Abstract
‘Oketsu’ is a pathophysiologic concept in Japanese traditional (Kampo) medicine, primarily denoting blood stasis/stagnant syndrome. Here we have explored plasma protein biomarkers and/or diagnostic algorithms for ‘Oketsu’. Sixteen rheumatoid arthritis (RA) patients were treated with keishibukuryogan (KBG), a representative Kampo medicine for improving ‘Oketsu’. Plasma samples were diagnosed as either having an ‘Oketsu’ (n = 19) or ‘non-Oketsu’ (n = 29) state according to Terasawa's ‘Oketsu’ scoring system. Protein profiles were obtained by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS) and hierarchical clustering and decision tree analyses were performed. KBG treatment for 4 or 12 weeks decreased the ‘Oketsu’ scores significantly. SELDI protein profiles gave 266 protein peaks, whose expression was significantly different between the ‘Oketsu’ and ‘non-Oketsu’ states. Hierarchical clustering gave three major clusters (I, II, III). The majority (68.4%) of ‘Oketsu’ samples were clustered into one cluster as the principal component of cluster I. The remaining ‘Oketsu’ profiles constituted a minor component of cluster II and were all derived from patients cured of the ‘Oketsu’ state at 12 weeks. Construction of the decision tree addressed the possibility of developing a diagnostic algorithm for ‘Oketsu’. A reduction in measurement/pre-processing conditions (from 55 to 16) gave a similar outcome in the clustering and decision tree analyses. The present study suggests that the pathophysiologic concept of Kampo medicine ‘Oketsu’ has a physical basis in terms of the profile of blood proteins. It may be possible to establish a set of objective criteria for diagnosing ‘Oketsu’ using a combination of proteomic and bioinformatics-based classification methods.
Keywords: proteinchip, surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS), keishibukuryogan (KBG)
Introduction
Concepts of ‘Oketsu’ and its Diagnosis
‘Oketsu’, blood stasis or stagnant syndrome, is a pathophysiologic concept unique to Japanese traditional (Kampo) medicine (1–3) and is thought to be closely related to several diseases such as climacteric disorder, rheumatoid arthritis (RA) (4), Behcet's disease (5), hyperuricemia and various inflammatory conditions (6). Recent reports have suggested that ‘Oketsu’ is correlated with a deterioration of erythrocyte deformability (7), an elevation in blood viscosity (8), acceleration of erythrocyte aggregation (9), autonomic nervous activity changes (10) and microcirculatory dysfunction (11). However, a precise concept of ‘Oketsu’ from the standpoint of western medicine has yet to be established. Doctors skilled in Kampo diagnosis and treatment are often able to improve the symptoms of these blood disorders, which may be considered to be untreatable by conventional western therapies. Therefore, if a scientific and objective standard for diagnosis and treatment of ‘Oketsu’ can be established, the public will be given greater access to the beneficial effects of Kampo medicine. Furthermore, elucidation of the concepts, theory and practice of Kampo medicines from a scientific and objective viewpoint could make a significant contribution to modern medicine. Although Terasawa et al. (12) have established criteria for diagnosing ‘Oketsu’ that are made up of indices of numerous symptom scores, an improved diagnostic system that eliminates descriptive definitions involving subjective judgments must be established. Therefore, it is very important to standardize the diagnostic criteria for ‘Oketsu’ objectively by using a universally acceptable scientific method.
Investigation of Biomarkers Using SELDI-TOF MS
Recent advances in mass spectrometry enable high-throughput profiling of the protein content of complex mixtures (13). For example, ProteinChip technology combined with surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS) allows the rapid detection of a wide range of proteins from a small amount of biologic material. Using this innovative technology, it is possible to detect changes in the expression profile of specific proteins of interest (14,15). Indeed, advances in mass spectrometry are accelerating the discovery of biomarkers for various disease states, especially for the detection of several tumors such as prostate, ovarian and breast cancers (16–18). Existing diagnostic biomarkers of tumors, such as ovarian cancer antigen 125 (CA125), are often inadequate for distinguishing benign from malignant neoplasm in clinical practice. However, three biomarkers discovered using the SELDI profiling system have significantly improved the detection of early stage ovarian cancer when used in combination with CA125 (16). The diagnosis of a disease using ‘multiple markers’ (i.e. ‘fingerprint analysis’) by employing a combination of comprehensive high-throughput profiling and bioinformatics-based discrimination methods is a promising approach, especially when a single gene aberration or protein cannot account for all phenotypic forms of the disease. As is the case with most human cancers, ‘Oketsu’ is subject to individual variation, multifactorial modification of the disease state and marked microheterogeneity of pathogenic molecules. Thus, confirming the diagnosis of a complex disease that cannot be verified by a single marker will be extremely useful in clinical practice.
The Aim of This Study
In the present study, we have attempted to establish ‘Oketsu’ biomarkers and/or diagnostic algorithms using SELDI-TOF MS. RA patients were chosen as subjects for this study because 53.8% of the RA patients have ‘Oketsu’ syndrome and a clinical relationship exists between the severity of Terasawa's ‘Oketsu’ score and Lansbury's activity index of RA (4). Keishibukuryogan (KBG), a representative medicine for the treatment of ‘Oketsu’ syndrome (19–21), was administered to 16 patients for 12 weeks and the activity of RA, severity of ‘Oketsu’ and proteomic pattern of plasma at 0, 4 and 12 weeks were evaluated. Clustering analysis and decision tree construction were performed to classify the ‘Oketsu’ and ‘non-Oketsu’ states. Our results confirm the need to establish objective criteria for diagnosing ‘Oketsu’ using a proteomic approach in combination with bioinformatics-based classification methods.
Methods
Drug: Keishibukuryogan
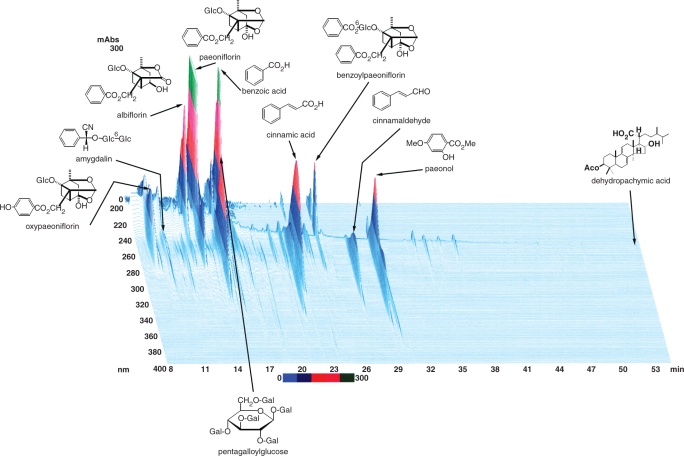
In Japan, more than 100 Kampo medicines including KBG have been approved as ethical medicines by the Ministry of Health, Labor and Welfare of Japan. KBG has been used for the treatment of gynecologic disorders such as uterine myoma, climacteric disorder, etc (22–25). KBG is comprised of five medical plants as follows: Cinnamomum cassia Blume (Cinnamomi Cortex), Poria cocos Wolf (Hoelen), Paeonia lactiflora Pallas (Paeoniae Radix), Prunus persica Batsch (Persicae Semen) and Paeonia suffruticosa Andrews (Moutan Cortex). These plants are registered in the Pharmacopoeia of Japan. In the traditional method of preparing Kampo formulas, which is even now being used in not just a few hospitals; the constituent herbs are cut into pieces, mixed according to the instructions in classical texts, decocted in hot water and ingested as a liquid extract. In recent years, technologic advances have made it possible to manufacture the extract as a spray- or a freeze-dried powder with high standards of quality, uniformity and stability. KBG is provided by Tsumura & Co. and several Kampo manufacturers in Japan. The ultraviolet-visible light (UV-VIS) profile of three-dimensional (3D) high performance liquid chromatography (HPLC) analysis of the most widely used KBG is shown in Fig. 1.
Figure 1.
Three-dimensional (3D) HPLC profile of KBG and UV spectra of its constituent crude drugs.
Subjects and Plasma Samples
We obtained plasma samples from 16 subjects with RA who visited the Department of Japanese Oriental Medicine, Toyama Prefectural Central Hospital. Informed consent was obtained from every subject prior to the study. All subjects met the American College of Rheumatology 1987 criteria for the diagnosis of RA (26). For evaluation of the ‘Oketsu’ state, we used the ‘Oketsu’ scoring system, with diagnostic criteria developed by Terasawa et al. (12) (Table 1). This ‘Oketsu’ scoring system consists of 17 inquiries with three scales of points that are determined after extensive multivariate analyses; the resultant score in this system has been reported to have a quantitative relationship with hemorheology data (7,8,20). Furthermore, analysis of relationship between this score and the diagnosis of ‘Oketsu’ by traditional Kampo doctors has revealed that predictive results are obtained depending on the classification of Oketsu syndrome as of a score of more than 20 points. Subjects were followed for 12 weeks at an out-patient clinic. After evaluation of the ‘Oketsu’ score (week 0), all of the subjects were treated with KBG; TJ-25, 2.5 g × 3/day for a period of 12 weeks. Some subjects received concomitant disease-modifying anti-rheumatic drugs (DMARD), non-steroidal anti-inflammatory drugs (NSAID), steroid or other Kampo medicines for RA. Furthermore, some subjects were given medicines to control other disease states. However, only subjects whose condition of disease and ‘Oketsu’ state were relatively stable were selected for the present study and they remained in a stable condition with no change in their other medicines or dosage during the period of study. Therefore, it is highly probable that the changes that occurred during the KBG treatment were caused by KBG, although future studies should be designed to evaluate the pure effect of KBG. We re-evaluated the ‘Oketsu’ score at 4 and 12 weeks after the commencement of KBG treatment. Plasma samples were obtained from each subject before treatment (week 0) and 4 and 12 weeks after treatment (i.e. a total of 48 plasma samples from 16 subjects). Samples were stored at −80°C until use.
Table 1.
Diagnostic criteria for ‘Oketsu’ syndrome
| Symptom | Score |
|
|---|---|---|
| Male | Female | |
| Dark-rimmed eyes | 10 | 10 |
| Areas of dark pigmentation of facial skin | 2 | 2 |
| Rough skin | 2 | 5 |
| Livid lips | 2 | 2 |
| Livid gingiva | 10 | 5 |
| Livid tongue | 10 | 10 |
| Telangiectasis/vascular spiders | 5 | 5 |
| Subcutaneous hemorrhage | 2 | 10 |
| Palmar erythema | 2 | 5 |
| Resistance and tenderness on pressure of the left para-umbilical region | 5 | 5 |
| Resistance and tenderness on pressure of the right para-umbilical region | 10 | 10 |
| Resistance and tenderness on pressure of the umbilical region | 5 | 5 |
| Resistance and/or tenderness on pressure of the ileo-cecal region | 5 | 2 |
| Resistance and/or tenderness on pressure of the sigmoidal region | 5 | 5 |
| Resistance and/or tenderness on pressure of the subcostal region | 5 | 5 |
| Hemorrhoids | 10 | 5 |
| Dysmenorrhea | – | 10 |
A total score larger than 20 is diagnosed as an ‘Oketsu’ state and that not exceeding 20 is diagnosed as a ‘non-Oketsu’ state. Mild symptoms are designated by half points.
Sample Preparation for Proteinchip Analysis of Plasma Unfractionated Plasma
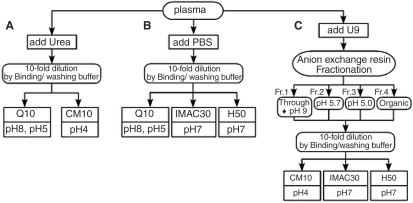
Figure 2A and B illustrates the preparation of unfractionated plasma samples. Plasma samples were thawed and diluted 1:10 by urea denaturing buffer (7 mol/l urea, 2 mol/l thiourea, 4% CHAPS, 1% DTT and 2% ampholyte) or PBS buffer.
Figure 2.
Flow charts of plasma processing for SELDI ProteinChip. (A) Unfractionated plasma (Urea) (B) Unfractionated plasma (PBS) (C) Anion exchange fractionation of plasma. Each plasma was divided into three aliquots and each aliquot was further processed according to the procedures denoted by (A), (B), (C). To cover as wide a range of peaks as possible, combinations of multiple ProteinChip previously optimized for each sample preparation were used.
Anion Exchange Fractionation of Plasma
Plasma samples were separated into four fractions in an anion exchange resin (Q Ceramic HyperD F resin: BIOSEPRA) in a 96-well filter plate (Nalge Nunc: 0.45 μm pore size) on a vacuum manifold as outlined in Fig. 2C. Plasma samples were thawed and spun at 20 000 g for 15 min at 4°C. Twenty microliters of plasma samples were added to 30 μl of U9 buffer (9 mol/l urea, 2% CHAPS and 50 mmol/l Tris-HCl, pH 9.0) and vortexed at 4°C for 20 min. Fifty microliters of the diluted plasma sample was applied to each well and 50 μl of U1 buffer (1 mol/l urea, 0.22% CHAPS and 50 mmol/l Tris-HCl, pH 9.0) was added. The plate was shaken at 4°C for 30 min. Proteins were eluted through the filter by washes with buffers of different pH. Firstly, after the flow-through was collected, the resins were incubated with 100 μl of wash buffer (50 mmol/l Tris-HCl, pH 9.0, 0.1% N-octylglucopyranoside, OGP) at room temperature for 10 min with shaking. The wash was collected as ‘fraction 1’ (flow-through + pH 9.0). This procedure was repeated two times with 100 μl each of appropriate buffer of decreasing pH (pH 5.7 and pH 5.0: 100 mmol/l sodium acetate, 0.1% OGP) to give ‘fraction 2’ and ‘fraction 3’. The final wash was performed with an organic buffer containing 33.3% isopropanol, 16.7% acetonitrile and 0.1% trifluoroacetic acid to give ‘fraction 4’.
Proteomic Profiling Using SELDI-TOF MS
Each prepared sample was diluted 1:10 by the appropriate binding/washing buffers (see below) and analyzed by four types of ProteinChip (Ciphergen Biosystems): CM10 (Weak Cation Exchange), Q10 (Strong Anion Exchange), IMAC30 (Immobilized Metal Affinity Capture coupled with copper) or H50 (Hydrophobic), as shown in Fig. 2. Each array surface was prepared according to the Ciphergen ProteinChip Applications guide (27). The binding/washing buffers for the ProteinChips were 50 mmol/l Tris-HCl, pH 8.0 and 50 mmol/l sodium acetate, pH 5.0 (for Q10), 100 mmol/l sodium acetate, pH 4.0 (for CM10), 50 mmol/l HEPES, pH 7.0 (for H50) or 100 mmol/l sodium phosphate-500 mmol/l NaCl, pH 7.0 (for CopperII-IMAC30). The energy-absorbing molecules (EAM), sinapinic acid (SPA) and α-cyano-4-hydroxycinnamic acid (CHCA), were applied to each spot in the ProteinChip array. Peptides and proteins in the m/z (mass/charge) 3000–10 000 range were ionized with CHCA, whereas those in the m/z 3000–30 000 range were ionized with SPA. For ionization with SPA, two different intensities of laser shot were used in order to acquire low (3000–10 000) and high (10 000–30 000)-mass spectra, which are designated as ‘SPA-low’ and ‘SPA-high’, respectively. The ProteinChip arrays were analyzed using a Ciphergen PBS IIc ProteinChip Reader, which were calibrated externally using molecular mass standards (Ciphergen Biosystems).
Analysis of Proteomic Data
All spectra were collected using the ProteinChip software version 3.1.1 (Ciphergen Biosystems). After smoothing, baseline subtraction and total ion current normalization, a proprietary-developed peak detection procedure (designated as ‘Cross Detector’) was performed for the spectral data. Firstly, peak screening was performed for all the spectra. This step provided a reliability score, representing ‘peak likelihood’, for each of the screened peaks by assessing the results obtained from plural runs of an identical peak screening procedure with different parameter values. Effective peak intervals were determined based on the cross-spectral density curve of the reliability score. All the peaks were then redefined so that each spectrum had one peak in each peak interval. The peaks obtained by the ‘Cross Detector’ algorithm were separately evaluated by the Mann–Whitney U-test to examine statistical significance of the difference in peak intensities between ‘Oketsu’ and ‘non-Oketsu’ states. To avoid harmful effects caused by multiple testing, we used the false discovery rate (FDR) for assessment of statistical significance. We adopted Benjamini and Hochberg's (28) FDR controlling procedure for FDR estimation and the criterion was set to FDR < 0.05.
Clustering and Decision Tree Classification
Spectral data were represented as a proteomic pattern characterized by the differentially expressed peaks. We applied a two-way hierarchical clustering method to those proteomic patterns using Spotfire Decision Site version 7.2, where the ‘weighted pair group method with arithmetic mean’ (WPGMA) and Tanimoto distance were used as a clustering method and similarity metrics, respectively. The decision tree analysis was performed as follows. Three decision trees on distinct strategies were constructed: (i) ordinary C5.0, (ii) PCA + C5.0 and (iii) C5.0 (high-factor-loading). Strategy (i), ordinary C5.0, generated a standard decision tree based on the C5.0 learning algorithm using all of the significantly differential peaks as candidates for splitters. Meanwhile, strategy (ii), PCA + C5.0, performed dimension reduction using Principal Component Analysis (PCA), which converted higher-dimensional space spanned by differential peaks into less-redundant and lower-dimensional space of several principal components. A C5.0 decision tree with the principal components for splitters was then generated. The third strategy (iii), C5.0 (high-factor-loading), also conducted PCA and selected fewer peaks that had high values of factor loading, which were obtained through the PCA computation. We selected the ten highest-factor-loading peaks for each of the principal components. A C5.0 tree was then constructed using the high-factor-loading peaks for splitters. We used Clementine version 7.2 to perform PCA and C5.0 tree construction.
Statistics
‘Oketsu’ scores are expressed as mean ± SD. Statistical analysis was determined by paired t-test with Bonferroni's correction for multiple comparisons.
Results
Prevalence of ‘Oketsu’ Syndrome and Improvement of ‘Oketsu’ By KBG
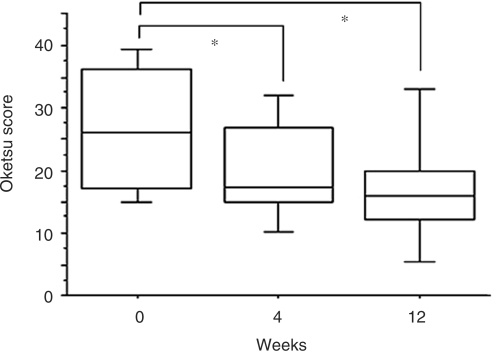
We chose to use the diagnostic criteria of ‘Oketsu’ by Terasawa et al. (12) (Table 1) because it constituted the most systematic method of scoring the symptoms. The oketsu score was evaluated at week 0 (pre-treatment) and 4 and 12 weeks after the commencement of KBG treatment. The pathophysiologic states of patients were classified into two categories according to the oketsu scores of the patients at the time of blood sample collection: a ‘non-Oketsu’ state (Oketsu score ≤ 20) and an ‘Oketsu’ state (Oketsu score > 20). Table 2 shows the age/sex distribution of 16 subjects and the oketsu scores at each time. Ten of 16 patients with RA (62.5%) were diagnosed as having ‘Oketsu’ at week 0. This percentage is consistent with the previously reported incidence of ‘Oketsu’ (53.8%) in RA (4). KBG treatment for 12 weeks decreased the Oketsu score in 11 of the 16 RA patients, including those in the ‘non-Oketsu’ group. Figure 3 shows the averaged values of the Oketsu score for all subjects. A significant decrease in the score was found after 4 and 12 weeks of KBG treatment, implying an improvement in the ‘Oketsu’ state diagnosed by Kampo medicine. For proteomic studies, 48 plasma samples obtained from 16 patients at three time points (0, 4 and 12 weeks) were independently assigned as ‘Oketsu’ (n = 19) or ‘non-Oketsu’ (n = 29) samples according to the diagnosis of ‘Oketsu’. Fifty-five SELDI profiles measured by 55 different conditions and pre-processing (Methods and Fig. 2) were obtained from each plasma sample using SELDI-TOF MS proteomics technology. The plasma from each subject was measured in duplicate on two separate ProteinChip arrays and treated as independent samples in the following analyses.
Table 2.
Oketsu score and age/sex distribution of subjects
| Subject number | Age | Sex |
Oketsu score |
Oketsu state/ non-Oketsu at 0 week | |||
|---|---|---|---|---|---|---|---|
| 0 week | 4 weeks | 12 weeks | 0W → 12W | ||||
| 1 | 56 | Female | 49 | 32 | 34 | ↓ | Oketsu |
| 2 | 67 | Female | 35 | 10 | 15 | ↓ | Oketsu |
| 3 | 65 | Female | 20 | 20 | 20 | → | non-Oketsu |
| 4 | 62 | Male | 10 | 10 | 10 | → | non- Oketsu |
| 5 | 68 | Female | 17 | 17 | 14.5 | ↓ | non- Oketsu |
| 6 | 64 | Female | 15 | 15 | 20 | ↑ | non- Oketsu |
| 7 | 63 | Female | 17 | 17 | 17 | → | non- Oketsu |
| 8 | 54 | Female | 27 | 17 | 10 | ↓ | Oketsu |
| 9 | 54 | Female | 25 | 12.5 | 5 | ↓ | Oketsu |
| 10 | 52 | Female | 25 | 25 | 15 | ↓ | Oketsu |
| 11 | 70 | Male | 32.5 | 17.5 | 17.5 | ↓ | Oketsu |
| 12 | 47 | Female | 15 | 15 | 5 | ↓ | non- Oketsu |
| 13 | 69 | Female | 37 | 27 | 15 | ↓ | Oketsu |
| 14 | 46 | Female | 38 | 27 | 19.5 | ↓ | Oketsu |
| 15 | 79 | Male | 30 | 30 | 25 | ↓ | Oketsu |
| 16 | 51 | Female | 39.5 | 39.5 | 44 | ↑ | Oketsu |
↓: decrease, →: no change, ↑: increase.
Figure 3.
Change in oketsu score after administration of KBG. The averaged values of oketsu score was evaluated at week 0 (pre-treatment) and 4 and 12 weeks after the commencement of KBG treatment. *P < 0.05 vs week 0, determined by paired t-test with Bonferroni's correction for multiple comparison.
Hierarchical Clustering of 55 SELDI Profiles Classifies ‘Oketsu’ and ‘non-Oketsu’ States
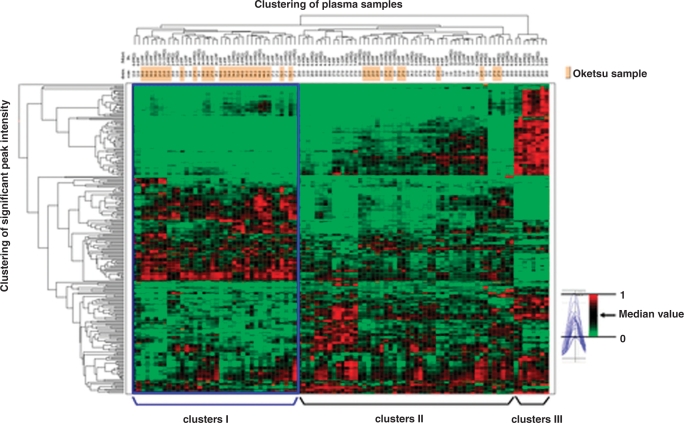
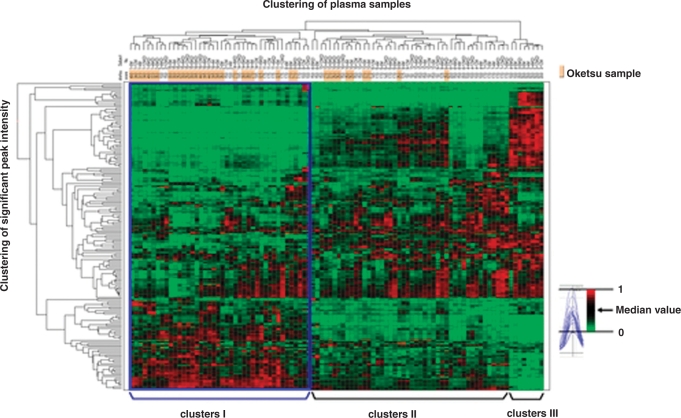
Spectral data were obtained using ProteinChip software version 3.1.1. Peak detection performed using Biomarker Wizard software (Ciphergen Biosystems) missed a considerable number of peaks that were found as specific peaks by manual inspection. We therefore developed a new algorithm of peak detection, termed ‘Cross Detector’, based on cross-parametric multiple screening and cross-spectral reliability density. The Cross Detector algorithm produced 2596 peaks in a mass range of 3000–30 000 from 5280 SELDI spectra. Our analysis found 266 peaks that were differentially expressed between the ‘Oketsu’ and ‘non-Oketsu’ plasma samples. Next, we performed two-way hierarchical clustering analysis (two-dimensional WPGMA), based on the 266 peaks and the ‘Oketsu’/’non-Oketsu’ classification. The clustering algorithm identified three major clusters (designated as ‘I’, ‘II’, ‘III’ in Fig. 4) in a longitudinal direction. Clusters I, II and III contained 38, 50 and 8 samples, respectively. Despite the difference in time points, the six samples obtained from each patient were clustered at a nearby cluster site. As indicated by the blue box, 68.4% (26/38) of the ‘Oketsu’ samples were found in cluster ‘I’. Cluster ‘II’ also contained some ‘Oketsu’ samples (12/38, 31.6%). The ‘Oketsu’ samples made up 68.4% and 24.0% of clusters ‘I’ and ‘II’, respectively. Intriguingly, all of the ‘Oketsu’ samples classified into cluster ‘II’ were derived from patients showing an improved ‘Oketsu’ state at 12 weeks. Our data suggest that the ‘Oketsu’ and ‘non-Oketsu’ states can be classified, at least to a certain extent, into different groups by clustering analysis of SELDI proteomic patterns of plasma proteins.
Figure 4.
Hierarchical clustering of 55 SELDI profiles. Clustering was based on the 266 differential peaks obtained from 55 SELDI profiles and ‘Oketsu’/ ‘non-Oketsu’ classification. The red (peak maximum = 1) or green (peak minimum = 0) color indicates the relative intensity, i.e. higher than or lower than the median value (black color), respectively.
Decision Tree Analysis to Construct a Predictive Model for ‘Oketsu’
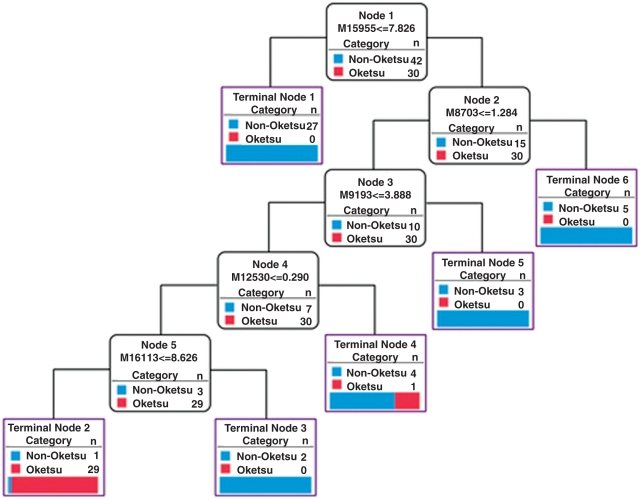
We next investigated whether or not a single peak in the SELDI profiles can be used as a marker to discern ‘Oketsu’ and ‘non-Oketsu’ plasma samples. However, no such peaks could be identified. We also performed multivariate analysis to identify a combination of peaks that could distinguish between the ‘Oketsu’ and ‘non-Oketsu’ states using a decision tree classification with the algorithm C5.0. In general, a decision tree should be created from a large number of samples and the generalization error should be estimated using a test set that is collected independently of the training set. However, this approach assumes that a large sample set is readily available. In the present study, because of limitations in sample acquisition, all 96 samples were divided into four data sets. Three data sets were used as the training set for the classifier and the remaining data set was used to evaluate the generalization error. The simulation by cross-validation was performed four times by replacing the test set. The 266 peaks described previously for the clustering were used to generate the decision tree. Table 3 shows the average values of estimated accuracy, sensitivity and specificity obtained from the four-fold cross-validation analyses. Analysis by C5.0 gave an estimated accuracy, sensitivity and specificity of 69.8%, 78.8% and 65.3%, respectively. The best example of the decision tree is shown in Fig. 5. The tree used five peaks (m/z 15 955, 8703, 9193, 12 530 and 16 113) to generate six terminal nodes and distinguished them with 97.2% accuracy in the training set. In the test set, the tree yielded 83.3% accuracy, 100.0% sensitivity and 75.0% specificity. Using the same data, we attempted to improve the accuracy, sensitivity and specificity of the analysis by using principal component analysis (PCA) combined with C5.0. Firstly, five principal components of PCA on the 266 peaks were used to construct the decision tree with the algorithm C5.0 (PCA + C5.0). Secondly, ten highest-factor-loading peaks obtained from PCA were used to construct the decision tree with C5.0 (C5.0 + high-factor-loading). However, the application of PCA to tree analysis did not improve the accuracy of the decision tree based on C5.0. Our results suggest that classification algorithms to distinguish the ‘Oketsu’ state from the ‘non-Oketsu’ state based on the SELDI protein profiling may be established provided that sufficient samples are analyzed.
Table 3.
A comparison of outcome of C5.0 with different features in the test set (55 SELDI profiles)
| classifier | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| C5.0 | 69.8 | 78.8 | 65.3 |
| PCA+C5.0 | 70.8 | 59.0 | 81.5 |
| C5.0 (high-factor-loading) | 65.7 | 74.2 | 62.0 |
PCA + C5.0: C5.0 tree constructed with five principal components obtained by PCA for splitters, C5.0 (high-factor-loading): C5.0 tree constructed with ten highest-factor-loading peaks for each of the principal components for splitters.
Figure 5.
Classification of ‘Oketsu’ and ‘non-Oketsu’ using the decision tree (C5.0) in the training set (55 SELDI profiles). If the answer to the question in a node of the tree is yes, proceed down to the left node; otherwise proceed down to the right node. M represents the m/z.
Reducing the Measurement Conditions (From 55 to 16) Gives a Similar Outcome
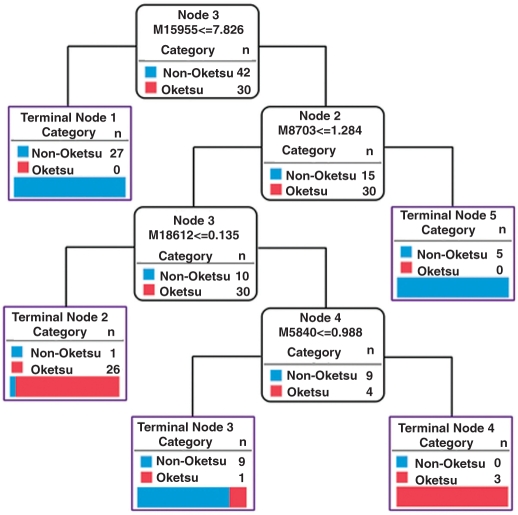
The measurement of 55 SELDI profiles from a single sample is both laborious and expensive. Therefore, we attempted to reduce the number of measurement conditions. A detailed examination of the 55 proteomic patterns and 266 detected peaks suggested that 16 SELDI profiles may be enough to obtain results compatible with those derived from the 55 profiles. The peak detection algorithm ‘Cross Detector’ produced 848 peaks from 1536 spectra of 16 SELDI profiles. We found 185 peaks that displayed significant differential expression between ‘Oketsu’ and ‘non-Oketsu’ states patients. Two-way hierarchical clustering analysis and classification decision tree analysis were performed using the methods described above. The result of two-way hierarchical clustering analysis was similar to that shown in Fig. 4 (73.7% of ‘Oketsu’ samples were clustered together (into ‘I’) as indicated by a blue box; Fig. 6). The accuracy, sensitivity and specificity of classification trees based on 16 SELDI profiles were no less than those based on the 55 profiles (Table 4). As shown in Fig. 7, the best example of the C5.0 decision tree used four peaks (m/z 15 955, 8703, 18 612 and 5840) to generate five terminal nodes. These were distinguished with 97.2% and 83.3% accuracy in the training set and the test set, respectively. It must be noted that the decision trees shown in Fig. 5 (55 profiles) and Fig. 7 (16 profiles) both used the same peaks, m/z 15 955 and 8703, for first and second splitter, respectively.
Figure 6.
Hierarchical clustering of 16 SELDI profiles. Clustering was based on the 185 differential peaks obtained from 16 SELDI profiles and ‘Oketsu’/’non-Oketsu’ classification. The red (peak maximum = 1) or green (peak minimum = 0) color indicates the relative intensity, i.e. higher than or lower than the median value (black color), respectively.
Table 4.
A comparison of outcome of C5.0 with different features in the test set (16 SELDI profiles)
| Classifier | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| C5.0 | 75.0 | 79.6 | 74.0 |
| PCA+C5.0 | 72.9 | 73.3 | 74.6 |
| C5.0 (high-factor-loading) | 66.7 | 62.7 | 71.2 |
PCA + C5.0: C5.0 tree constructed with five principal components obtained by PCA for splitters, C5.0 (high-factor-loading): C5.0 tree constructed with ten highest-factor-loading peaks for each of the principal components for splitters.
Figure 7.
Classification of ‘Oketsu’/’non-Oketsu’ using the decision tree (C5.0) in the training set (16 SELDI profiles). If the answer to the question in a node of the tree is yes, proceed down to the left node; otherwise, proceed down to the right node. M represents the m/z.
Discussion
Kampo medicine is a traditional Japanese medicine. It originates from a traditional Chinese medicine, but has developed into a unique form in Japan. Traditional Chinese and Kampo medicines have special kinds of medical terminology for the diagnosis and treatment of diseases and ‘Oketsu’ is one of the pathophysiologic concepts that have been most frequently used in Kampo medicine. Ancient Chinese medical texts describe a disorder of the blood circulation causing various symptoms, such as blood stasis, reduced blood flow and cessation of flow. This phenomenon is called ‘Oketsu’ in Japanese or ‘Yu Xue’ in Chinese. However, the meaning of ‘Oketsu’ differs slightly from that of ‘Yu Xue’ and Oketsu has gained a more important position in Kampo medicines in Japan. The main cause of the stagnation in blood flow leading to ‘Oketsu’ syndrome is thought to be insufficient exercise or sleep, mental or physical stress, constipation or a high-calorie diet. At the present time, it may be not possible to translate ‘Oketsu’ accurately in terms of Western medicine. However, ‘Oketsu’ is known to be related to disorders in the peripheral microcirculation, including excessive sensitivity to cold, menstrual disorder and infertility, as well as refractory diseases, such as RA, systemic lupus erythematosus (SLE), disseminated intravascular coagulation (DIC) and various allergic responses (4,6). Because ‘Oketsu’ is also a concept denoting ‘the preparatory state for developing recognizable diseases’, the diagnosis and treatment of ‘Oketsu’ not only will improve the symptoms of the manifest diseases, but also may prevent the development of these diseases. Kampo medicines are formulations consisting of crude drugs that have been used in Japan for thousands of years for patients with a wide variety of disorders. Nowadays, more than 100 Kampo medicines have been approved as ethical medicines by the Ministry of Health, Labour and Welfare of Japan. The drugs are manufactured on a modern industrial scale in which the quality and quantity of ingredients are standardized under strict guidelines. KBG is thought to be one of the most important prescriptions for improving ‘Oketsu’ syndrome. KBG has been used for the treatment of various types and stages of diseases (6,22–25), from the stage prior to disorder to chronic intractable diseases that are untreatable even by a wide array of therapeutic strategies of modern Western medicine.
In an investigation of the effect of a Kampo medicine (e.g. KBG) on the ‘symptom’ (e.g. ‘Oketsu’ state) diagnosed by Kampo medicine, it is difficult to determine clearly the detailed efficacies and mechanisms of action of the medicine, because there are often multiple active ingredients and components and target sites, and mutual interactions of these multiple components. As described previously, disease concepts such as ‘Oketsu’ are the composite of different and multiple symptoms that can be influenced by environmental and genetic factors. Therefore, the molecular alterations induced in the ‘Oketsu’ state by KBG, in addition to its efficacy, would be mediated by complicated set of multimodal and multifactorial events. Conventional drug research has mainly focused on the mode of action of a relatively small number of molecules. Recent advances in “omics” technology have resulted in the development of extremely high-throughput and comprehensive techniques for functional genomics, proteomics and metabolomics. A powerful array of tools is available for investigating the intricate interplay of different signal-transduction pathways in the cell. It is conceivable that these technologies may offer a crucial contribution to clarification of the concepts and efficacy of Kampo medicine(s) in terms of molecular biology.
In the present study, we have investigated the expression of plasma protein in patients in the ‘Oketsu’ state by using a proteomics approach. We used the patients’ plasma to analyze protein expression for three main reasons. Firstly, collection of blood samples is performed routinely in medical practice and is minimally invasive. Secondly, the ‘Oketsu’ state is considered to be closely related to hemorheological and hemostasis abnormalities. Increases in fibrinogen level, erythrocyte aggregation and blood viscosity in ‘Oketsu’ syndrome and their amelioration by KBG treatment have been reported (8,9,11,19–21). Thirdly, the abnormalities in hemostasis suggest that there are proteomic changes in the coagulation-fibrinolytic system in which a massive proteolysis cascade plays a critically important role. A cascade of this type may be involved in ‘Oketsu’ syndrome. Therefore, we focused our attention on the expression of proteins in the plasma rather than the serum, which is devoid of most of the proteins involved in the coagulation-fibrinolytic system. Thus, the adoption of proteomic analysis of plasma in ‘Oketsu’ patients would be reasonable both practically and theoretically.
The present study demonstrated that the ‘Oketsu’ and ‘non-Oketsu’ states were classified into different clusters in terms of the plasma protein levels. Hierarchical clustering, a common method of the clustering analysis family continuously aggregates similar objects or ‘clusters’ into a larger cluster and eventually generates a single cluster that contains all objects. Therefore, the classification is constructed in a tree-like manner and is represented by a so-called ‘dendrogram’. The similarity between objects is defined by distance metrics such as Euclidean distance, Manhattan distance and lots of other proposed types of metrics. The more similar the objects/clusters are in the earlier stage, the more they are merged. In the present study, the objects to be classified (plasma samples) were represented as a proteomic pattern characterized by differentially expressed peaks, that is, an N-dimensional vector whose element values represent peak intensities. The similarity between two samples was calculated as the Tanimoto distance between two vectors of proteomic patterns. By the successive aggregation of similar samples, we obtained sample classification (the upper horizontal row in the color map of Fig. 4). Meanwhile, the given proteomic patterns provided another set of ‘objects’ to be classified, namely, peaks. The profile of each peak was composed by the peak intensities of 96 samples and the similarity of the profiles among 266 peaks was calculated as described above. By the successive aggregation of similar peaks, we obtained peak classification (the left vertical row in the color map of Fig. 4). Such simultaneous clustering of an identical data set vertically and horizontally is called ‘two-way clustering’, an approach that has been reported to have potentially high performance in the identification of local groups of genes and/or proteins with similar expression patterns (29). Our results indicate that a characteristic proteomic pattern may exist in the plasma of ‘Oketsu’ patients, although a larger-scale clinical trial will be necessary to validate this finding. Furthermore, the hierarchical clustering is a heuristic approach but may not provide any enough “evidence” in principle.
In general, statistical analysis of the world of ‘omics’ is now being developed. Although there are examples of statistical analyses of hierarchical clustering, the practical significance of such analyses is a complex and controversial issue. At present, ‘omics’ approaches are best-suited to create a new assumption/hypothesis and many researchers have pointed out that validation studies should be conducted in separate follow-up studies. In this case, validation studies with bigger cohorts, methodologic improvement and strictly defined protocols will be absolutely necessary, however, should future studies be undertaken.
Next, to develop a diagnostic criterion for ‘Oketsu’, a decision tree analysis was performed. A decision tree is a decision support tool used to identify the strategy most likely to reach a goal. In the present case, the algorithm searches and selects a certain SELDI peak as a classifier that most efficiently splits ‘Oketsu’ samples (>20 ‘Oketsu'scoring) from ‘non-Oketsu’ samples (≤20 ‘Oketsu’ scoring). Usually, the first classifier cannot give a complete classification. Instead, there is an ‘Oketsu’ -sample-rich group containing a smaller number of ‘non-Oketsu’ samples and a ‘non-Oketsu’-sample-rich group containing a smaller number of ‘Oketsu’ samples. For each group, the algorithm repeats the search for another (second) classifier. The combination of the first and second classifiers thus gives a more accurate classification. This process can be repeated until the tree classifies all of the samples correctly. Thus, the constructed decision tree gives the most efficient method of the classification of the given samples and in the next step; the tree can be used as a predictive model. When a new sample with SELDI data but without ‘Oketsu’ scores is given, it can be predicted whether it is ‘Oketsu’ or ‘non-Oketsu’ by examining the SELDI data in reference to the decision tree. The decision tree construction in the present study, however, remains a preliminary trial that should be improved by large-scale examination. Further studies with independently collected samples will be needed to verify the above points. Collectively, our finding at least addresses the possibility that this ancient and apparently very speculative concept ‘Oketsu’ might have a physical basis and that objective criteria for diagnosing ‘Oketsu’ can be established using a combination of ProteinChip technology and bioinformatics tools.
Several concerns have been raised related to the present approach. Firstly, various researchers have pointed out that, in not a few cases, SELDI-TOF protein patterns in serum are not reproducible across different experiments for reasons such as differences in base line correction, sample preparation protocol, spectrum analysis method, mass calibration method, etc (30). In preceding studies, we have tested the effect of the ProteinChip lot, sample processing, measures taken at different times and analytical methods and constructed highly standardized protocols. These protocols contain a reliable peak detection algorithm (‘Cross Detector’) and robust methods for selecting significant peaks (‘Peak Separability Analysis’), which are now patent pending and will be described in a future paper. Thus, we think that it is possible that objective criteria for diagnosing ‘Oketsu’ can be established using a combination of ProteinChip technology and bioinformatics tools. Ray et al. (31), by standardizing methodology on sample processing, reference specimen, acquisition procedure of mass data, analytical methods, etc. have also demonstrated the potential of the SELDI platform (ProteinChip) for reproducible and consistent analysis of serum/plasma across multiple sites.
Secondly, the availability and reproducibility of ‘profiles’ is highly dependent on the measuring equipment. Extensively standardized equipment, well-validated protocols and their assured execution are necessary. In contrast, measuring the concentration of the proteins identified usually requires relatively low-cost, convenient and highly reproducible means such as enzyme-linked immunosorbent assay and enzymatic biochemical analysis. In fact, we have conducted such a study in a narrower set of subjects with clear-cut symptoms. In that study, we have identified certain predictive biomarker proteins for the beneficial effects of KBG in RA (manuscript in preparation). These markers may be used to improve the responder rate for KBG in RA that is closely related to ‘Oketsu’. It may not be hopeful to identify a single or small number of candidates as markers for the whole ‘Oketsu’ syndrome, because it is related to a very broad array of disorders. However, the identified accumulation of such studies targeted or limited to subjects with subsets of ‘Oketsu’ syndrome will largely contribute to delineate the whole picture of ‘Oketsu’.
In summary, the present study raises the possibility that objective criteria for diagnosing ‘Oketsu’ can be established using a combination of ProteinChip technology and bioinformatics tools. Further studies will be required to identify suitable ‘Oketsu’ biomarkers in detail. Traditional medicine and complementary and alternative medicine (CAM), which are mainly based on an empirical approach, have not been fully accepted in the framework of modern medicine. Problems preventing the utilization of traditional medicines have been a lack of sufficient scientific evidence of their efficacy, standard diagnosis and treatment protocols and communication procedures through scientifically acceptable language. The present study may open the way to elucidating these traditional medical concepts from the viewpoint of modern science and medicine and may thus lead to the integration of traditional medicine/CAM and modern medicine.
Acknowledgements
This study was supported by a grant for CLUSTER (Co-operative Link of Unique Science and Technology for Economy Revitalization) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. This study is dedicated to the memory of the late Mr Mizushima (INTEC Web and Genome Informatics Corporation).
References
- 1.Terasawa K. Evidence-based Reconstruction of Kampo Medicine: Part I-Is Kampo CAM? Evid Based Complement Alternat Med. 2004;1:11–6. doi: 10.1093/ecam/neh003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terasawa K. Evidence-based Reconstruction of Kampo Medicine: Part II-The Concept of Sho. Evid Based Complement Alternat Med. 2004;1:119–23. doi: 10.1093/ecam/neh022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terasawa K. Evidence-based Reconstruction of Kampo Medicine: Part-III-How Should Kampo be Evaluated? Evid Based Complement Alternat Med. 2004;1:219–22. doi: 10.1093/ecam/neh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Matsuda H, Shimada Y, Kita T, Itoh T, Terasawa K. Correlation between activity of rheumatoid arthritis and severity of ‘Oketsu’ syndrome. J Trad Med. 1998;15:123–6. [Google Scholar]
- 5.Caca I, Nazaroglu H, Unlu K, Cakmak SS, Ari S, Sakalar YB. Color doppler imaging of ocular hemodynamic changes in Behcet's disease. Jpn J Ophthalmol. 2004;48:101–5. doi: 10.1007/s10384-003-0024-0. [DOI] [PubMed] [Google Scholar]
- 6.Tsukamoto Y, Nakajima S, Kiyohara S, Nasu M. Effects of Kampo medicines on atopic dermatitis and complement system. J Med Pharmaceut Soc WAKAN-YAKU. 1993;10:135–40. [Google Scholar]
- 7.Hikiami H, Kohta K, Sekiya N, Shimada Y, Itoh T, Terasawa K. Erythrocyte deformability in ‘Oketsu’ syndrome and its relations to erythrocyte viscoelasticity. J Trad Med. 1996;13:156–64. [Google Scholar]
- 8.Terasawa K, Toriizuka K, Tosa H, Ueno M, Hayashi T, Shimizu M. Rheological studies on ‘Oketsu’ syndrome I. The blood viscosity and diagnostic criteria. J Med Pharmaceut Soc WAKAN-YAKU. 1986;3:98–104. [Google Scholar]
- 9.Kohta K, Hikiami H, Terasawa K, Hamazaki T, Itoh T, Tosa H. Hemorheological studies of ‘Oketsu’ syndrome - Erythrocyte aggregation in ‘Oketsu’ syndrome. J Med Pharmaceutl Soc WAKAN-YAKU. 1992;9:221–8. [Google Scholar]
- 10.Shibahara N, Sekiya N, Sakai S, Goto H, Kita T, Shimada Y, et al. Correlation between ‘Oketsu’ syndrome and autonomic nervous activity - a diachronic study on the same subjects. J Trad Med. 2002;19:81–6. [Google Scholar]
- 11.Terasawa K, Itoh T, Morimoto Y, Hiyama Y, Tosa H. The characteristics of the microcirculation of bulbar conjunctiva in ‘Oketsu’ syndrome. J Med Pharmaceut Soc WAKAN-YAKU. 1988;5:200–5. [Google Scholar]
- 12.Terasawa K, Shinoda H, Imadaya A, Tosa H, Bandoh M, Satoh N. The Presentation of Diagnostic Criteria for “Yu-xie” (Stagnated Blood) Conformation. Int J OrientMed. 1989;14:194–213. [Google Scholar]
- 13.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 14.Issaq HJ, Veenstra TD, Conrads TP, Felschow D. The SELDI-TOF MS approach to proteomics: protein profiling and biomarker identification. Biochem Biophys Res Commun. 2002;292:587–92. doi: 10.1006/bbrc.2002.6678. [DOI] [PubMed] [Google Scholar]
- 15.Yang SY, Xiao XY, Zhang WG, Zhang LJ, Zhang W, Zhou B, et al. Application of serum SELDI proteomic patterns in diagnosis of lung cancer. BMC Cancer. 2005;5:83. doi: 10.1186/1471-2407-5-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Bast RC, Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64(16):5882–90. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 17.Qu Y, Adam BL, Yasui Y, Ward MD, Cazares LH, Schellhammer PF, et al. Boosted decision tree analysis of surface-enhanced laser desorption/ionization mass spectral serum profiles discriminates prostate cancer from noncancer patients. Clin Chem. 2002;48:1835–43. [PubMed] [Google Scholar]
- 18.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–304. [PubMed] [Google Scholar]
- 19.Hikiami H, Goto H, Hattori N, Sakakibara I, Shimada Y, Terasawa K. Comparative efficacy of Keishi-bukuryo-gan and Pentoxifylline on RBC deformability in patients with ‘Oketsu’ syndrome. Phytomedicine. 2003;10:459–66. doi: 10.1078/094471103322331395. [DOI] [PubMed] [Google Scholar]
- 20.Kohta K, Hikiami H, Shimada Y, Matsuda H, Hamazaki T, Terasawa K. Effects of Keishi-bukuryo-gan on erythrocyte aggregability in patients with multiple old lacunar infarction. J Med Pharmaceut Soc WAKAN-YAKU. 1993;10:251–9. [Google Scholar]
- 21.Itoh T, Terasawa K, Kohta K, Shibahara N, Tosa H, Hiyama Y. Effects of Keishi-bukuryo-gan and Trapidil on the microcirculation in patinets with cerebro-spinal vascular disease. Journal of Medical and Pharmaceutical Society for WAKAN-YAKU. 1992;9:40–6. [Google Scholar]
- 22.Ushiroyama T, Ikeda A, Sakuma K, Ueki M. Comparing the effects of estrogen and an herbal medicine on peripheral blood flow in post-menopausal women with hot flashes: hormone replacement therapy and gui-zhi-fu-ling-wan, a Kampo medicine. Am J Chin Med. 2005;33:259–67. doi: 10.1142/S0192415X05002813. [DOI] [PubMed] [Google Scholar]
- 23.Suehiro T, Matsumata T, Shikada Y, Sugimachi K. The effect of the herbal medicines dai-kenchu-to and keishi-bukuryo-gan on bowel movement after colorectal surgery. Hepatogastroenterology. 2005;52:97–100. [PubMed] [Google Scholar]
- 24.Ishikawa H, Ohashi M, Hayakawa K, Kaneko S, Hata M. Effects of guizhi-fuling-wan on male infertility with varicocele. Am J Chin Med. 1996;24:327–31. doi: 10.1142/S0192415X96000396. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto S, Yoshino H, Shirahata Y, Shimodairo K, Okamoto R. Pharmacotherapeutic effects of kuei-chih-fu-ling-wan (keishi-bukuryo-gan) on human uterine myomas. Am J Chin Med. 1992;20:313–7. doi: 10.1142/S0192415X92000333. [DOI] [PubMed] [Google Scholar]
- 26.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 27.ProteinChip C. ProteinChip Applications Guide Volume 2 [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 29.Madeira SC, Oliveira AL. Biclustering algorithms for biological data analysis: a survey. IEEE/ACM Trans Comput Biol Bioinform. 2004;1:24–45. doi: 10.1109/TCBB.2004.2. [DOI] [PubMed] [Google Scholar]
- 30.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–85. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 31.Rai AJ, Stemmer PM, Zhang Z, Adam BL, Morgan WT, Caffrey RE, et al. Analysis of Human Proteome Organization Plasma Proteome Project (HUPO PPP) reference specimens using surface enhanced laser desorption/ionization-time of flight (SELDI-TOF) mass spectrometry: multi-institution correlation of spectra and identification of biomarkers. Proteomics. 2005;5:3467–74. doi: 10.1002/pmic.200401320. [DOI] [PubMed] [Google Scholar]