]
- 43.Horsburgh, C. R., Jr., Cohn, D. L., Roberts, R. B., Masur, H., Miller, R. A., Tsang, A. Y., and Iseman, M. D. (1986) Antimicrob. Agents Chemother. 30 955–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yakrus, M. A., and Good, R. C. (1990) J. Clin. Microbiol. 28 926–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst, R. K., Guina, T., and Miller, S. I. (1999) J. Infect. Dis. 179 Suppl. 2, S326–S330 [DOI] [PubMed] [Google Scholar]
- 46.Ernst, R. K., Yi, E. C., Guo, L., Lim, K. B., Burns, J. L., Hackett, M., and Miller, S. I. (1999) Science 286 1561–1565 [DOI] [PubMed] [Google Scholar]
- 47.Hajjar, A. M., Ernst, R. K., Tsai, J. H., Wilson, C. B., and Miller, S. I. (2002) Nat. Immunol. 3 354–359 [DOI] [PubMed] [Google Scholar]
- 48.Ojha, A. K., Varma, S., and Chatterji, D. (2002) Microbiology 148 3039–3048 [DOI] [PubMed] [Google Scholar]
- 49.Mukherjee, R., Gomez, M., Jayaraman, N., Smith, I., and Chatterji, D. (2005) Microbiology 151 2385–2392 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.