Abstract
A-kinase anchoring proteins (AKAPs) target protein kinase A (PKA) to a variety of subcellular locations. Conventional AKAPs contain a 14-18-amino acid sequence that forms an amphipathic helix that binds with high affinity to the regulatory (R) subunit of PKA type II. More recently, a group of dual specificity AKAPs has been classified on the basis of their ability to bind the PKA type I and the PKA type II isozymes. In this study we show that dual specificity AKAPs contain an additional PKA binding determinant called the RI Specifier Region (RISR). A variety of protein interaction assays and immunoprecipitation and immunolocalization experiments indicates that the RISR augments RI binding in vitro and inside cells. Cellular delivery of the RISR peptide uncouples RI anchoring to Ezrin leading to release of T cell inhibition by cAMP. Likewise, expression of mutant Ezrin forms where RI binding has been abrogated by substitution of the RISR sequence prevents cAMP-mediated inhibition of T cell function. Thus, we propose that the RISR acts in synergy with the amphipathic helix in dual specificity anchoring proteins to enhance anchoring of PKA type I.
The second messenger cAMP is frequently utilized in mammalian cells to regulate a variety of physiological processes. Cyclic AMP is generated at the plasma membrane in response to the occupancy of G-protein-coupled receptors. This ultimately leads to the stimulation of adenylyl cyclases, the enzymes that produce cAMP. The newly synthesized cAMP diffuses into the cell where it is available to activate a variety of effector proteins. These include protein kinase A (PKA)4 (reviewed in Ref. 1), cAMP-regulated ion channels (2), and Epac guanine nucleotide exchange factors (3). Activation of the PKA holoenzyme occurs upon binding of cAMP to the regulatory (R) subunits. This promotes dissociation of the active catalytic (C) subunits from the tetrameric complex and results in the phosphorylation of substrates in the vicinity of the active kinase (4, 5). PKA holoenzymes are classified as either type I or type II on the basis of their R subunit composition (RI or RII) (6). Four genes encode R subunits (RIα, RIβ, RIIα, and RIIβ). These proteins have distinct physical properties and affinities for cAMP (1). Because PKA is a broad specificity serine/threonine protein kinase that regulates a wide range of cellular processes, additional mechanisms have evolved to influence the selectivity of PKA action (7). Specificity in PKA action is maintained in part by interaction with protein kinase A anchoring proteins (AKAPs). This family of structurally diverse but functionally related scaffolding proteins targets PKA and other signaling proteins toward distinct substrates. These protein-protein targeting interactions contribute to spatial and temporal regulation of second messenger signaling events (reviewed in Refs. 7, 8).
The AKAP family now includes more than 50 members when including splice variants (7, 8). Although most of the AKAPs were initially identified on the basis of their ability to bind PKA type II inside cells, it is now recognized that several of these anchoring proteins such as D-AKAP1, D-AKAP2, AKAP220, Ezrin, Merlin, and PAP7 have a dual specificity as they also bind PKA type I (9-14). Other AKAPs are reported to selectively bind RI such as AKAPCE, myosin, and α4 integrins (15-17). However, only two of these dual specificity proteins, the mitochondrial protein PAP7 and Ezrin (12, 18), have been shown to preferentially interact with PKA type I in situ (15-17).
Conventional AKAPs contain a conserved amphipathic helix of 14-18 residues that forms the PKA-anchoring domain (19-21). This region inserts into a hydrophobic groove formed by the R dimer (22, 23). The RII subunits dimerize at the N terminus in an antiparallel fashion forming an X-type four-helix bundle that is necessary for AKAP binding. RI contains a structurally similar docking and dimerization domain (20, 22-29). However, additional residues at the extreme N terminus in RI are believed to fold back onto a putative four-helix bundle and might contribute additional binding determinants for the RI dimer (29).
Peptides patterned on the conserved amphipathic helix are effective inhibitors of PKA anchoring inside cells. As a result they are very useful tools to assess the role of anchored PKA in the control of cAMP-responsive events. The Ht31 peptide, derived from AKAP-Lbc, is considered to be the prototypic anchoring disrupter peptide (30, 31). A more potent dual-specific anchoring disrupter peptide called AKAP-IS was designed by bioinformatics (32). In 2006, the crystal structure of AKAP-IS in complex with the docking and dimerization domain of RIIα was solved (26). We took advantage of this information to develop a high affinity and RII isoform-specific anchoring disrupter peptide called SuperAKAP-IS. Similar technologies were used to generate an RI-selective anchoring disrupter peptide termed RIAD (33). Both reagents can selectively be used as tools to delineate functional roles for the anchored PKA type I and anchored PKA type II isozymes inside cells.
However, other mechanisms that do not involve the amphipathic helix may also serve to anchor PKA. For example, pericentrin binds to the regulatory subunits of PKA (type II) through a binding domain of 100 amino acids (34), whereas α4 integrins interact directly with the intact PKA (type I) holoenzyme via its cytoplasmic tail (17). Interestingly, none of these AKAPs contain the conserved amphipathic α-helix.
In T cells, PKA type I phosphorylation of serine 364 in the C-terminal Src kinase (Csk) activates the enzyme to block T cell activation (35). This proceeds through a mechanism that involves Csk-mediated suppression of the tyrosine kinase Lck. PKA type I is found at lipid rafts in resting T cells as well as in the soluble fraction (35). Activation of the T cells involves a rapid polarization of the cell and the redistribution of PKA type I (36). We have recently shown that the dual-specific AKAP Ezrin targets PKA type I to the immunological synapses during these events (37). In addition to binding PKA, Ezrin also associates with EBP50, a linker protein that contacts Csk-binding protein (Cbp)/phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG). Thus, Ezrin, EBP50, and Cbp/PAG form a scaffold that assembles the negatively regulatory cAMP-PKA type I-Csk pathway in T cell lipid rafts (37).
In this study we document a comparison of Ezrin to other R-selective AKAPs and identify an additional region outside the amphipathic helix that facilitates RI binding. Bioinformatic analyses suggest that such regions are present in a majority of dual specificity AKAPs. We have called this region the RI Specifier Region (RISR). In vitro binding studies show that the RISR is important for Ezrin interaction with RI. Cell-based experiments suggest that mutations in the RISR of Ezrin that perturb RI anchoring alter the suppression of T cell signaling through a cAMP-PKA type I-Csk pathway.
EXPERIMENTAL PROCEDURES
Sequence Analysis—The amino acid sequences of the dual specificity AKAPs Ezrin (NP_003370), Merlin (NP_000259), PAP7 (NP_073572), D-AKAP1 (CAA66000), and D-AKAP2 (O88845) were aligned using the multiple sequence alignment feature of the DNAstar Lasergene® software. MEME software was used for consensus sequence generation (38). MEME settings included one motif per sequence, and a motif length of 24 amino acids was specified.
Cell Cultures—Jurkat cells expressing the SV40 large T antigen (Jurkat TAg) were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 1 mm pyruvate, and 1× nonessential amino acids (complete medium). HEK293 and HEK293T cells were grown in Dulbecco's modified Eagle's medium (complete medium). Human peripheral blood was obtained from healthy donors (Ullevaal University Hospital Blood Centre, Oslo, Norway), and primary T cells were purified by negative selection as described previously (39). Briefly, leukocytes were separated from peripheral blood mononuclear cell using gradient centrifugation (Lymphoprep; Axis-shield PoC AS). T cells were isolated from the resulting cell suspension by negative selection using magnetic beads coated with anti-CD14 and anti-CD19 (Dynal AS, Invitrogen). All cells were cultured at 37 °C with 5% CO2.
Constructs—MEMEcons (QEALQREEESKRRQEEAEQR) and MEMEcons* (QEALQREEESKRRQERQSSA) sequences were cloned via TOPO cloning technology (Invitrogen) and thereafter into the EcoRI sites of pEGFP-C1 vector (Clontech). Bovine RIαΔ1-37, RIαΔ1-42, RIαΔ1-52, and RIαΔ1-59 mutants were cloned into pRSET-B. All constructs were confirmed by sequencing.
Immunoprecipitation—HEK293 cells at 50-80% confluency were transfected with 5-10 μg of plasmid DNA per 10-cm dish using Lipofectamine™ (Invitrogen). Cells were lysed 24 h after transfection in lysis buffer (20 mm Hepes, pH 7.5, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100) with protease inhibitors (Complete Mini EDTA-free tablets; Roche Diagnostics). Immunoprecipitations were carried out using antibodies against PKA RIα (BD Transduction Laboratories). Immunocomplexes were washed three times in lysis buffer containing 0.1-1% Triton X-100 before SDS-PAGE and immunoblotting with the indicated antibody (Ab). siRNA Duplexes and siRNA-insensitive Constructs—The 21-nucleotide siRNA duplex targeting human Ezrin mRNA (Ez799, 5′-cccuuggacugaauauuuaug-3′;5′-uaaauauucaguccaagggca-3′) and a triple G/C-mutated control (Ez799M3, 5′-cgcuucgagugaauauuuaug-3′;5′-uaaauauucacucgaagcgca-3′) have been described previously (37) and were synthesized in-house. Ezrin clones insensitive to siRNA Ez799 were generated by introducing three nucleotide switches, C798G, T781G, and G787C (upper primer GCTTGGAGTTGATGCGCTGGGACTCAATATTTATGAGAAAGATGA; lower primer TCATCTTTCTCATAAATATTGAGTCCCAGCGCATCAACTCCAAGC) to avoid recognition by siRNA Ez799 in coexpression experiments.
Protein Expression and Purification—Bovine RIα (full-length and deletion mutants) and murine RIIα protein were expressed in Escherichia coli BL21 by isopropyl 1-thio-β-d-galactopyranoside induction (4 h) and purified on cAMP-agarose beads. Human RIα was affinity-purified and subsequently biotinylated as described previously (40). Expression and purification of GST-D-AKAP1 were as described earlier (40, 41). Truncated (278-474 and 278-404) Ezrin wild type, R389A-, or K359A/K360A/R389A-substituted protein fused to GST were expressed in E. coli BL21 cells, induced using 0.4 mm isopropyl 1-thio-β-d-galactopyranoside, and purified on glutathione-Sepharose (Sigma).
Peptide Synthesis—Peptides used for ligand proximity assay (AlphaScreen) and IL-2 assays were synthesized on an Intavis MultiPep robot (Intavis Bioanalytical Instruments AG) and verified by high performance liquid chromatography. Peptides sequences were as follows: RIAD (LEQYANQLADQIIKEATE), RISR (ESKRRQEEAEQRK), RISR-RIAD (ESKRRQEEAEQRKLEQYANQLADQIIKEATE), RISR-Ht31 (ESKRRQEEAEQRKLIEEAASRIVDAVIEQV), RISR(Q6P/R12P)-Ht31 (ESKRRPEEAEQPKLIEEAASRIVDAVIEQV), Ht31 (LIEEAASRIVDAVIEQV), Arg9-RISR (RRRRRRRRRESKRRQEEAEQRK), and Arg9-RISR(Q6P/R12P) (RRRRRRRRRESKRRPEEAEQPK).
Autospot Peptide Array—Peptide spots were synthesized with Fmoc (N-(9-fluorenyl)methoxycarbonyl) protection chemistry on cellulose membranes using a Multipep automated peptide synthesizer (Intavis Bioanalytical Instruments AG) as described (42).
AlphaScreen Assay—For characterization of biomolecular interactions using AlphaScreen (amplified luminescence proximity homogeneous assay), biotinylated RIα and GST-D-AKAP1 were conjugated to streptavidin-coated donor beads and anti-GST-coated acceptor beads (PerkinElmer Life Sciences), respectively. The assay was carried out in the dark using 384-well white opaque plates (PerkinElmer Life Sciences) as described previously (40). Briefly, 20 nm GST-D-AKAP1 was incubated with 20 nm biotinylated RIα in the presence of different concentrations of peptide for 15 min on ice in a total of 15 μl of assay buffer (25 mm Hepes, pH 7.4, 100 mm NaCl, and 0.1% BSA). Five μl of acceptor beads were added and incubated on ice for 30 min. Subsequently, 5 μl of donor beads were added and incubated at room temperature for 90 min before reading on an EnVision™ multiplate reader (PerkinElmer Life Sciences). Both bead types were added at 20 μg/ml final concentration. IC50 values were estimated by nonlinear regression analysis using SigmaPlot (SPSS Inc.).
R Overlay—R overlays were conducted as described (19), using 32P-labeled recombinant murine RIIα (43) or recombinant bovine RIα (A98S), substituted to allow autophosphorylation (44). Briefly, the membrane with immobilized peptide or protein was blocked in Blotto (5% (w/v) nonfat dry milk and 0.1% BSA in Tris-buffered saline (TBS)). Purified recombinant R (4 μg) was radiolabeled with purified catalytic subunit (C) of PKA (0.02 μg/μl) and [γ-32P]ATP (1.4 μCi μl-1) in 50 mm MOPS, 50 mm NaCl, 2 mm MgCl2, and 1 mm dithiothreitol, pH 6.8, and separated from free [32P]ATP by gel filtration (G-50-Sepharose). Specific activity was quantified by liquid scintillation counting (1600TR Tri-Carb, Packard Instrument Co.). All overlays were performed overnight at room temperature using 1 × 106 cpm/ml TBS, 0.01% Tween 20 (TBS-T). For competition assays, soluble peptide was added to the radiolabeled RIα and incubated for 2 h before the spotted peptides were added. The membranes were washed five times in TBS-T, and the signal was detected by autoradiography.
Solid Phase Pulldown—Lysate from 40 × 106 Jurkat TAg cells was incubated overnight at 4 °C with RISR or RISR(R12P) peptide synthesized in triplicate on cellulose membrane. The membranes were subsequently washed twice (20 min) in lysis buffer (50 mm Hepes, pH 7.4, 10 mm NaPPi, 0.1% Triton X-100, 50 mm NaF, 100 mm NaCl, 5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 mm Na3VO4) with protease inhibitors (Complete Mini, EDTA-free tablets, Roche Applied Science) and twice in high salt lysis buffer (1 m NaCl). PKA R subunit bound to the spotted peptides were eluted by boiling in SDS-PAGE sample buffer and analyzed by Western blotting using mouse anti-RIα and mouse anti-RIIα (BD Transduction Laboratories).
Deletional Mapping of RISR Binding Area in RI—RISR peptides synthesized in triplicate on membranes were incubated with equal amounts of different N-terminally truncated RIα proteins (25 nm) in binding buffer (20 mm Tris-HCl, pH 7.4, 300 mm NaCl, 1 mm EDTA, 0.01% Triton X-100, 0.2 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol) with protease inhibitors (Complete Mini, EDTA-free tablets, Roche Diagnostics). Assays were performed as described above, and binding was determined using mouse anti-RIα (BD Transduction Laboratories).
GST Pulldown—Purified GST or GST-Ezrin fragments and RIα subunit were incubated at a 1:1 ratio (100 nm each) in 100 μl of pulldown buffer (20 mm Tris-HCl, pH 7.4, 300 mm NaCl, 1 mm EDTA, 0.2% Triton X-100, 0.2 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol) with protease inhibitors (Complete Mini, EDTA-free tablets, Roche Diagnostics) for 30 min at 4 °C. Fifty μl of 50% glutathione-Sepharose slurry (Sigma) equilibrated in binding buffer were added to the proteins and incubated with rotation for 2 h at 4 °C. Glutathione-Sepharose beads were subsequently washed three times with 500 μl of binding buffer and bound proteins eluted by boiling in SDS-PAGE sample buffer. Eluted proteins were analyzed by Western blotting using mouse-anti-RIα (BD Transduction Laboratories) and anti-GST horseradish peroxidase conjugate (Amersham Biosciences).
Circular Dichroism—CD spectra of 24-mer peptides were obtained in the presence of 0-50% (v/v) trifluoroethanol (TFE) at 20 °C using a Jasco J-810 spectropolarimeter (Jasco International Co., Ltd.) as described previously (45). Each sample was scanned five times at 20 nm/min (bandwidth 1 nm, and response time 1 s), over a 190-260 nm wavelength range.
Transfection of Peripheral Blood T Cells, Peptide Loading, and Stimulation—Purified T cells were either transfected with 1 μg of DNA and 500 nm siRNA according to the manufacturer's instructions using the human T cell nucleofector kit (Amaxa Biosystems) and incubated for 48 h in complete medium or loaded with 25 μm Arg9-RISR peptides for 2-3 h before stimulation. A total of 2 × 105 cells was subsequently incubated for 15 min with or without 8-(4-chlorophenylthio)-cAMP, at different concentrations and thereafter stimulated with anti-CD3/anti-CD28-coated beads (1.6 beads/cell) (Dynabeads® CD3/CD28 T cell expander) for 16 h. Supernatants were harvested and IL-2 levels determined by ELISA according to the manufacturer's instructions (R & D Systems).
Immunofluorescence Analysis—For immunofluorescence analysis, transfected T cells were washed in PBS and settled for 30 min on poly(l-lysine)-coated coverslips (Sigma). Transfected HEK293T cells were grown on coverslips for 48 h. At room temperature, cells were fixed with 3% paraformaldehyde in PBS for 15 min, permeabilized with 0.1% Nonidet P-40/PBS for 5 min, and then blocked for 30 min with 2% BSA, 0.01% Tween 20/PBS (PBST-BSA). Primary antibodies anti-VSV-G (1:100, Roche Diagnostics) or anti-human RIα clone 4D7 (1:250) in PBST-BSA were added for 30 min. Cells were then incubated with fluorochrome-conjugated secondary antibodies (Alexa Fluor 488 goat anti-mouse IgG1, Alexa Fluor 546 goat anti-mouse IgG2a, and Alexa Fluor 555 goat anti-mouse IgG (1:500), Molecular Probes) in PBST-BSA for 30 min before being mounted with glass coverslips using fluorescent mounting medium (DakoCytomation). Confocal microscopy was performed with a Zeiss LSM 510 META confocal microscope with a Plan-Apochromat 63 × 1.4 oil differential interference contrast objective lens, using laser excitation at 488 and 546 nm. Pictures were obtained using sequential scanning, and the exposure settings and gain of laser were kept the same for each condition. Colocalization was determined using the freely available ImageJ software (rsb.info.nih.gov). Images were opened in ImageJ, and a membrane segment was manually outlined in each image for which colocalization was determined by defining “pixels containing red (Alexa 546) and green (Alexa 488)” AND “pixels containing red (Alexa 546) OR green (Alexa 488)” as Boolean variables and counting the number of pixels true for green and red divided by the number of pixels true for green or red using the Process/Image calculator tool of the software.
Statistical Analysis—Data are presented as mean ± S.E. and were analyzed by Mann-Whitney U test. Differences with two-sided p < 0.05 were considered significant.
RESULTS
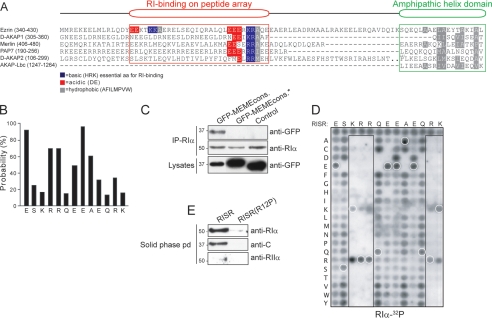
Identification and Characterization of an Additional R-binding Motif in Dual Specificity AKAPs—Following identification of Ezrin as the dual specificity AKAP for PKA type I in T cells, detailed mapping studies were initiated with fragments of the protein (37) to further define the RI and RII binding surfaces. In vitro binding experiments showed that the Ezrin 278-474 fragment bound RI and RII as assessed by overlay assays (Fig. 1A, two middle panels, 1st lane). However, a smaller fragment of Ezrin (residues 278-404) only retained the ability to bind RI (Fig. 1A, compare two middle panels, 2nd lane). Because this region lacks the amphipathic helix (schematic illustration, top panel), we reasoned that additional RI-binding determinants must reside in this segment of the protein. To define this region more precisely, a family of 160 overlapping immobilized 20-mer peptides (offset by two residues) were synthesized on a peptide array. These peptides were screened for RI binding to reveal a region of about 30 amino acids in Ezrin that was distinct from the amphipathic helix and contained two clusters of basic side chains (residues 356-375 and residues 378-397, Fig. 1B).
FIGURE 1.
Identification of an additional R-binding motif in the dual specificity AKAP, Ezrin. A, indicated GST-Ezrin fusion proteins containing either the RI-binding region together with the R-binding amphipathic helix domain (amino acids 278-474) or the RI-binding region of Ezrin alone (amino acids 278-404) were subjected to SDS-PAGE and RI-32P or RII-32P overlay. B, peptide array substitution analysis of PKA RI-binding to Ezrin. Wild type or substituted peptides (substituted amino acids marked in boldface) were spot-synthesized on solid phase and subjected to RI-32P or RII-32P overlay. Note: alanine substitution of Ezrin Lys-359/Lys-360 or Ezrin Arg-381 led to loss of RI binding. C, indicated GST-Ezrin proteins (amino acids 278-474) with mutations as in B were tested for interaction with RIα by GST pulldown (pd) using glutathione-Sepharose beads. GST served as a negative control. The relative binding intensities to RIα were quantified by densitometry (mean ± S.E.; n = 3).
Further analysis of these sequences using the same approach showed that removal of basic side chains at positions 359, 360, and 381 impaired the interaction with RI (Fig. 1B). Substitution of Arg-381, Lys-359, and Lys-360 for alanine reduced RI binding, but more dramatic effects were obtained when combinations of these side chains were replaced. Control experiments confirmed that RII did not bind to peptides encompassing this RI-binding enhancer region of Ezrin (Fig. 1B).
These results were confirmed when a GST pulldown assay was used to show that incorporating the amino acid substitutions identified in Fig. 1B reduced RI binding by 80-90% (Fig. 1C, 1st lane versus 2nd and 3rd lanes). Control experiments established that the GST protein alone did not interact with RI. Collectively, these results suggest that the RI-binding enhancer region and the amphipathic helix binding domain are necessary for RI binding in vitro. Sequence alignment of the dual specificity AKAPs Ezrin, D-AKAP1, D-AKAP2, PAP7, and Merlin identified a region of homology upstream of the amphipathic helix (Fig. 2A). Further analyses revealed that almost all anchoring proteins previously denoted as dual specificity AKAPs contain such a region. In contrast, classical RII-binding proteins such as AKAP-Lbc and AKAP79/150 did not. Peptide array analyses showed that the RI-binding enhancer regions bound radiolabeled RI in a solid phase binding assay (Fig. 2A, binding indicated by area with red frame).
FIGURE 2.
Characterization of the additional R-binding motif RISR. A, sequence alignment of the region N-terminal to the R-binding amphipathic helix domain in the dual specificity AKAPs Ezrin, D-AKAP1, Merlin, PAP7, and D-AKAP2. Green box, conserved R-binding amphipathic helix domain with AKAP-Lbc included; red box, RI binding detected on peptide array; reverse background letters, functionally conserved (blue, basic; red, acidic; gray, hydrophobic) residues among dual specificity AKAPs. B, using the MEME algorithm, a position-dependent scoring matrix representing the probability that an amino acid is found at a given position divided by the frequency that this amino acid is found in the nonredundant protein data base was calculated. C, coimmunoprecipitation (IP) of the GFP-fused D-AKAP homology region consensus (cons.) wild type or consensus where the last five amino acids EAEQR were changed to RQSSA (denoted *) with PKA-RIα as detected by anti-GFP immunoblotting. D, two-dimensional array of 260 RI-binding consensus peptide derivatives (as 13-mers) was synthesized where each residue in the native peptide (given by their single-letter codes above each array) was replaced with all the 20 amino acids (given by their single-letter codes to the left of each array). The 1st row in the array corresponds to the RISR, the consensus sequence determined from the alignment of dual specificity AKAPs. Binding of 32P-labeled RIα was detected by autoradiography. White circles denote peptides in the array that corresponds to the native RISR sequence. The positions of important amino acids in RISR are indicated by boxes. E, RISR solid phase pulldown (pd). RISR peptides synthesized in triplicate on membranes were incubated in Jurkat TAg cell lysate overnight, and immobilized proteins were thereafter eluted, and the levels of PKA-RIα, PKA-RIIα, and PKA-C were analyzed by immunoblotting. Anti-RIα and anti-RIIα blots shown are both from 3-min exposures. C and E are representative of three independent experiments. D represents five independent experiments.
The aligned RI-binding enhancer regions in these anchoring proteins were analyzed using the MEME algorithm to produce a consensus sequence (38). The MEME algorithm generates a position-dependent scoring matrix by systematically calculating the probability that an amino acid would be found at a given position (Fig. 2B). The optimal sequence was Glu-Ser-Lys-Arg-Arg-Gln-Glu-Glu-Ala-Glu-Gln-Arg-Lys in the context of a 24-mer. Peptides of this sequence bound RI as assessed by the overlay assay (Fig. 2D, upper row of peptides). Next, a consensus sequence fused to GFP was expressed inside cells to determine whether this region was sufficient to bind RI. Immunoblot analysis showed that RIα coprecipitated with the GFP-MEME fusion protein (Fig. 2C). RIα was not detected in control immune complexes formed with GFP fused to a control peptide (negative control) or GFP alone (Fig. 2C). The levels of RIα in the immune complexes and the level of GFP in the cell lysates were evaluated as loading controls (Fig. 2C, middle and lower panels). The 13-amino acid peptide minimal binding region ESKRRQEEAEQRK was termed the RI Specifier Region (RISR) as it appeared to enhance binding to RI when compared with RII in cells containing both RI and RII (Fig. 1A and Fig. 2, C and E).
Further characterization of RISR was performed by a two-dimensional peptide array study. A total of 260 peptide derivatives was synthesized in which each residue in the peptide (given by its single-letter code above the array, Fig. 2D) was substituted with all the 20 naturally occurring amino acids. The peptides were analyzed for solid phase binding of RI-32P and detected by autoradiography (Fig. 2D).
Substitution of positively charged side chains at positions Lys-3, Arg-4, Arg-5, Arg-12, and Lys-13 reduced RI binding (boxes in Fig. 2D). All RI binding was lost in peptides with a proline in these positions. An R12P substitution (ESKRRQEEAEQPK) or Q6P/R12P double substitutions (ESKRRPEEAEQPK) were chosen as negative control peptides in future studies.
Some spots appeared to be stronger than the native sequence in the two-dimensional array. However, introducing these substitutions in new peptides gave no apparent effect on RI binding.
To analyze the RISR peptide with respect to binding of endogenously expressed PKA, we performed a solid phase immobilization assay. The RISR peptide and the negative control peptide RISR(R12P) were immobilized on membranes and incubated with Jurkat TAg cell lysate. Bound proteins were eluted with SDS loading buffer, and the presence of PKA subunits was assessed by immunoblotting using specific antibodies. As shown in Fig. 2E, PKA-RIα and PKA-C subunits were pulled down by the RISR peptide (1st lane) but not by the negative control peptide (2nd lane). In comparison, little or no PKA-RIIα was pulled down by the RISR peptide.
The RISR sequence is rich in α-helix forming side chains such as alanine and glutamic acid (46). The helicity of RISR as measured by CD was 53% in aqueous solutions and 91% in the presence of 50% TFE in the context of a 24-amino acid sequence (Table 1). It is well known that 50% TFE induces and stabilizes α-helical structures (47-49). We conclude that RISR is capable of forming a helical configuration that augments binding of PKA to the conventional amphipathic helix.
TABLE 1.
Helical content of 24-mer peptide sequences obtained by CD measurements
| Peptide | Solvent | α-Helical content |
|---|---|---|
| % | ||
| Ezrin | Water | 53 |
| 50% TFE | 89 | |
| Merlin | Water | 34 |
| 50% TFE | 93 | |
| PAP7 | Water | 80 |
| 50% TFE | 96 | |
| RISR | Water | 53 |
| 50% TFE | 91 |
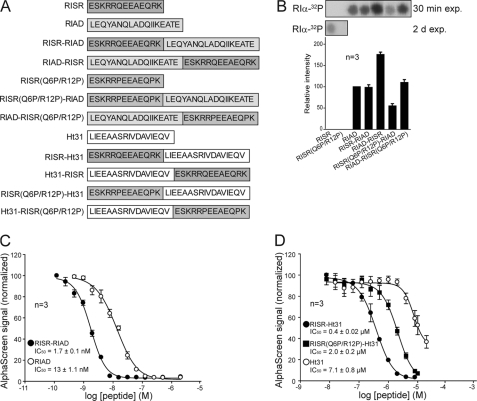
Mapping of the PKA RIα Region Binding to RISR—Reciprocal mapping studies were initiated to define the binding surface on RI that interacts with RISR. The ability of solid phase-bound RISR peptide to immobilize a family of N-terminally truncated RIα proteins in solution at equal concentration (25 nm) was examined. Binding to the RI anchoring disrupter (RIAD) peptide, which is patterned after the amphipathic helix, was analyzed in parallel (33). N-terminally truncated versions of the R subunit (RIαΔ1-11 and RIαΔ1-15) bound RIAD more avidly than full-length RIα (Fig. 3A, bottom panel) as reported previously (33). However, RIAD binding was lost with RIαΔ1-24, a form of the R subunit that lacks a majority of the docking domain (25) (Fig. 3A, bottom panel).
FIGURE 3.
Mapping of the PKA RIα region binding to RISR. A, solid phase pulldown (pd). RISR peptides synthesized in triplicates on membranes were incubated in a binding buffer with equal amounts of indicated N-terminally truncated RIα proteins (25 nm). Bound protein was eluted and subjected to immunoblot analysis to determine the level of various N-terminally truncated RIα proteins bound. Two different antibodies to RI raised against either the full-length protein or a fragment covering amino acids 225-381 were used with identical results. B, RISR and RIAD were spot-synthesized on solid phase, and RI binding was analyzed by 32P-radiolabeled RIα overlay in the absence (top row) or the presence of 1 μm of the indicated peptides. w/o, without. C, RIAD and RISR-binding regions in PKA-RIα as identified in A. Helical region, helix I, and II in RI are labeled according to Ref. 25. A and B are representative of six and four independent experiments, respectively.
RISR bound wild type RIα and the RIαΔ1-11 and RIαΔ1-15 proteins with lower intensity but bound the RIαΔ1-24 form more tightly (Fig. 3A, upper panel). This would suggest that RISR interacts with determinants located downstream of side chains that are necessary for binding to the amphipathic helix. Confirmation of this notion was provided by evidence that the RIαΔ1-37 truncation also binds immobilized RISR (Fig. 3A, upper panel). However, upon further truncation using RIαΔ1-42, RISR binding was lost. Hence, our data suggest that RISR interacts with unique determinants on RI that are located C-terminal in the docking and dimerization domain. Furthermore, RISR bound equally well to cAMP-free as to cAMP-saturated RI indicating that cAMP binding does not affect interaction with the RISR (data not shown).
Our evidence that the RISR and RIAD-binding sites in RIα were nonoverlapping was further substantiated by the finding that excess RISR peptide in solution competed for RI binding to immobilized RISR but was unable to uncouple interactions with RIAD (Fig. 3B). Reciprocal experiments showed that excess RIAD peptide blocked RIAD interaction with RI but had no effect on RI binding to RISR. Our data suggest that the amphipathic helix binding domain and RISR have distinct binding sites in RIα and may bind concomitantly (Fig. 3C).
RISR Motif Serves to Enhance and Specify R Binding to AKAPs—Because RISR may bind RIα concomitantly with the amphipathic helix, we examined the effect of RISR on R binding in combination with the amphipathic helix R-binding regions of the RI-selective RIAD and the RII-selective Ht31. Fig. 4A shows a schematic illustration of the various combinations of RISR with the RIAD and Ht31 amphipathic helix binding domains that were tested. R binding was analyzed by RIα-32P overlay of immobilized peptides (Fig. 4B). In these solid phase assays, the relative binding of RIAD to RI was almost 100-fold stronger than that of RISR. When RISR was combined with RIAD, however, binding was increased by 75% for RIAD-RISR, whereas no effect was observed by addition of RISR in the order RISR-RIAD. RISR(Q6P/R12P) exhibited no RI binding, and RISR(Q6P/R12P)-RIAD and RIAD-RISR(Q6P/R12P) were used as controls to exclude the possibility that the increase in RI binding was only because of the increase in peptide length. Thus, a synergetic effect on RI affinity was achieved by combination of the RISR and RIAD peptide motifs. These data indicate that combining RISR and the amphipathic helix enhances RI binding.
FIGURE 4.
RISR motif serves as an enhancer for R binding. A, schematic illustration of the various combinations of RISR with the RIAD and Ht31 amphipathic helix binding domains. B, peptides were synthesized on membrane, and R binding was analyzed by RIα-32P overlay. The relative R binding of the combined peptides versus RIAD was quantified by densitometry of autoradiogram (mean ± S.E.; n = 3). exp, exposure; 2D exp, two-dimensional exposure. C and D, concentration-dependent competition of RIα and GST-D-AKAP1 interaction (20 nm each) by peptides RISR-RIAD (•) and RIAD (○) (C) or RISR-Ht31 (•), RISR(Q6P/R12P)-Ht31 (▪) and Ht31 (○) (D) in an amplified ligand proximity assay (AlphaScreen). IC50 values were calculated by nonlinear regression analysis using SigmaPlot. Data represent mean ± S.E. from three independent experiments performed in duplicate.
We have previously reported the binding properties of the dual specificity D-AKAP1 using an amplified luminescence ligand proximity assay (AlphaScreen) (40). Therefore, we evaluated the effect of RISR-peptide combinations in solution using this RI competition binding platform to circumvent the possibility that steric hindrance had introduced artifacts into the solid phase assay, resulting in loss of binding, e.g. for the peptide RISR-RIAD. In the AlphaScreen assay, RISR that has comparably low RI affinity alone was not able to affect the D-AKAP1-RI interaction (data not shown), whereas both RIAD and Ht31 competed RI binding to D-AKAP1 with IC50 values of 13 ± 1.1 nm and 7.1 ± 0.8 μm, respectively. When the RISR sequence was combined with either peptide, the IC50 decreased by 7.6-fold (RIAD) or 17.8-fold (Ht31) showing that RISR works as an enhancer for RI binding (Fig. 4, C and D). To rule out the possibility that the decrease in IC50 was not only because of variation in peptide length, the RISR(Q6P/R12P) control peptide with two proline substitutions was also combined with Ht31. This appeared to increase modestly affinity as the IC50 changed from 7.1 to 2.0 μm for Ht31 versus RISR(Q6P/R12P)-Ht31, respectively. However, as the IC50 of RISR-Ht31 was still almost 1 order of magnitude lower than that of the control, the synergistic effect of RISR appeared to be specific (Fig. 4D). Furthermore, to examine the contribution of the endogenous RISR to the affinity of Ezrin for RI and RII, we did in-solution binding with increasing concentrations of RI and RII to wild type and K359A/K360A/R381A-substituted GST-Ezrin without the RISR and GST pulldown as in Fig. 1C. We found that the presence of the RISR increased the affinity of Ezrin for RI by 1.71 ± 0.21-fold, whereas the affinity for RII was decreased by 0.71 ± 0.057-fold (mean ± S.E., n = 3-4; data not shown). On total this means that there is a 2-fold or more change in relative affinity for RI over RII upon introduction of RISR, which brings the KD values for the Ezrin-RI and RII interactions into the same range.
Disruption of the RISR Motif in Ezrin Alters RIα Localization Inside Cells—A combined strategy of RNA interference and rescue with modified Ezrin forms was used to evaluate the role of the RISR motif in the subcellular anchoring of RI. These experiments were performed in two phases. First of all, HEK293T cells were transfected with siRNA to deplete the cells of endogenous Ezrin (37). Second, the cells were transfected with mammalian expression vectors encoding VSV-G-tagged Ezrin that is refractory to functional siRNA. Mutations in the RISR motif were introduced to yield Ezrin forms with substitutions K359A, K360A, and R381A. Changes in the subcellular location of RIα-GFP were assessed by the detection of GFP fluorescence, and the cellular distribution of VSV-G-tagged Ezrin was evaluated with antibodies against the epitope tag. siRNA-resistant Ezrin wild type and RIα-GFP colocalized in a membrane-proximal compartment that was consistent with earlier observations (Fig. 5, A-E) (37). However, when siRNA-resistant Ezrin (K359A/K360A/R381A) mutated in the RISR motif was coexpressed with RIα-GFP, a reduction in colocalization was observed in the membrane-proximal compartment (Fig. 5, F-J). Quantification of colocalization by identification of double-positive pixels as in Fig. 5, E and J, indicated a reduction in colocalization by ∼35-40% (1.5-fold decrease, n = 20) when the native RISR motif in Ezrin was mutated (Fig. 5K).
FIGURE 5.
Disruption of Ezrin RISR leads to displacement of RIα localization in HEK293T cells. A-J, HEK293T cells were transfected with Ezrin siRNA, plasmids encoding RIα-GFP and siRNA-resistant (denoted by *) Ezrin* wild type-VSV-G (A-E) or Ezrin* (K359A/K360A/R381A)-VSV-G (F-J) and subjected to immunofluorescence analysis using monoclonal mouse Ab to VSV-G and Alexa Fluor-conjugated secondary Ab. Merged images show overlapping subcellular distribution that appears yellow (A, B, F, and G). Images B-E and images G-J (scale bar, 1 μm) are magnifications of images A and F (scale bar, 5 μm), respectively. Images E and J show only double-positive pixels (Dbl. pos. pxls.) calculated by defining green and red pixels as Boolean variables using the “AND” connection in the Process/Image calculator tool of ImageJ. K, colocalization analysis of 20 membrane segments from different HEK293T cells transfected with Ezrin* wild type (wt)-VSV-G or Ezrin* (K359A/K360A/R381A)-VSV-G. Colocalization ratios were calculated from double-positive pixels/all-stained pixels (mean ± S.E.; n = 20).
Next, endogenous Ezrin was knocked down by siRNA in human peripheral blood T cells, followed by introduction of siRNA-resistant, VSV-G-tagged Ezrin wild type or K359A/K360A/R381A-substituted Ezrin defective in the RISR. Localization of Ezrin and endogenous RIα was examined by double immunofluorescence staining for VSV-G and RIα. Although some RIα is known to be in the cytoplasm in T cells, we recently reported that a fraction of RIα colocalizes with peripherally located Ezrin in the vicinity of the cell membrane that serves to scaffold the PKA type I-Csk immunomodulating pathway in T cells (37). Consistent with our earlier observations, wild type Ezrin colocalized with RIα in the membrane-proximal compartment (Fig. 6, A-E), whereas Ezrin (K359A/K360A/R381A) did not appear to colocalize with RIα to the same extent (Fig. 6, F-J). Indeed, quantification of colocalization of endogenous PKA RIα and Ezrin appeared to be reduced by almost 50% (1.8-fold decrease, n = 40) in primary T cells when the native RISR motif in Ezrin was knocked out by alanine substitution of amino acids Lys-359, Lys-360, and Arg-381 (Fig. 6K). This confirms the results seen in GST-pulldown experiments (Fig. 1D) and suggests that the RISR motif may have functional significance for RI binding to AKAPs in situ.
FIGURE 6.
Ezrin RISR contributes to membrane colocalization of endogenous RIα in T cells. A-J, primary T cells were transfected with Ezrin siRNA and plasmids encoding Ezrin* wild type-VSV-G (A-E) or Ezrin* (R381A/K359A/K360A)-VSV-G (F-J) and subjected to immunofluorescence analysis using monoclonal mouse Abs to VSV-G (IgG 1k) and RIα (IgG 2a) and isotype-specific Alexa Fluor-conjugated secondary Ab. Merged images show overlapping subcellular distribution that appears yellow (A, B, F, and G). Images B-E and images G-J (scale bar, 1 μm) are magnifications of image A and F (scale bar, 3 μm), respectively. Images E and J show only double-positive pixels (Dbl. pos. pxls.) calculated by defining green and red pixels as Boolean variables using the “AND” connection in the Process/Image calculator tool of ImageJ. K, colocalization analysis of 40 membrane segments from different T cells transfected with Ezrin* wild type (wt)-VSV-G or Ezrin* (K359A/K360A/R381A)-VSV-G. Colocalization ratios were calculated from double-positive pixels/all-stained pixels (mean ± S.E.; n = 40).
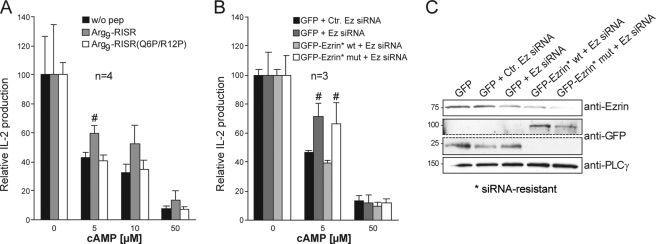
cAMP-mediated Inhibition of TCR-induced IL-2 Production Is Abrogated When RIα Interaction with the RISR Domain Is Disrupted—cAMP inhibits TCR-signaling events such as IL-2 production. This occurs through the modulation of a kinase cascade inhibitory pathway where PKA type I phosphorylates Ser-364 in the tyrosine kinase Csk to stimulate its catalytic activity (35). Elevated Csk activity next inhibits the Src family tyrosine kinase Lck leading to a suppression of T cell activation. Ezrin functions as the anchoring protein that brings PKA type I in proximity to Csk by forming a supramolecular signaling complex together with the Csk-binding protein PAG and the linker protein EBP50 (37). Because PKA type I anchoring contributes to the modulation of this kinase cascade, we wanted to assess whether disruption of the Ezrin-PKA interaction by the RISR peptide would interfere with cAMP-mediated inhibition of T cell function. We generated cell-permeable versions of RISR and the control peptide RISR(QP6/RP12) by introducing nine arginine residues to the N termini of the peptides. Normal peripheral blood CD3+ T cells were incubated with these peptides for 2-3 h. Cells were either left untreated, stimulated with CD3/CD28, or pretreated with increasing concentrations of the cAMP analog before CD3/CD28 stimulation. Twenty hours post-activation, levels of IL-2 secreted to the media were measured by ELISA. The Arg9-RISR peptide partially reversed cAMP/PKA-mediated inhibition of IL-2 production, whereas the Arg9-RISR(Q6P/R12P) control peptide had no effect (Fig. 7A).
FIGURE 7.
cAMP-mediated inhibition of TCR-induced IL-2 production is abrogated in cells loaded with RISR peptide or upon replacement of wild type Ezrin with Ezrin* K359A/K360A/R381A. A, effect of RISR on cAMP-mediated inhibition of IL-2 secretion was analyzed in primary T cells. RISR(Q6P/R12P) was used as a negative control. After peptide loading (2 or 3 h), cells were pretreated (15 min) with 8-CPT-cAMP (0, 5, or 50 μm) and either kept unstimulated or stimulated for 16 h with anti-CD3/anti-CD28-coated beads to induce IL-2 production and secretion. Subsequently, supernatants were harvested and assessed for IL-2 by ELISA. Levels of IL-2 secreted are shown relative to those of anti-CD3/anti-CD28-stimulated cells in the absence of 8-CPT-cAMP (mean ± S.E.; n = 4). # denotes two-sided p < 0.05 compared with control by Mann-Whitney U test. w/o pep, without peptide. B, effect of expression of Ezrin* K359A/K360A/R381A on cAMP-mediated inhibition of IL-2 secretion. Primary T cells were transfected with siRNA against Ezrin (Ez), either siRNA-insensitive (*) Ezrin* wild type (wt) or K359A/K360A/R381A-substituted Ezrin* and left for 48 h before stimulation and analysis as in A. Levels of IL-2 secretion are shown relative to those of anti-CD3/anti-CD28-stimulated cells in the absence of 8-CPT-cAMP (mean ± S.E.; n = 3). # denotes two-sided p < 0.05 compared with control by Mann-Whitney U test. mut, mutant. C, Ezrin knockdown and expression of GFP-Ezrin* in one representative experiment (n = 3) corresponding to data shown in B. Area between dashed lines in immunoblots showing anti-GFP was removed. Anti-GFP blots shown are from a single gel and exposure.
Parallel experiments were then conducted in the context of the Ezrin protein. We examined the effect of replacing endogenous Ezrin with the Ezrin with disrupted RISR on cAMP-mediated inhibition of T cell function using the RNA interference/rescue strategy described above. Human peripheral blood T cells were transfected with control siRNA or functional siRNA to remove endogenous Ezrin before siRNA-resistant wild type or K359A/K360A/R381A-substituted GFP-Ezrin was introduced. Forty eight hours post-transfection, cells were treated with increasing concentrations of cAMP analog, activated as above, and IL-2 levels were measured. Consistent with our earlier observations (37), knockdown of endogenous Ezrin changed the sensitivity of IL-2 production to cAMP and partially reversed cAMP-mediated inhibition at lower doses of cAMP (Fig. 7B, compare dark gray bar with black bar with control siRNA at 5 μm cAMP). Introduction of siRNA-resistant wild type Ezrin restored the sensitivity to cAMP (Fig. 7B, light gray bars), whereas introduction of K359A/K360A/R381A-substituted Ezrin where the native RISR motif had been disrupted did not (Fig. 7B, compare open bars to light gray bars). Under optimal conditions (1000 nm siRNA and 48 h post-transfection), we achieved 80% knockdown of Ezrin at the protein level (Fig. 7C). Control blots with anti-GFP showed that all constructs were expressed and that wild type and mutant siRNA-resistant GFP-Ezrin were expressed at the same level (Fig. 7C). In conclusion, disruption of PKA type I binding to the native RISR motif in Ezrin either by peptide loading to compete the interaction or by replacement of endogenous Ezrin with RISR-defective Ezrin reduces the sensitivity to cAMP-mediated inhibition of T cell function indicating that RISR has functional significance in Ezrin.
DISCUSSION
In this study we demonstrate that dual specificity AKAPs contain an additional PKA-binding region. By substitution analyses, biochemical interaction studies, and immunolocalization, we show that the RISR that resides in Ezrin preferentially facilitates the localization of PKA type I in T cells. Furthermore, we demonstrate that RISR-facilitated targeting of PKA type I is important for the sensitivity of cAMP-mediated inhibition of T cell function. By peptide array substitution and deletion studies, we also define a consensus RISR that independently binds RI and serves to confer specificity and affinity to RI interaction with AKAPs.
When RISR was coupled to either RIAD or Ht31, a synergistic effect of combining both sequences in the same peptide was observed on binding to RI both by solid phase immobilization assays and in solution competition assays. Thus, AKAPs that contain both regions have a higher affinity for RI than AKAPs that only contain an amphipathic helix. This additional region may have evolved to facilitate PKA type I anchoring, which appears to be a more dynamic process than PKA type II anchoring. These differences could relate to how the R-binding helix associates with the docking and dimerization domains of RI and RII. Surface plasmon resonance, fluorescence polarization, and equilibrium dialysis experiments suggest that RI binds AKAP peptides with a lower affinity and a faster off-rate than RII. Consequently, the AKAP-PKA type I holoenzyme complex is rather dynamic (19, 32, 41). The presence of an additional binding surface such as the RISR would provide a mechanism for multisite contact with the RI subunit that may stabilize the formation of the anchored PKA type I complex. In contrast the RII interaction with the amphipathic helix is robust enough to favor the maintenance of an anchored PKA holoenzyme complex inside cells (7).
The importance of RISR in binding to PKA type I was illustrated by the fact that substitutions in the Ezrin RISR led to reduced binding to RI both in vitro and in situ even though the amphipathic helix was intact. In contrast, the Ezrin RISR displayed little or no binding to RII and did not seem to enhance binding of RII to the amphipathic PKA binding domain either. Ezrin associates with Cbp/PAG and EBP50 and targets PKA type I to this supramolecular complex, thereby positioning PKA in close proximity to Csk for phosphorylation and regulation of Csk activity (37). Thus, it was relevant to use Ezrin as a model to analyze the functional effect of RISR in an siRNA knockdown and rescue approach. By substituting amino acids in the RISR of Ezrin, PKA type I was efficiently displaced from this complex, and sensitivity to cAMP-mediated inhibition of T cell function was reduced resulting in comparably higher levels of IL-2 secreted. Furthermore, by addition of arginine-coupled RISR peptides to cells, a similar disruption of the Ezrin-PKA type I interaction was observed as when the RISR of Ezrin was substituted. This perturbation of the PKA type I interaction with Ezrin in the Ezrin-Cbp-PAG-EBP50 complex upon the delivery of RISR reduced the sensitivity to cAMP-mediated inhibition of T cell signaling. In conclusion, these experiments confirmed that the RISR is important for PKA type I binding to the dual specificity AKAP Ezrin. Further studies will be necessary to investigate the functional role of the RISR in other AKAPs containing this region. For example, the dual specificity AKAP PAP7 has been shown to bind RI in vivo (12) and contain a putative RISR. PAP7-mediated hormonal control of steroid biosynthesis has been shown to implicate RI (33), and it would be interesting to assess the functional significance of RISR in this context in future studies.
AKAPs are known to be functionally related but structurally unrelated proteins that share a 14-18-amino acid amphipathic α-helical domain (19-21). Our biochemical studies suggest that the RISR also is of α-helical nature as proline substitutions abolish binding. This is further confirmed by CD measurements. Structural analyses suggest that the pseudo-symmetry of the helical axis in the AKAP PKA binding domain aligns with that of the four-helix bundle formed by the R subunit dimer. We believe that RISR binds to RI via a different mechanism. Several pieces of data support this notion. First of all, it appears that RISR binds to RI outside the area contacted by the amphipathic helix domain as evident from interaction studies with truncated RI proteins. Second, RISR does not display any internal symmetry that is a defined characteristic of the conventional PKA anchoring domain (26, 28). Consequently, it is likely that D-AKAPs contact the symmetrically arranged RI dimer with the alignment and symmetry coordinated by the amphipathic helix domain. This could suggest that RISR contacts only one of the RI protomers in the dimer. In support of this, we could not find any indication of duplications and inversions of RISR in the majority of dual specificity AKAPs, with the possible exception of Ezrin and Merlin. Interestingly, RISR appeared to localize N-terminal to the amphipathic helix binding domain in AKAPs found to contain this region, although positioning the consensus RISR C-terminal to the helix was also permissible in synthetic peptides.
RISRs can be detected in a number of dual specificity AKAPs such as Ezrin, Merlin, PAP7, D-AKAP1, and D-AKAP2. This could imply that this region is an evolutionally conserved motif that has been incorporated into these AKAPs to facilitate binding of PKA type I rather than PKA type II. Whether an AKAP signal complex contains PKA type I or type II may have functional consequences as PKA type I activates at 2-4-fold lower concentrations of cAMP than PKA type II. Consequently, RISR-facilitated recruitment of PKA type I rather than PKA type II to a specific AKAP complex could increase the sensitivity of the particular physiological process regulated by the signal complex to cAMP.
AKAPs with unconventional PKA-binding regions, such as pericentrin and α4 integrins (17, 34), do not contain RISR-like sequences and appear to have distinct types of binding sites. This indicates a greater diversity in PKA-binding sites than earlier recognized. It is interesting to speculate that AKAPs containing these unconventional binding sites might also be members of subclasses that could prove to contain yet unrecognized members. Some AKAPs such as AKAP220, AKAP350/450/cg-Nap, and BIG2 also have two or three R-binding sites spread throughout the protein sequence, but the functional significance of this multiple anchoring and the possible interdependency of the sites has not been investigated (14, 50-52).
In summary, our data show that RISR is an additional PKA-binding region in dual specificity AKAPs, and this region has functional significance for cAMP sensitivity of processes regulated by PKA bound to the dual specificity AKAP Ezrin.
Acknowledgments
We are grateful to Jorun Solheim, Gladys M. Tjørhom, and Guri Opsahl for technical assistance; Ola Blingsmo for peptide synthesis; and Espen Harbitz at the Institute of Molecular Biosciences at University of Oslo for assistance with CD analysis. We thank Dr. Cathrine R. Carlson for contributions during the early phases of this work.
This work was supported by grants (to K. T.) from the Norwegian Functional Genomics Programme, The Research Council of Norway, The Norwegian Cancer Society, Novo Nordic Foundation, the European Union Grant 037189, thera-cAMP and Grants (to J. D. S.) from the National Institutes of Health (DK 54441) and Foundation Leducq. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PKA, protein kinase A; AKAP, A-kinase anchoring protein; RISR, RI specifier region; TFE, trifluoroethanol; RIAD, RI anchoring disrupter; Csk, C-terminal Scr kinase; Lck, lymphocyte-specific protein-tyrosine kinase; EBP50, Ezrin-Radixin-Moesin-binding phosphoprotein 50; siRNA, small interfering RNA; PAG, phosphoprotein associated with glycosphingolipid-enriched microdomains; HEK, human embryonic kidney; GFP, green fluorescent protein; GST, glutathione S-transferase; BSA, bovine serum albumin; TBS, Tris-buffered saline; Ab, antibody; TCR, T cell receptor; MOPS, 4-morpholinepropanesulfonic acid; VSV, vesicular stomatitis virus.
References
- 1.Tasken, K., and Aandahl, E. M. (2004) Physiol. Rev. 84 137-167 [DOI] [PubMed] [Google Scholar]
- 2.Kaupp, U. B., and Seifert, R. (2002) Physiol. Rev. 82 769-824 [DOI] [PubMed] [Google Scholar]
- 3.de Rooij, J., Zwartkruis, F. J., Verheijen, M. H., Cool, R. H., Nijman, S. M., Wittinghofer, A., and Bos, J. L. (1998) Nature 396 474-477 [DOI] [PubMed] [Google Scholar]
- 4.Taylor, S. S., Buechler, J. A., and Yonemoto, W. (1990) Annu. Rev. Biochem. 59 971-1005 [DOI] [PubMed] [Google Scholar]
- 5.Dell'Acqua, M. L., and Scott, J. D. (1997) J. Biol. Chem. 272 12881-12884 [DOI] [PubMed] [Google Scholar]
- 6.Skalhegg, B. S., and Tasken, K. (2000) Front. Biosci. 5 D678-D693 [DOI] [PubMed] [Google Scholar]
- 7.Wong, W., and Scott, J. D. (2004) Nat. Rev. Mol. Cell Biol. 5 959-970 [DOI] [PubMed] [Google Scholar]
- 8.Jarnaess, E., and Tasken, K. (2007) Biochem. Soc. Trans. 35 931-937 [DOI] [PubMed] [Google Scholar]
- 9.Gronholm, M., Vossebein, L., Carlson, C. R., Kuja-Panula, J., Teesalu, T., Alfthan, K., Vaheri, A., Rauvala, H., Herberg, F. W., Tasken, K., and Carpen, O. (2003) J. Biol. Chem. 278 41167-41172 [DOI] [PubMed] [Google Scholar]
- 10.Huang, L. J., Durick, K., Weiner, J. A., Chun, J., and Taylor, S. S. (1997) J. Biol. Chem. 272 8057-8064 [DOI] [PubMed] [Google Scholar]
- 11.Huang, L. J., Durick, K., Weiner, J. A., Chun, J., and Taylor, S. S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 11184-11189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, H., Degenhardt, B., Tobin, D., Yao, Z. X., Tasken, K., and Papadopoulos, V. (2001) Mol. Endocrinol. 15 2211-2228 [DOI] [PubMed] [Google Scholar]
- 13.Liu, J., Rone, M. B., and Papadopoulos, V. (2006) J. Biol. Chem. 281 38879-38893 [DOI] [PubMed] [Google Scholar]
- 14.Reinton, N., Collas, P., Haugen, T. B., Skalhegg, B. S., Hansson, V., Jahnsen, T., and Tasken, K. (2000) Dev. Biol. 223 194-204 [DOI] [PubMed] [Google Scholar]
- 15.Angelo, R., and Rubin, C. S. (1998) J. Biol. Chem. 273 14633-14643 [DOI] [PubMed] [Google Scholar]
- 16.Kussel-Andermann, P., El-Amraoui, A., Safieddine, S., Hardelin, J. P., Nouaille, S., Camonis, J., and Petit, C. (2000) J. Biol. Chem. 275 29654-29659 [DOI] [PubMed] [Google Scholar]
- 17.Lim, C. J., Han, J., Yousefi, N., Ma, Y., Amieux, P. S., McKnight, G. S., Taylor, S. S., and Ginsberg, M. H. (2007) Nat. Cell Biol. 9 415-421 [DOI] [PubMed] [Google Scholar]
- 18.Dransfield, D. T., Bradford, A. J., Smith, J., Martin, M., Roy, C., Mangeat, P. H., and Goldenring, J. R. (1997) EMBO J. 16 35-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carr, D. W., Stofko-Hahn, R. E., Fraser, I. D., Bishop, S. M., Acott, T. S., Brennan, R. G., and Scott, J. D. (1991) J. Biol. Chem. 266 14188-14192 [PubMed] [Google Scholar]
- 20.Hausken, Z. E., Coghlan, V. M., Hastings, C. A., Reimann, E. M., and Scott, J. D. (1994) J. Biol. Chem. 269 24245-24251 [PubMed] [Google Scholar]
- 21.Hausken, Z. E., and Scott, J. D. (1996) Biochem. Soc. Trans. 24 986-991 [DOI] [PubMed] [Google Scholar]
- 22.Newlon, M. G., Roy, M., Morikis, D., Hausken, Z. E., Coghlan, V., Scott, J. D., and Jennings, P. A. (1999) Nat. Struct. Biol. 6 222-227 [DOI] [PubMed] [Google Scholar]
- 23.Newlon, M. G., Roy, M., Morikis, D., Carr, D. W., Westphal, R., Scott, J. D., and Jennings, P. A. (2001) EMBO J. 20 1651-1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banky, P., Huang, L. J., and Taylor, S. S. (1998) J. Biol. Chem. 273 35048-35055 [DOI] [PubMed] [Google Scholar]
- 25.Banky, P., Newlon, M. G., Roy, M., Garrod, S., Taylor, S. S., and Jennings, P. A. (2000) J. Biol. Chem. 275 35146-35152 [DOI] [PubMed] [Google Scholar]
- 26.Gold, M. G., Lygren, B., Dokurno, P., Hoshi, N., McConnachie, G., Tasken, K., Carlson, C. R., Scott, J. D., and Barford, D. (2006) Mol. Cell 24 383-395 [DOI] [PubMed] [Google Scholar]
- 27.Hausken, Z. E., Dell'Acqua, M. L., Coghlan, V. M., and Scott, J. D. (1996) J. Biol. Chem. 271 29016-29022 [DOI] [PubMed] [Google Scholar]
- 28.Kinderman, F. S., Kim, C., von Daake, S., Ma, Y., Pham, B. Q., Spraggon, G., Xuong, N. H., Jennings, P. A., and Taylor, S. S. (2006) Mol. Cell 24 397-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leon, D. A., Herberg, F. W., Banky, P., and Taylor, S. S. (1997) J. Biol. Chem. 272 28431-28437 [DOI] [PubMed] [Google Scholar]
- 30.Carr, D. W., Hausken, Z. E., Fraser, I. D., Stofko-Hahn, R. E., and Scott, J. D. (1992) J. Biol. Chem. 267 13376-13382 [PubMed] [Google Scholar]
- 31.Diviani, D., Soderling, J., and Scott, J. D. (2001) J. Biol. Chem. 276 44247-44257 [DOI] [PubMed] [Google Scholar]
- 32.Alto, N. M., Soderling, S. H., Hoshi, N., Langeberg, L. K., Fayos, R., Jennings, P. A., and Scott, J. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4445-4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlson, C. R., Lygren, B., Berge, T., Hoshi, N., Wong, W., Tasken, K., and Scott, J. D. (2006) J. Biol. Chem. 281 21535-21545 [DOI] [PubMed] [Google Scholar]
- 34.Diviani, D., Langeberg, L. K., Doxsey, S. J., and Scott, J. D. (2000) Curr. Biol. 10 417-420 [DOI] [PubMed] [Google Scholar]
- 35.Vang, T., Torgersen, K. M., Sundvold, V., Saxena, M., Levy, F. O., Skalhegg, B. S., Hansson, V., Mustelin, T., and Tasken, K. (2001) J. Exp. Med. 193 497-507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skalhegg, B. S., Tasken, K., Hansson, V., Huitfeldt, H. S., Jahnsen, T., and Lea, T. (1994) Science 263 84-87 [DOI] [PubMed] [Google Scholar]
- 37.Ruppelt, A., Mosenden, R., Gronholm, M., Aandahl, E. M., Tobin, D., Carlson, C. R., Abrahamsen, H., Herberg, F. W., Carpen, O., and Tasken, K. (2007) J. Immunol. 179 5159-5168 [DOI] [PubMed] [Google Scholar]
- 38.Bailey, T. L., and Elkan, C. (1994) Proc. Int. Conf. Intell. Syst. Mol. Biol. 2 28-36 [PubMed] [Google Scholar]
- 39.Aandahl, E. M., Aukrust, P., Skalhegg, B. S., Muller, F., Froland, S. S., Hansson, V., and Tasken, K. (1998) FASEB J. 12 855-862 [DOI] [PubMed] [Google Scholar]
- 40.Stokka, A. J., Gesellchen, F., Carlson, C. R., Scott, J. D., Herberg, F. W., and Tasken, K. (2006) Biochem. J. 400 493-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herberg, F. W., Maleszka, A., Eide, T., Vossebein, L., and Tasken, K. (2000) J. Mol. Biol. 298 329-339 [DOI] [PubMed] [Google Scholar]
- 42.Frank, R. (2002) J. Immunol. Methods 267 13-26 [DOI] [PubMed] [Google Scholar]
- 43.Hausken, Z. E., Coghlan, V. M., and Scott, J. D. (1998) Methods Mol. Biol. 88 47-64 [DOI] [PubMed] [Google Scholar]
- 44.Durgerian, S., and Taylor, S. S. (1989) J. Biol. Chem. 264 9807-9813 [PubMed] [Google Scholar]
- 45.Birkemo, G. A., Mantzilas, D., Luders, T., Nes, I. F., and Nissen-Meyer, J. (2004) Biochim. Biophys. Acta 1699 221-227 [DOI] [PubMed] [Google Scholar]
- 46.Levitt, M. (1978) Biochemistry 17 4277-4285 [DOI] [PubMed] [Google Scholar]
- 47.Jasanoff, A., and Fersht, A. R. (1994) Biochemistry 33 2129-2135 [DOI] [PubMed] [Google Scholar]
- 48.Lehrman, S. R., Tuls, J. L., and Lund, M. (1990) Biochemistry 29 5590-5596 [DOI] [PubMed] [Google Scholar]
- 49.Sonnichsen, F. D., Van Eyk, J. E., Hodges, R. S., and Sykes, B. D. (1992) Biochemistry 31 8790-8798 [DOI] [PubMed] [Google Scholar]
- 50.Li, H., Adamik, R., Pacheco-Rodriguez, G., Moss, J., and Vaughan, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1627-1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi, M., Mukai, H., Oishi, K., Isagawa, T., and Ono, Y. (2000) J. Biol. Chem. 275 34592-34596 [DOI] [PubMed] [Google Scholar]
- 52.Witczak, O., Skalhegg, B. S., Keryer, G., Bornens, M., Tasken, K., Jahnsen, T., and Orstavik, S. (1999) EMBO J. 18 1858-1868 [DOI] [PMC free article] [PubMed] [Google Scholar]