Abstract
The assembly of just a single kinetochore at the centromere of each sister chromatid is essential for accurate chromosome segregation during cell division. Surprisingly, despite their vital function, centromeres show considerable plasticity with respect to their chromosomal locations and activity. The establishment and maintenance of centromeric chromatin, and therefore the location of kinetochores, is epigenetically regulated. The histone H3 variant CENP-A is the key determinant of centromere identity and kinetochore assembly. Recent studies have identified many factors that affect CENP-A localization, but their precise roles in this process are unknown. We build on these advances and on new information about the timing of CENP-A assembly during the cell cycle to propose new models for how centromeric chromatin is established and propagated.
Introduction
The centromere, with its unique chromatin composition (Fig. 1a), is an essential locus that allows chromosomes to associate with spindle microtubules and segregate chromosomes to daughter nuclei during cell division. Because multiple spindle attachment points (kinetochores) per chromatid result in chromosome missegregation in most organisms, this complex machinery needs to be assembled at a single site on each chromosome. Failures of correct kinetochore assembly, microtubule attachment or sister-chromatid cohesion can result in chromosome loss and gain events that can have highly detrimental effects. In somatic cells (during mitotic division) such failures can drive tumour formation1, and defective chromosome segregation in meiosis has a profound effects on human fertility — a high proportion of recognized human pregnancies are aneuploid2. Clearly, a complete picture is required of how the locations of centromeres, and thus kinetochores, are specified in order to fully understand the processes that allow the accurate inheritance of genetic material. This information should also aid the design of artificial chromosomes, which are potentially useful as therapeutic vectors3.
Figure 1. Centromere structure and organization.

a | Centromeric chromatin underlies the kinetochore, which contains inner and outer plates that form microtubule-attachment sites. Pericentromeric heterochromatin flanks centromeric chromatin, and contains a high density of cohesin, which mediates sister-chromatid cohesion. b | Schematic depiction of centromeric DNA in humans and mice, Drosophila, Schizosaccharomyces pombe, Candida albicans and Saccharomyces cerevisiae. c | Model of the three-dimensional structure of mitotic centromeres. Data from mammalian and Drosophila cells suggest that centromeric chromatin is folded so that nucleosomes that are modified by dimethylation of lysine 4 of histone H3 (H3K4me2) form a discrete domain internal to CENP-A nucleosomes, which are presented on the chromosome surface, allowing kinetochore assembly and association with spindle microtubules. CDE,centromere DNA element; H2AZ, histone H2AZ; imr, innermost repeats.
Normally, centromeres are faithfully propagated at the same site from one cell and organismal generation to the next. The site chosen for centromere and kinetochore formation in some organisms, such as the budding yeast, Saccharomyces cerevisiae, seems to be specified by recruitment of specific proteins to a short DNA sequence of less than 200 bp4 (Fig. 1b). However, in most eukaryotes the kinetochore forms on only a subset of the long arrays of repetitive DNA associated with centromeres5, 6, 7, 8 (Fig. 1b). Thus, these ‘regional’ centromeres do not seem to be specified by the primary DNA sequence; instead, epigenetic regulation seems to have a dominant role.
The evidence for epigenetic regulation of most centromeres is compelling (reviewed in Refs 9,10,11,12,13). The presence of endogenous centromeric repeats does not guarantee kinetochore formation. For example, human and fly chromosomes that contain two separated blocks of centromeric DNA (structural dicentric chromosomes) segregate normally, because only one site is able to recruit centromere proteins and form a kinetochore, whereas the other is epigenetically inactivated14, 15, 16, 17, 18. Most strikingly, kinetochores can form in flies and humans at sites that lack any sequences that are homologous to those at normal centromeres; once established, these ‘neocentromeres’ propagate centromere function faithfully19, 20, 21, 22, 23. In the fission yeast, Schizosaccharomyces pombe, plasmids with minimal centromeric DNA establish functional centromeres stochastically, but once the functional state is attained it is propagated faithfully24. Remarkably, the engineered deletion of an endogenous fission yeast centromere allows the assembly and maintenance of a centromere at other novel chromosomal locations25. It is paradoxical that centromere function and many associated proteins are essential and conserved, yet centromere specification is not hard-wired to the DNA sequence and displays dramatic plasticity. One explanation arises from considering the speed with which satellite repeats change during evolution; centromere plasticity might be necessary for karyotype evolution and might ultimately contribute to speciation9, 10, 26, 27, 28.
The key issues that centromere biologists are currently grappling with are determining the molecular mechanisms that specify and faithfully propagate centromere identity at a single site, how these events are coordinated with the cell cycle, and how the need for centromere stability is balanced with plasticity. We still lack a comprehensive mechanistic understanding of how centromere identity and propagation are accomplished and epigenetically regulated. However, there have been significant and exciting advances in our understanding of the unique composition and organization of centromeric chromatin, and in the identification of molecules that affect its formation and function. Here, we summarize this information, mainly from analyses in budding yeast, fission yeast, fruitflies and mammals, and propose models based on these findings. We will focus on a key determinant of centromere identity and kinetochore formation, the centromere-specific histone H3 variant, CENP-A (centromere protein A; also known as CenH3), and our current understanding of the molecules and mechanisms involved in CENP-A assembly into centromeric chromatin.
Centromeric chromatin is unusual
What determines the site of kinetochore assembly if DNA sequence is not the primary trigger? Epigenetic regulation of centromere formation suggests that chromatin composition or organization has a key role. In general, the packaging of chromosomal DNA in the nucleus in an organized manner allows for multiple levels of regulation of functions such as transcription and replication. Nucleosomes are inherently inhibitory to the transcription and replication machinery, and thus they need to be ‘opened’ to allow the passage of polymerases. In addition, a large number of post-translational modifications, including acetylation, methylation, ubquitylation, sumoylation and phosphorylation, act to attract or exclude factors that are correlated with different functional chromatin states29.
In addition to the canonical histones (H2A, H2B, H3 and H4), histone variants such as H2AZ, H2AX or H2AV (depending on the organism) and H3.3 are associated with distinct regions of the genome. In general, H2A variants in yeast are found at promoters and might aid nucleosome disassembly during induction30, 31, and H2AZ is also associated with ‘boundary’ elements that demarcate chromosomal domains32. H2AZ has also been implicated in centromere structure in mice33. The H3.3 variant replaces H3 following transcriptional activation, and its deposition is coincident with elongation34, 35. The inclusion of H2AZ in nucleosomes containing H3.3 renders them highly unstable, but promotes the stability of nucleosomes containing H3 (Ref. 36). Unlike canonical histones, the deposition of which is coupled to DNA replication and requires the chromatin assembly factor (CAF) complex, histone variants are assembled into nucleosomes by a replication-independent exchange mechanism that relies on other protein complexes, such as HIRA for H3.3 and SWR1 for H2AZ37, 38. Replacement of H3 with H3.3 independently of replication allows histone modifications that repress transcription to be erased, and thereby aids transcriptional activation and propagation of the active state.
In all eukaryotes, kinetochores are assembled on distinctive chromatin that contains CENP-A14, 39, 40, 41, 42, 43, 44, 45, 46 (Fig. 1b). CENP-A is a constitutive chromatin component, underlying the kinetochore throughout the cell cycle. It is specific to centromeres and forms a higher-order structure that lies at the heart of the mitotic kinetochore (see below). CENP-A proteins are clearly required for kinetochore assembly in all eukaryotes studied: CENP-A depletion results in mislocalization of most kinetochore proteins, whereas depletion of most kinetochore proteins has no effect on CENP-A localization41, 44. In addition, overexpression of CENP-A results in its mislocalization to normally non-centromeric regions and the formation of ectopic kinetochores47. These observations suggest that CENP-A is both a structural and functional foundation for the kinetochore, and that it lies at or close to the apex of the pathway that is responsible for kinetochore formation. However, as will be discussed below, recent studies have identified proteins that are co-dependent with CENP-A for centromere localization, suggesting that CENP-A is not alone at the top of the hierarchy.
In S. cerevisiae, complete centromere function is specified by only 125 bp of DNA, which is composed of three centromere DNA elements (CDEs): CDEI, CDEII and CDEIII (reviewed in Ref. 4). The 15 bp of CDEIII is most important as it attracts a complex containing sequence-specific DNA-binding proteins (Ndc10, Cep3, Ctf13 and Skp1). This complex dictates the assembly of only a single nucleosome, containing the CENP-A homologue CENP-ACse4, over the middle AT-rich CDEII element39, 48(Fig. 1b). By contrast, human, mouse, rice and Drosophila centromeres contain blocks of CENP-A nucleosomes that are interspersed with blocks of histone H3-containing nucleosomes7, 49 (Fig. 1b). The length of DNA encapsulated in this unique structure is approximately 500–1500 kb in humans and 200–500 kb in flies. The entire region seems to be folded in mitotic chromosomes so that all CENP-A is on the surface of the chromosome, with H3 residing underneath (Fig. 1c).
Canonical histones are highly conserved and contain sites for a plethora of post-translational modifications, particularly in their N-terminal tails29. These modifications are correlated with different functional states, such as transcriptional activity or silencing, and are involved in chromatin assembly and disassembly. Surprisingly, although regional centromeres are universally embedded in pericentric heterochromatin, the interspersed H3 domains within centromeres themselves are modified in a pattern that is distinct from both euchromatin and heterochromatin50 (Fig. 1b,c). These domains contain the H3K4me2 modification (dimethylation of lysine 4 of histone H3), which is normally associated with open (active) chromatin, but they do not contain the acetylated residues that also usually mark open chromatin. They also lack the H3K9me modification (methylation of lysine 9 of histone H3) that is associated with ‘silent’ chromatin and with the flanking pericentromeric heterochromatin. In addition, H2AZ is present in the interspersed H3 domains of mouse centromeres33. Even in S. pombe, centromeres seem to adhere to this arrangement. The central kinetochore domain, composed of inner repeats and a central core, is packaged in approximately 10 kb of chromatin that is composed mainly of the CENP-A homologue CENP-ACnp1 (Ref. 51) and that seems to contain some histone H3K4me2 chromatin52 (Fig. 1b).
The function of the interspersed H3 domains, with their distinct histone modifications, is not known. They might contribute to CENP-A recruitment, or participate in assembling the cylindrical three-dimensional structure of centromeric chromatin in mitotic chromosomes, in which CENP-A-containing nucleosomes are located on the poleward side of each chromatid and the interspersed H3 blocks reside in the interface between sister centromeres7 (Fig. 1c). This arrangement could ensure that CENP-A is exposed on opposite sides of cohesed sister centromeres, which would promote the formation of sister kinetochores that interact with microtubules from opposite poles, and thus promote bi-orientation. A similar model has recently been proposed for the single S. cerevisiae CENP-A nucleosome, which lies at the apex of a C-loop53 (Fig. 1c). S. pombe might also contain a similar substructure to coordinate multiple microtubule attachments54. The H3K4me2 modification on the interspersed H3 domains might also be indicative of a transcription-coupled process that acts to ‘proofread’ CENP-A chromatin domains, evicting H3 and replacing it with CENP-A (see below).
CENP-A structure and nucleosome organization
Like all histones, H3 and CENP-A contain globular histone fold domains (HFDs) that are composed of three alpha helices separated by loops45, 55. The helices of H3 intertwine with those of H4 to form a tetrameric [H3–H4]2 structure — the kernel of the ‘canonical’ nucleosome. Given the high structural conservation observed for canonical histones, it is striking that the N-terminal tails of CENP-A from different organisms are highly divergent both in length and in amino-acid sequence9. For example, the tails are 120 and 130 amino acids in budding yeast and Drosophila CENP-A, respectively, but are only 45 amino acids in humans and 20 amino acids in fission yeast. This suggests that the tails evolve rapidly along with organism-specific CENP-A-interacting proteins, and that there might be distinct but overlapping mechanisms and proteins used to chaperone and incorporate CENP-A in different organisms27. Surprisingly little is known about post-translational modifications of CENP-A and whether they are functionally important.
In fission yeast, micrococcal nuclease (MNase) digestion analysis showed that the central kinetochore domain at all three centromeres adopts an unusual chromatin structure56, 57. Centromeres display a smeared nucleosomal pattern, in contrast to the canonical 150 bp ladder seen in bulk chromatin, suggesting less regular spacing with respect to DNA sequences. Likewise, the smaller regional centromeres of the yeast Candida albicans also display an atypical nucleosomal pattern58. However, in vitro assembled CENP-A chromatin utilizing human CENP-A in place of H3 forms octamers with H4, H2A and H2B and does not generate an unusual MNase pattern59. The causes of the unusual nucleosome pattern in some organisms is currently unknown, but it is presumably a property conferred by the replacement of H3 with CENP-A; the DNA ends might be more accessible, and in some organisms might lead to less precise termini.
Recent analyses show that CENP-A is structurally distinct from H3 in that it forms a more rigid and compact interface with H4 in both tetramers and nucleosomes55, 60. The region responsible for this difference is in the first loop (L1) and second alpha helix (2), which corresponds to CENP-A residues that are involved in centromere recruitment (see below) (Fig. 2a). Analyses of in vitro assembled CENP-A nucleosomes suggest that they are more prone to disassembly than normal H3 nucleosomes, which might aid the removal of CENP-A nucleosomes from non-centromeric loci61. Thus, physical properties of [CENP-A–H4]2 and canonical [H3–H4]2 tetramers, and presumably the resulting nucleosomes, are distinct and might contribute to functional differences.
Figure 2. CENP-A structure and nucleosome composition.
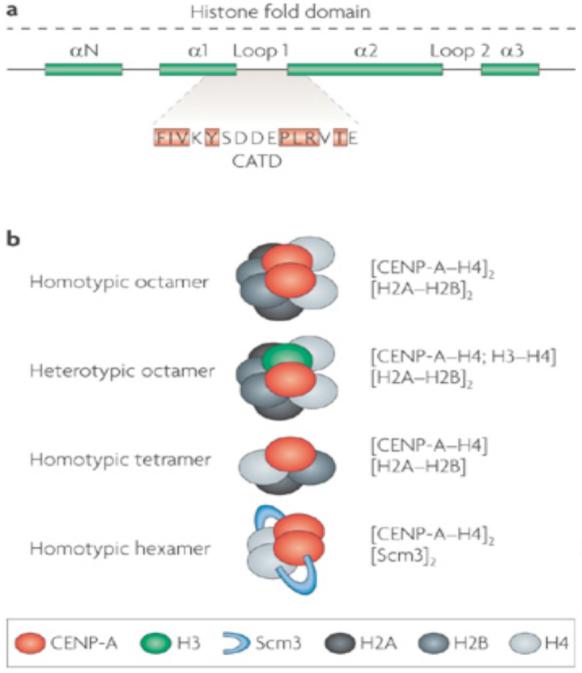
a | The histone fold domain of histone H3 proteins is composed of four alpha helical domains (N and 1–3). Loop 1 separates 1 and 2. The CENP-A targeting domain (CATD) contains key differences from H3, and is sufficient for localization to centromeres when substituted into canonical H3 (the amino acids highlighted in orange are required in Drosophila). The CATD forms a rigid conformation in combination with H4, and this distinguishes CENP-A–H4 from H3–H4 heteromers. b | In non-centromeric regions, canonical histone H3 assembles into octameric nucleosomes composed of two H2A, H2B, H3, and H4 histone subunits. In centromeric chromatin, CENP-A can assemble into homotypic octamers, in which both H3 subunits are replaced by CENP-A, or into heterotypic octamers, which contain one canonical H3 and one CENP-A subunit. In Drosophila, CENP-A has been reported to form half-nucleosomes, that is, homotypic tetramers containing one subunit each of H2A, H2B, H4 and CENP-ACID (the Drosophila homologue of CENP-A). In Saccharomyces cerevisiae, Scm3 has been reported to replace the H2A–H2B dimers, forming an unusual hexamer containing two Scm3, CENP-A and H4 subunits. Additional forms of CENP-A nucleosomes can be envisaged by the replacement of one or both H2A subunits with H2A variants.
Recent studies suggest that the composition of CENP-A nucleosomes is also unusual. The single CENP-ACse4 nucleosome in budding yeast seems to lack H2A–H2B, and instead contains a non-histone protein Scm3, that might form a hexamer with the [CENP-ACse4–H4]2 tetramers62 (Fig. 2b). In addition, a recent report indicates that the Drosophila CENP-A homologue, CENP-ACID, might exist as a CENP-ACID–H4–H2A–H2B tetramer or half-nucleosome in interphase Drosophila S2 cells63 (Fig. 2b). However, [H3–H4]2 tetramers form the stable core of canonical octameric nucleosomes, and a heterotypic tetramer is highly unusual. It is possible that CENP-A nucleosomes are octamers in vivo, and that the observed recovery of tetramers from Drosophila cells results from the propensity of CENP-A octamers to disassemble61, the absence from CENP-A of residues that can form crosslinks, or from active assembly–disassembly processes that take place during interphase64.
Despite these reports of unusual CENP-A nucleosomes, the affinity purification of CENP-A nucleosomes from human and fly cells suggest that they mainly exist as octamers that contain [CENP-A–H4–H2A–H2B]2 (Refs 7, 65), and that CENP-A octamers can form in vitro59, although some hybrid or intermediate CENP-A–H3 nucleosomes might also exist (Fig. 2b). In addition, H2B is detected in the central domain of fission yeast centromeres, and mutations in H2B reduce CENP-A levels in the central domain66. Although the effect of H2B mutations could be indirect, these analyses suggest that H2B might be an integral component of fission yeast CENP-A nucleosomes; however, the possibility that some incomplete or Scm3-containing CENP-A nucleosomes are also present has not been excluded. It is also possible that the composition and structure of CENP-A nucleosomes varies during the cell cycle, for example, during S phase, interphase and mitosis. Clearly, we need more detailed information about the components and properties of CENP-A nucleosomes, and whether they change during the cell cycle.
Cell-cycle timing of CENP-A deposition
Parental canonical H3 nucleosomes are distributed randomly to daughter strands67. Typically, assembly factors such as ASF1 and CAF collaborate to mediate deposition of new H3-containing nucleosomes along DNA as it is synthesized during S phase (reviewed in Refs 68, 69). Nascent nucleosomes are thought to be deposited on both the leading and lagging strands, enabling the propagation of parental chromatin structures to both daughter cells70.
Centromeric DNA replicates in mid to late S phase in Drosophila and human cells71, 72, and in very early S phase in S. pombe73. Based on the behaviour of canonical H3 nucleosomes during replication, it was assumed that half of the pre-existing CENP-A nucleosomes are segregated to each chromatid. However, in S. cerevisiae all pre-existing CENP-A is evicted and replaced by new CENP-A during S phase, perhaps leading to temporary kinetochore disruption and orientation-switching74. Recent studies in human cells have shown that CENP-A nucleosomes are extremely stable, and segregate to daughter chromatids during each S phase75, providing a mechanism for continuity of CENP-A and centromere inheritance (Fig. 3a). However, as the amount of CENP-A at each centromere is diluted twofold during each S phase, at some point during the cell cycle the CENP-A ‘gaps’ must be filled in or replenished with nucleosomes containing newly synthesized protein.
Figure 3. Cell-cycle regulation of CENP-A assembly.
a | Pre-existing nucleosomes are distributed equally to sister-chromatids at the replication fork. The chromatin assembly factors CAF and ASF1 coordinate the deposition of new histone H3 and H4, and assemble new nucleosomes behind the replication fork. CENP-A density at centromeres drops by one half following replication, most probably owing to CENP-A-nucleosome segregation at the fork. The resulting nucleosome ‘gaps’ could be ‘filled’ with new H3 nucleosomes during S phase, or might be maintained as gaps until new CENP-A nucleosomes are assembled. CENP-A nucleosomes could also be split during replication, forming half-nucleosomes (not shown). b | Pulse–chase experiments in human cells show that new CENP-A is assembled at centromeres starting in telophase and continuing through G1. Thus, the amount of CENP-A at each centromere is highest at the end of G1, and is halved during S phase owing to the distribution of CENP-A to both sister chromatids. Cellular CENP-A protein levels are highest in G2, but new CENP-A assembly does not initiate until telophase. The amount of CENP-A in centromeres does not return to maximal levels until the end of G1, suggesting that kinetochore assembly occurs at centromeres containing half the maximal amount of CENP-A. MCM, minichromosome maintenance helicase; PCNA, proliferating cell nuclear antigen; DNA Pol, DNA polymerase.
CENP-A nucleosome assembly is replication-independent71, but until recently it was unclear whether new CENP-A nucleosomes are assembled during the S, G2, M or G1 phase of the cell cycle. In S. pombe, CENP-A levels reach a maximum before the canonical histones in early S phase, and newly synthesised CENP-A can be deposited both during S phase and in G2 (Ref. 76) (note that G1 is exceedingly short in fission yeast, with cells proceeding almost directly from mitosis into S phase). The simplest explanation is that CENP-A deposition in S phase is replication-coincident, rather than replication-coupled, so that one mechanism operates to allow incorporation of new CENP-A during both S and G2 (Ref. 51). However, the GATA-like Ams2 protein is required to boost histone levels in S phase but is also centromere-associated and affects CENP-A assembly at centromeres77, 78. The interplay between Ams2, canonical histone levels and CENP-A assembly at centromeres is complex; there might be both normal S phase and G2 ‘salvage’ pathways76. Elegant fluorescent CENP-A quench–pulse–chase approaches in human cells have been used to analyse the timing of centromere localization of newly synthesized CENP-A. Surprisingly, CENP-A assembly was shown to occur in late mitosis (telophase) and throughout G1 (Ref. 75) (Fig. 3b). In Drosophila, CENP-ACID assembly occurs during anaphase of mitosis in the syncytial embryonic nuclear divisions79, which cycle between S and M phases without G1 or G2 phases.
The assembly of nucleosomes during mitosis and the G1 phase is an unprecedented finding, and has a significant impact on our understanding of the mechanics of centromere propagation. First, these results suggest that the amount of CENP-A at centromeres is halved during most of the cell cycle, from the time of replication in S phase to the end of mitosis (Fig. 3b). This raises questions about the composition of centromeric chromatin during this period, which includes the time of kinetochore formation and function, and what happens to CENP-A chromatin during replication. Do the CENP-A ‘gaps’ generated during DNA replication and nucleosome segregation in S phase contain H3 nucleosomes, which are replaced or exchanged at the end of mitosis or in G1, or are they nucleosome-free, as observed at transcription start sites in many organisms (Fig. 3a)? Are CENP-A nucleosomes ‘split’ during replication, forming half-nucleosomes that are reconstituted as octamers only during late mitosis or in G1? In addition, the timing of CENP-A assembly coincides with the completion of the primary function of the centromere, namely kinetochore formation, attachment to the spindle and segregation of chromatids. This raises the intriguing possibility that accomplishing centromere function serves as a signal for initiating centromere assembly12, 27, 80. Although mass disruption of microtubules by nocodazole treatment does not disrupt assembly of newly synthesized CENP-A75, it is possible that the link is more complex. For example, the signal for CENP-A assembly might require attachment and bi-orientation in early mitosis, plus detachment at the end of mitosis.
Regulators of CENP-A deposition
The incorporation of histones into chromatin involves numerous events, including histone gene transcription and translation, protein modifications, import into the nucleus, and nucleosome assembly, remodelling or exchange (Fig. 4a). Each of these steps provides an opportunity for regulation of CENP-A localization to centromeres by contributing factors, including the properties of CENP-A and the activity of CENP-A-interacting components (Box 1). Two key questions that need to be addressed to understand the molecular mechanisms responsible for epigenetic propagation of centromere identity are: what properties of CENP-A are necessary and sufficient to direct deposition of new CENP-A specifically to centromeres following replication, and what are the trans-acting factors responsible for centromere-specific assembly of CENP-A? acids of loop 1 and the 2 helix from human CENP-A in place of the equivalent region of the H3 HFD localized to centromeres55(Fig. 2a). Surprisingly, centromere localization of fusion proteins containing this CENP-A targeting domain (CATD) occurred even after CENP-A depletion by small-interfering RNA (siRNA). Although it is possible that low levels of CENP-A remained at centromeres after siRNA depletion, these results suggest that pre-existing CENP-A need not be present at centromeres for recruitment of new CENP-A. Similar studies of chimeric histones showed that the Drosophila CENP-ACID-targeting domain is even smaller, consisting of the C-terminus of 1, all of loop 1 and the N-terminus of 2 (15 amino acids)83. Single amino-acid substitutions within this region disrupted centromere localization (Fig. 2a). The CATD lies in the more rigid, compact interface with H4 that resides in the nucleosome core55 (Fig. 2a). However, it is unlikely that interactions between the CATD and H4 are sufficient for targeting, as H4 is deposited throughout the genome. Thus, other proteins that regulate centromere-specific localization probably recognize the CATD or the CATD:H4 interface.
Figure 3. Cell-cycle regulation of CENP-A assembly.
a | Nucleosome assembly pathways. Human and Saccharomyces cerevisiae proteins that are thought to function in these pathways are shown. Newly synthesized histone H3 and H4 are acetylated by a histone acetylase (HAT) and form tetramers in the cytoplasm. Chaperones, such as the human RbAp46/48 and NASP, facilitate H3–H4 import into the nucleus in combination with an importin protein, such as the S. cerevisiae Kap123. Replication-coupled assembly into chromatin is mediated in humans by the chromatin assembly factors ASF1 and CAF (which includes RbAp46/48), and acetyl group (Ac) modifications are removed by histone deacetylases. The protein complex HIRA mediates transcription-coupled assembly of H3–H4 into chromatin. Nucleoplasmin (in humans) and Nap1 (in S. cerevisiae) act as chaperones for H2A–H2B dimers and, in S. cerevisiae, Kap114 mediates their nuclear import. NASP and RbAp46/48 also associate with CENP-A, and might participate in its nuclear import. Specific post-translational modifications might also play a part in CENP-A nuclear import and deposition. The factors that mediate CENP-A assembly into chromatin are unknown. However, the facilitates chromatin transcription (FACT) remodelling complex associates with CENP-A, and RbAp46/48 can mediate CENP-A nucleosome assembly in vitro and also associates with the MIS18 complex, which is recruited to centromeres in telophase and through G1. Defects in histone gene transcription, translation, modification or import could affect the ability to assemble intact CENP-A chromatin and result in loss of CENP-A from centromeres (Box 1). b | Factors involved in CENP-A localization. Pericentromeric heterochromatin facilitates the establishment CENP-A on naked DNA templates in Schizosaccharomyces pombe, and the alpha satellite-binding protein CENP-B facilitates de novo centromere formation in human cells. Normally, propagation of centromere identity after DNA replication requires assembly of newly synthesized CENP-A at pre-existing locations. Factors that are physically associated with CENP-A include the nucleosome associated complex (NAC) and the MIS18 complex, RbAp46/48, NASP and FACT, among others. Some NAC components are reciprocally required for the centromere localization of CENP-A, demonstrating interdependency at the top of the propagation pathway. MIS18 components are known to function upstream of CENP-A, as their localization is not disrupted in CENP-A mutants. Whether these and/or other factors are specifically required for CENP-A assembly into chromatin is currently unknown. Other kinetochore components, such as MIS12, have been shown to localize independently of CENP-A. Components of the CENP-A distal (CAD) complex are not required for CENP-A localization, but CENP-A is required to recruit CAD and kinetochore components that assemble the inner and outer plates and microtubule attachment sites.
Box 1. Processes affecting CENP-A localization: terminology matters.
Terminology affects the way we think about the factors that regulate biological processes. Many components have been identified in genetic screens or biochemical purifications, and depletion of these components results in loss of CENP-A from centromeres. These are often referred to as CENP-A ‘loading’ or ‘assembly’ factors, which implies a specific role in centromeric nucleosome deposition. However, lack of CENP-A at centromeres could result from disrupting events that occur before the deposition of CENP-A into sub-kinetochore chromatin, as well as defects in subsequent maintenance.
We suggest that ‘CENP-A localization factor’ be used as a general term for molecules that, when depleted, cause a reduction in CENP-A at centromeres, and that ‘CENP-A deposition factor’ should be used only for components shown to be required for deposition of newly synthesized CENP-A protein into sub-kinetochore chromatin. Bona fide charging factors would include specific chaperones that guide CENP-A to centromeres, and assembly or exchange factors that promote its incorporation into nucleosomes (see main text). CENP-A localization and deposition factors might interact physically with CENP-A itself. This is not a strict criterion as they could be required for obligatory events that must occur in parallel, such as H3 nucleosome disassembly and eviction.
Decreased CENP-A at centromeres could also occur indirectly as a result of defects in the processes listed below. Such components should be classified as CENP-A influencing factors. The regulation of these events is important; however, we suggest that the terminology used to classify factors reflect their specific molecular functions. This will facilitate a more informed understanding of the many events required for the formation and maintenance of sub-kinetochore chromatin.
CENP-A expression
The levels of mature CENP-A mRNA are cell-cycle regulated, and defective transcription would result in insufficient CENP-A being available for incorporation. CENP-A mRNA might be subject to post-transcriptional regulation to ensure that it is not overproduced, and aberrant RNA processing and turnover might affect CENP-A protein levels.
Protein and mRNA turnover
An inappropriate ‘glut’ of histones or CENP-A is detrimental to cells. Association of histones with chaperones inhibits histone gene transcription and triggers degradation of histone mRNA. Abnormal protein turnover could indirectly affect CENP-A localization at centromeres.
Histone gene expression
The levels of other histone and non-histone components can affect centromeric CENP-A levels at centromeres, For example, H4 is necessary for formation of CENP-A–H4 heteromers before incorporation. In addition, excess H3 can compete with CENP-A for the available H4 pool.
Histone modifications and nuclear import
Cytoplasmic post-translational histone modifications affect import into the nucleus and their assembly into chromatin. Little is known about post-translational modification of CENP-A, but perturbing modification components (for example, histone acetylation and deacetylation) could affect the import of CENP-A nucleosome components and interactions with chaperones and assembly factors. Similarly, defective import mechanisms could result in less CENP-A being available for deposition.
H3 nucleosome disassembly
The assembly of CENP-A into nucleosomes might involve the disassembly and replacement of H3 nucleosomes. Loss of the factors that are required to disassemble H3 nucleosomes, such as remodelling complexes or post-translational modifications (for example, acetylation of histone H3 on lysine 56, H3K56Ac) might indirectly cause loss of CENP-A. Although identifying such factors would contribute to understanding the mechanism of CENP-A deposition, they might not provide specificity.
Many proteins have been identified by biochemical purifications as being physically associated with CENP-A, and factors required for centromere localization of CENP-A have been identified by genetic screens and subsequent analyses. An effect on CENP-A localization has been used to suggest that a factor has a direct role in assembly or loading of CENP-A nucleosomes. However, it is important to define the precise roles of the factors required for CENP-A localization with respect to the multiple steps and molecular processes involved in histone expression and transport, and in nucleosome assembly and maintenance (Box. 1; Fig. 4a).
The S. cerevisiae Ndc10 protein is a component of the CBF3 DNA complex that binds the 15 bp CDEIII centromere element and that is absolutely required to specify centromere function. The Scm3 protein has been shown to form a hexameric complex with a [CENP-ACse4–H4]2 tetramer62 (see Fig. 2b and above), and interacts with the AT-rich CDEII element84, 85. Scm3, Ndc10 and CENP-ACse4 are interdependent for centromere localization. It is unclear whether Scm3 is required for incorporation of newly synthesized CENP-ACse4 into chromatin and/or maintenance of CENP-ACse4 at centromeres. It is also unknown whether [CENP-ACse4–H4]2 tetramers are initially associated with H2A and H2B tetramers, which are then replaced by Scm3. Nevertheless, ‘point’ centromere assembly seems to require sequence-specific binding through CBF3 and Ndc10, through association of Scm3 with CENP-ACse4–H4 and CDEII, and through Ndc10–Scm3 binding.
Progress has also been made in identifying trans-acting proteins that are required for CENP-A localization in eukaryotes which contain more complex and epigenetically determined centromeres. The S. pombe proteins Mis6 and Ams2 are required for CENP-ACnp1 localization77, 86, but it is not clear whether they directly mediate CENP-ACnp1 deposition at centromeres or act indirectly to maintain CENP-ACnp1 localization (Boxes 1, 2). Additional proteins that affect CENP-ACnp1 localization have been isolated (Mis15–18)87, two of which have a conserved role in localizing human CENP-A. Components of the human MIS18 complex, consisting of MIS18A, MIS18B and MIS18-binding protein (MIS18BP; KNL-2 in worms), are particularly interesting factors required for centromere formation. The human MIS18 complex collaborates with the general histone H3–H4 chaperone RbAp46/48, which is also a component of the CAF complex (see below) and only localizes to centromeres in the period between late anaphase to telophase and through G1 in humans89. This is intriguing in light of recent evidence that human CENP-A assembly occurs between telophase and the following G1 phase (Refs 75, 79). M18BP1 and KNL-2 depletion experiments suggest that the MIS18 complex might ‘license’ centromere formation, analogous to the way in which licensing factors permit replication-origin activation89. Similarly Mis16, a fission yeast RbAp46/48 homologue, and Mis18 dissociate from centromeres from early mitosis until late anaphase/telophase. The association of Mis18 might permit CENP-A assembly in all subsequent stages, but this might only normally occur when there is sufficient CENP-A and associated H4 in early S phase, which in fission yeast immediately follows telophase and is coincident with centromere replication51. In fission yeast the tetratricopeptide repeat (TPR)-domain protein Sim3, which is homologous to the histone-binding proteins NASP (human) and N1/N2 (Xenopus), has been shown to be distributed throughout the nucleus but also associates with CENP-ACnp1 and is required for its localization at centromeres. In Xenopus, N1/N2 is required to store free H3–H4 as tetramers in the cytoplasm; therefore, in fission yeast, Sim3 might act to escort CENP-A to centromeres and prevent its misincorporation at other sites90.
Box 2. Factors that can affect CENP-A localization.
The assembly of chromatin is a complex process. The assembly of CENP-A into chromatin must require chaperones that deliver newly synthesized histones to the nucleus, and chromatin assembly or exchange complexes (Fig. 4a). There might be redundant contributions made by different factors and pathways (Box 1). Furthermore, different disassembly, assembly and exchange factors might act at distinct steps of the process leading to CENP-A incorporation, and thus these steps need to be distinguished.
Chaperones are required to escort CENP-A to centromeres. These may or may not be concentrated at centromeres. Some chaperones might not only deliver histones but could also participate in the assembly process. For example, in fission yeast Sim3 acts as a CENP-A chaperone.
Assembly factors can be divided into those that disassemble or transfer nucleosomes and those that take part in the de novo assembly of CENP-A nucleosomes. As there is a possible overlap between CENP-A and H3 assembly factors, these may or may not be specifically localized at centromeres. Such factors (including NAP1 and FACT) might be required for H2A and H2B assembly into or disassembly from H3–CENP-A nucleosomes (Fig. 4a).
Specific CENP-A amino acids (those in the histone fold domain) are distinct from the amino acids in histone H3 and are sufficient to target CENP-A to the centromeres when placed in canonical H3. It is not known what factors recognize CENP-A but not H3.
Histone modifiers can also be considered as factors that are required for localization. Because many events involving histones are stimulated or inhibited by post-translational modifications, it is likely that the enzymes responsible for a particular histone modification will exert an influence on CENP-A chromatin assembly.
Propagation factors might be involved after CENP-A nucleosomes are established at a locus, as CENP-A nucleosomes can self-propagate through multiple cell divisions rendering establishment factors non-essential.
Establishment factors are required to trigger the assembly of CENP-A nucleosomes on naive centromeric DNA (for example, neocentromeres). These might be associated with centromeric DNA or with pericentromeric chromatin.
Recent biochemical purification studies in human and chicken cells have identified two major centromere-associated complexes65, 91. CENP-B, -C, -H, -M, -N, -T and -U were found to be associated with one or a few CENP-A nucleosomes, forming the nucleosome-associated complex (NAC)65 (Fig. 4b). Other proteins isolated in these studies did not associate directly with CENP-A, and instead formed a more distal complex called CAD (CENP-A distal). NAC proteins are needed to recruit CAD components, which in turn are required for assembling subsets of outer kinetochore proteins during mitosis. Some NAC components, including CENP-I, -K and -M (the homologue of Mis6) were shown to be required for localization of a CENP-A–GFP fusion protein, which was under the control of a constitutive promoter, suggesting a role in the incorporation of newly synthesized CENP-A91. Surprisingly, CENP-A is also required to localize NAC to centromeres, indicating reciprocal dependency in the initial stages of centromeric chromatin assembly (Fig. 4b).
Factors required for centromere propagation were recently identified in Drosophila using an unbiased, genome-wide RNAi screen for defects in CENP-ACID localization92. A novel constitutive centromere protein (CAL1) and CENP-C were shown to be essential for the assembly of newly synthesized CENP-ACID. CENP-ACID, CAL1 and CENP-C co-immunoprecipitate and are mutually dependent for centromere localization and function. As observed for MIS18 and MIS18BP in humans, levels of CAL1 are reduced on metaphase centromeres and increase coincident with CENP-A loading in late anaphase to telophase. In addition, cyclin A and the anaphase-promoting complex (APC) inhibitor RCA1 (Emi1 in vertebrates) were identified as regulators of centromere propagation and CENP-ACID localization. Cyclin A is centromere-localized, and cyclin A and RCA1 couple centromere assembly to the cell cycle through regulation of the Fzr (also known as Cdh1) subunit of the APC. These findings identify essential components of the epigenetic machinery that ensures proper specification and propagation of the centromere, as well as components involved in coordinating centromere inheritance with cell division.
A major gap in our understanding of centromere propagation is the identity of molecules that directly mediate CENP-A deposition into chromatin. Newly synthesized canonical H3 nucleosomes are assembled behind the replication fork by ATP-dependent activities that are mediated by complexes, such as CAF, and by ASF1 as a subunit of the replication coupling assembly factor (RCAF)68, 69. Although deposition of CENP-A can be replication-independent71, 75, 76, 90, 93, it is currently unclear whether CENP-A nucleosomes are assembled de novo on naked DNA, or by exchange–replacement mechanisms (Fig. 3). In Drosophila, the CAFp55 protein (homologues include RbAp46/RbAp48 in mammals and Mis16 in S. pombe) copurified with tandem affinity purification (TAP)-tagged CENP-ACID, and was demonstrated to assemble CENP-ACID nucleosomes on plasmids in vitro94, but their role in CENP-ACID assembly in vivo has not been determined. By contrast, neither CAF nor HIRA were associated with human CENP-A-containing chromatin or with the NAC complex65. This could imply that assembly of centromeric chromatin does not require these histone chaperones, at least in human cells. Alternatively, histone H3 interspersed with CENP-A in centromeric regions might be deposited by CAF or HIRA, and might be required for subsequent CENP-A assembly via replacement–exchange or remodelling mechanisms.
De novo establishment of CENP-A chromatin
Although centromeres are normally propagated faithfully at the same site, new centromeres can be established de novo in the absence of pre-existing CENP-A or other guiding marks (Fig. 4b). Examples include the neocentromere formation observed in flies and humans, and the movement of centromere locations that occurs during evolution20, 21. Ectopic kinetochores can also be established in Drosophila after CENP-ACID overexpression and mislocalization to normally non-centromeric, euchromatic locations47. It is possible that establishment pathways act to renew the site of CENP-A chromatin and kinetochore assembly when a cell suffers major catastrophes (that is, stress that disrupts centromere–kinetochore function). The processes that establish new CENP-A chromatin also might provide backup to and be redundant with propagation mechanisms, to ensure that a centromere is assembled at a particular chromosomal location.
De novo establishment of centromeres has been demonstrated experimentally by introducing naked DNA templates containing sequences associated with centromere function in vivo into cells and assessing their ability to assemble a kinetochore. Active kinetochores can clearly be established on naive DNA transferred into budding yeast, fission yeast, and plant and mammalian cells95, 96, 97, 98, 99. However, in mammalian cells and fission yeast this process can be inefficient, perhaps owing to size requirements that are dictated by the need for sufficient sister-chromatid cohesion to resist pulling forces, and/or a lack of particular contextual cues provided by surrounding chromatin100. In addition, it seems that the establishment of centromeres on artificially introduced centromeric DNA is not possible in some organisms, such as the pathogenic yeast C. albicans58. The inefficiency or lack of de novo centromere assembly might be a consequence of the normal processes that restrict centromere propagation to one site per chromosome, or differences in organisms in their ability to assemble appropriate chromatin templates on naked DNA.
Recent analyses in mammalian cells indicate that the CENP-B protein, which binds a specific 17-bp element within a subset of alpha-satellite monomers, promotes the assembly of CENP-A into chromatin and, therefore, kinetochore formation100. In addition, targeting of transcriptional repressing or activating complexes can inactivate centromeres101. In fission yeast, RNA interference (RNAi)-directed heterochromatin is important, as it allows CENP-ACnp1 chromatin to be established on adjacent central core DNA, but it is not required for its maintenance102 (Fig. 4b). The RNAi components Dicer and argonaute, and the heterochromatin protein 1 (HP1) counterpart Swi6, are also required for establishment of CENP-ACnp1 in this system. This suggests that in fission yeast the centromeric heterochromatin flanking the central domain provides a platform that mediates de novo CENP-A assembly on the nearby central core, although the mechanistic link remains to be determined.
The specific properties of the central core that might aid CENP-A deposition are also unknown; for example, the GATA-like transcription factor Ams2 is enriched in the central core and might contribute to deposition, but central core DNA alone is not sufficient to attract CENP-A nucleosomes77. Intriguingly, the use of flanking lox sites and Cre recombinase to delete a fission yeast centromere results in the establishment of new functional centromeres in subtelomeric regions. DNA that is homologous to centromeric central core DNA cannot be detected in these regions, suggesting that chromatin structure or composition is the major determinant, rather than primary DNA sequence. Consistent with this model, telomeric regions are known to be assembled in heterochromatin, and depletion of RNAi or heterochromatin factors reduces the efficiency of neocentromere establishment25.
Non-coding transcripts and centromere formation
Small and large non-coding RNAs (for example, siRNAs, roX, Xist and NoRC) have roles in epigenetic regulation through the recruitment of chromatin-modifying activities103, 104, 105, 106. There are tantalizing observations that suggest that transcription of centromeric DNA or of other non-coding RNAs could similarly affect centromere formation: RNAase-sensitive electron-dense material is present at newt lung-cell kinetochores107; transcripts homologous to centromeric DNA have been detected in mammals108 and plants and are processed into siRNAs in plants109, 110; and active protein-coding genes can reside within centromeric chromatin111, 112. Strikingly, RNAs derived from centromeric retrotransposons and CentC centromeric repeats are enriched in CENP-A chromatin immunoprecipitates in maize113. Moreover, transcripts from human alpha-satellite DNA are associated with CENP-C and the inner centromere protein INCENP, and the addition of recombinant alpha satellite RNA to permeabilized human cells is required to target exogenous CENP-C to centromeres114. Finally, low levels of RNA polymerase II enrichment have been observed at the central domain in S. pombe115.
The relationship between RNAs derived from centromeric DNA and kinetochore assembly and/or architecture remains to be determined. Many chromatin modifications and nucleosome disassembly, reassembly and remodelling events are associated with transcription69, 116. Dimethylation or trimethylation of histone H3 on lysine 4 (H3K4me2 or H3K4me3) is associated with the 5′ regions of active genes and attracts chromatin factors such as NURF or Chd1, which reposition nucleosomes117. The H3K4me2 modification is found within the interspersed H3 blocks that are present in human, fly and plant centromeric chromatin50, 52. In S. pombe the central domain contains H3K4me2 (Ref. 52), and the Chd1 homologue, Hrp1, is required for normal CENP-A chromatin levels118. Thus, it is possible that transcription of centromeric DNA, rather than the RNA sequences, promotes CENP-A deposition (Fig. 5a). Intriguingly, the facilitates chromatin transcription (FACT) remodelling complex co-purifies with human CENP-A nucleosomes65, 119. FACT is required for efficient transcription through chromatin and it acts to dissociate H2A–H2B dimers from nucleosomes, thus aiding nucleosome disassembly in front of, and reassembly behind, the advancing RNA polymerase II69, 116, 117. Despite its association with CENP-A, there is no functional analysis that directly implicates FACT in CENP-A deposition. It is possible that the function of FACT with respect to CENP-A has nothing to do with transcription; FACT also interacts with DNA polymerase and might aid replication through CENP-A nucleosomes and mediate their random distribution to daughter strands69.
Figure 5. Possible roles for transcription of centromeric repeats.

a | Transcription of centromeric chromatin might facilitate the replacement of histone H3 with CENP-A following replication (for example, in the subsequent G1 phase in human cells). The FACT (facilitates chromatin transcription) complex translocates nucleosomes from in front of RNA polymerase II (RNAPII) to behind it during transcription. FACT associates with CENP-A nucleosomes, suggesting that the act of transcription might be involved in the replacement of H3 with CENP-A, analogous to the transcription-coupled replacement of H3 with H3.3, which is mediated by HIRA. b | Non-coding RNAs that are homologous to centromeric DNA, that is, nascent transcripts or processed small RNAs (not shown), could have an active role in recruiting specific chromatin remodelling and/or assembly factors that direct CENP-A nucleosome deposition to centromeres. The deposition of H3 or the formation of nucleosomal gaps in centromeric chromatin during S phase could trigger transcription of centromeric DNA, and restoration of full levels of CENP-A nucleosomes could terminate assembly by blocking transcription.
Alternatively, the RNAs produced from centromeric DNA might facilitate formation of CENP-A chromatin (see above). Centromeric RNAs might help to localize factors required for CENP-A deposition, analogous to the siRNA-mediated recruitment of histone-modifying enzymes103, 104 (Fig. 5b). They might also contribute to the formation of essential protein complexes, as observed for dosage compensation (the MSL complex in flies)106. Finally, nascent transcripts could hybridize with their centromeric templates to form R-loops120. Such unusual centromeric RNA–DNA hybrids could elicit an unconventional DNA-damage response, resulting in repair-coupled chromatin remodelling and deposition of CENP-A. Indeed, base-excision repair has been implicated in CENP-A chromatin assembly in Xenopus extracts121.
The presence of RNA at kinetochores and the association of RNAs that are homologous to centromeric DNA with centromeric chromatin proteins is intriguing. However, it remains to be determined whether the act of transcription or the non-coding transcripts themselves play a direct part in establishing or maintaining CENP-A chromatin. An important question to address is when centromeric transcription and transcripts are produced during the cell cycle with respect to the timing of CENP-A loading.
Models for propagation of centromere identity
What are the possible mechanisms responsible for propagating centromere identity? We believe that the central issue is to understand how new CENP-A is recruited to and incorporated into replicated centromeres. Without direct evidence for specific models, a range of molecules and mechanisms need to be considered. The basic facts described above that need to be accommodated into any model are: centromeric chromatin contains both CENP-A and H3 nucleosomes (Fig. 1); assembly occurs from late mitosis through to G1 (Fig. 3b); part of the CENP-A HFD (that is, the CATD) is necessary and sufficient for targeting (Fig. 2); and localization of new CENP-A and maintenance of existing CENP-A can be differentially regulated (Fig. 4b). Here we discuss three general models and the types of proteins that could be involved, based on these findings. These models are neither mutually exclusive nor encompass all possibilities, but serve as a useful framework for future analyses.
Deposition by CENP-A-specific proteins that recognize replicated CENP-A chromatin
Although we do not know exactly what happens at CENP-A chromatin during replication (octamer or half-nucleosome segregation, H3 deposition, or nucleosome gaps), the end result is the same: some type of nucleosome that contains CENP-A (Fig. 2b) is segregated to both daughter chromatids75 (Fig. 3a). Chromatin-loading factors that specifically recognize CENP-A could deposit new CENP-A nucleosomes onto each chromatid in the same region during telophase and through G1, replenishing the CENP-A content11, 72. Such a cyclical mechanism could account for the faithful propagation of centromeres at a single site, and the faithful propagation of neocentromere identity23, 122, 123.
If H3 nucleosomes are deposited during S phase in ‘gaps’ formed by CENP-A nucleosome segregation (Fig. 6a), then CENP-A deposition during telophase and through G1 could involve replacement or exchange of H3 nucleosomes. In fission yeast, replacement of H3 with CENP-A could be coincident with replication or occur subsequently in G2 (Refs 51, 76). A precedent for this type of mechanism comes from H3.3 incorporation into sites of gene expression34, 35, 36. In this case, CAFs would be required for H3 deposition, perhaps only during S phase, and segregated CENP-A would provide the specificity signal for new deposition by nucleosome-exchange factors, analogous to the HIRA complex, which would be recruited during loading. Transcription of centromeric DNA might also be involved in the eviction of H3 and its replacement with CENP-A (Fig. 5a).
Figure 6. Models for regulation of CENP-A propagation during the cell cycle.
a | In human cells, new CENP-A is deposited at centromeres in G1 to restore maximum occupancy at centromeres before replication. In S phase, following replication, half of these CENP-A nucleosomes remain with each sister chromatid, and the spaces are either temporarily filled with H3-containing nucleosomes or remain as nucleosomal gaps until the end of mitosis (M). Successful kinetochore assembly, microtubule attachment, bi-orientation and segregation of chromosomes to opposite spindle poles during mitosis might somehow authorise CENP-A deposition in the following G1 phase. Restoration of CENP-A levels at centromeres might occur by recruitment of CENP-A assembly factors that deposit new CENP-A in gaps, or by exchange factors that mediate replacement of H3 nucleosomes with CENP-A nucleosomes. b | Specificity of CENP-A deposition mediated by the interspersed H3 blocks. Following replication of centromeric chromatin, a distinct combination of post-translational modifications in the interspersed H3 blocks (for example, dimethylation of lysine 4 of histone H3, H3K4me2) might specify the recruitment of factors that are required for the replacement of H3 with CENP-A exchange or the deposition of new CENP-A nucleosomes into nucleosomal gaps. c | Specificity of CENP-A deposition mediated by CENP-A. CENP-A octamers might split during replication so that half-nucleosomes composed of single H2A, H2B, CENP-A and H4 subunits are retained on each sister chromatid through G2 and metaphase. In telophase and through G1, half-nucleosomes might attract specific assembly factors that rebuild complete CENP-A octamers, thus restoring centromeric chromatin integrity. Alternatively, CENP-A octamers might be segregated intact at the replication fork, and might specify recruitment of factors that mediate assembly of CENP-A nucleosomes into adjacent gaps, or replacement of adjacent H3 nucleosomes deposited in S phase (not shown).
Alternatively, if segregation of existing CENP-A nucleosomes to daughter strands during replication leaves ‘nucleosome gaps’, either segregated CENP-A or the gaps themselves could serve as a signal for new CENP-A assembly. This mechanism would require de novo assembly factors and/or activities, rather than those associated with H3 exchange (Fig. 6a). Assembly complexes could be centromere-specific, with unique components that are specific for the deposition of CENP-A nucleosomes, for example, the MIS18 complex. Alternatively, general assembly factors that are not centromere-specific might mediate CENP-A recruitment, for example, the RbAp46/48 histone chaperone among others. If so, ‘adaptor’ proteins might be required to recruit general assembly factors and/or CENP-A specifically to centromeres. The propagation of the epigenetic state of heterochromatin provides a paradigm for this mechanism, in which HP1 serves as an adaptor protein that binds both methylated H3K9 and the histone-lysine methyltransferase Su(var)3-9 (Ref. 124). By extension, non-coding RNAs generated from centromeric DNAs could be involved in recruitment of CENP-A assembly factors (Fig. 5b).
CENP-A assembly or exchange mediated by the interspersed H3 domains
A second model is suggested by the observation that the interspersed H3 nucleosomes display a modification pattern that is distinct from euchromatin and from the flanking heterochromatin50. The modifications that are present in the interspersed H3 domains could recruit specific CENP-A assembly proteins to the region (Fig. 6b). Proteins that could be required in these mechanisms include CAFs (for assembling interspersed H3), H3 modification enzymes or binding proteins, and remodelling or exchange complexes such as HIRA or NURF. Specificity for centromeric chromatin could be accomplished by unique factors or combinations of ‘common’ proteins or components of complexes.
Regulation by intrinsic properties of CENP-A nucleosomes
A third model postulates that interactions between molecules in CENP-A nucleosomes provide the signal for new CENP-A recruitment. H3 dimers remain stably associated during segregation of nucleosomes to replicated daughter chromatids70. Although CENP-A is also segregated to daughter chromatids75, CENP-A nucleosomes might be processed differently from H3 nucleosomes, as suggested by evidence that CENP-ACID may be in a half-nucleosome during interphase in Drosophila63, and that [CENP-A–H4]2 tetramers are less stable than [H3–H4]2 tetramers55. It is possible that CENP-A octamers are split during replication in S phase to form CENP-A–H4–H2A–H2B tetramers, or just CENP-A–H4 dimers, and that incorporation of new CENP-A during telophase and through G1 involves reconstitution of octamers (Fig. 6c). The splitting of nucleosomes during replication would require novel proteins, which could be specific to CENP-A. CENP-A recruitment might still require additional proteins, such as histone chaperones or proteins that bind CENP-A, perhaps through the CATD (Fig. 2a), or other centromeric chromatin components.
Conclusions
In the last few years many CENP-A-interacting proteins and factors that affect its localization have been identified. Some of these associate with CENP-A in complexes and are enriched at centromeres, whereas others are more broadly distributed. Some of these factors must influence the process of chromatin remodelling to ensure that CENP-A is replenished and propagated at the same location on a chromosome following replication. Surprisingly, in human cells new CENP-A is deposited at centromeres in late mitosis (that is, telophase) and in the G1 phase of the cell cycle. In addition, other proteins that are required for its deposition (RbAp46/48 and the MIS18 complex) show the same temporal pattern of localization.
The challenge for the future is to understand the events and signals that permit the deposition of CENP-A during this period and prevent its incorporation at other times. What factors mediate CENP-A nucleosome transfer from in front of a replication fork to the nascent strands behind? Are CENP-A nucleosomes split so that both daughter strands receive a CENP-A half-nucleosome in S phase? Is H3 initially deposited in centromeric chromatin in S phase and subsequently replaced with CENP-A, or are gaps created that are subsequently filled with CENP-A nucleosomes? What are the key CENP-A assembly and exchange factors? How are broadly distributed generic CAFs adapted to provide specificity to CENP-A deposition: by temporal regulation (for example, only CENP-A is deposited during mitosis or G1) or by formation of complexes with unique, CENP-A-specific components? Further dissection is also required to unravel the mechanism by which flanking centromeric heterochromatin influences the establishment of CENP-A chromatin and kinetochore assembly. It is also possible that the function of a centromere in mediating successful bipolar orientation and segregation influences the site of CENP-A deposition. Determining how the assembly of centromeric chromatin is integrated with cell-cycle progression is key to elucidating the regulatory signals. The next decade should allow a more complete understanding of how this unique form of chromatin is assembled and propagated at centromeres.
Acknowledgements
The authors thanks members of their laboratories for comments and suggestions. R.C.A. is a Wellcome Trust Principal Research Fellow and his research is supported by the Wellcome Trust (065061/Z). G.H.K.'s work on centromeres is supported by the National Institutes of Health (R01 GM066272).
Biographies
ROBIN C. ALLSHIRE
Robin Allshire received his B.A. in genetics from Trinity College, Dublin, Ireland, in 1981 before completing his Ph.D. with Chris Bostock at the Medical Research Council (MRC) Mammalian Genome Unit, University of Edinburgh, UK, in 1985. He spent 4 years as a postdoctoral researcher with Nicholas Hastie at the MRC Human Genetics Unit, Edinburgh. From 1989–1990 he was an independent visiting scientist at Cold Spring Harbor Laboratory, New York. In 1990 he set up his laboratory at the MRC Human Genetics Unit, Edinburgh, to study chromosome segregation using fission yeast as a model organism. In 2002 he was awarded a Wellcome Trust Principal Research Fellowship and moved his laboratory to the Wellcome Trust Centre for Cell Biology at the University of Edinburgh, UK. His laboratory focuses on centromeres and how they assemble the machinery required to segregate chromosomes during cell division. His laboratory discovered that genes are silenced when placed within fission yeast centromeres and this has allowed his team to identify and investigate components of these distinctive structures.
GARY H. KARPEN
Gary Karpen received his B.A. in biology from Brandeis University, Waltham, Massachusetts, in 1978 and his Ph.D. from the Department of Genetics, University of Washington, Seattle, in 1987. His thesis with Charles Laird demonstrated that a single ribosomal DNA gene was sufficient to form a nucleolus. Work on the structure and function of a Drosophila minichromosome was initiated during 4 years as a postdoctoral fellow with Allan Spradling at the Carnegie Institute, Baltimore, Maryland. He started his own laboratory at the Salk Institute, La Jolla, California, in 1991, and moved to the Lawrence Berkeley National Laboratory, University of California at Berkeley, in 2003. His laboratory studies the determinants of centromere identity, genome-wide analysis of histone modifications and chromosomal proteins, heterochromatin genomics and function, and DNA repair of repeated sequences.
References
- 1.Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nature Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 3.Monaco ZL, Moralli D. Progress in artificial chromosome technology. Biochem. Soc. Trans. 2006;34:324–327. doi: 10.1042/BST20060324. [DOI] [PubMed] [Google Scholar]
- 4.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 5.Spence JM, et al. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J. 2002;21:5269–5280. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell AR, Jeppesen P, Nicol L, Morrison H, Kipling D. Epigenetic control of mammalian centromere protein binding: does DNA methylation have a role? J. Cell Sci. 1996;109:2199–2206. doi: 10.1242/jcs.109.9.2199. [DOI] [PubMed] [Google Scholar]
- 7.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc. Natl Acad. Sci. USA. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Dawe RK, Henikoff S. Centromeres put epigenetics in the driver's seat. Trends Biochem. Sci. 2006;31:662–669. doi: 10.1016/j.tibs.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: cyclical stories and forking paths. Nature Rev. Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 12.Mellone BG, Allshire RC. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 2003;13:191–198. doi: 10.1016/s0959-437x(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 13.Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 14.Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- 15.Warburton PE, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan BA, Willard HF. Stable dicentric X chromosomes with two functional centromeres. Nature Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- 18.Agudo M, et al. A dicentric chromosome of Drosophila melanogaster showing alternate centromere inactivation. Chromosoma. 2000;109:190–196. doi: 10.1007/s004120050427. [DOI] [PubMed] [Google Scholar]
- 19.Alonso A, et al. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 2007;8:R148. doi: 10.1186/gb-2007-8-7-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 21.Choo KH. Domain organization at the centromere and neocentromere. Dev. Cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 22.Lo AW, et al. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 2001;11:448–457. doi: 10.1101/gr.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nature Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 24.Steiner NC, Clarke L. A novel epigenetic effect can alter centromere function in fission yeast. Cell. 1994;79:865–874. doi: 10.1016/0092-8674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 25.Ishii K, et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 26.Bulazel KV, Ferreri GC, Eldridge MD, O'Neill RJ. Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biol. 2007;8:R170. doi: 10.1186/gb-2007-8-8-r170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik HS, Henikoff S. Conflict begets complexity: the evolution of centromeres. Curr. Opin. Genet. Dev. 2002;12:711–718. doi: 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- 28.Murphy TD, Karpen GH. Centromeres take flight: alpha satellite and the quest for the human centromere. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Li B, et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl Acad. Sci. USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raisner RM, et al. Histone variant H2A.Z. marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 33.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc. Natl Acad. Sci. USA. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 35.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nature Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 36.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nature Rev. Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 37.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 38.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 39.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 40.Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 41.Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nature Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl Acad. Sci. USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-Alike protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer DK, O'Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heun P, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl Acad. Sci. USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan H, Jiang J. Rice as a model for centromere and heterochromatin research. Chromosome Res. 2007;15:77–84. doi: 10.1007/s10577-006-1104-z. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nature Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castillo AG, et al. Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet. 2007;3:e121. doi: 10.1371/journal.pgen.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cam HP, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nature Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 53.Yeh E, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregan J, et al. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr. Biol. 2007;17:1190–1200. doi: 10.1016/j.cub.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 56.Polizzi C, Clarke L. The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J. Cell Biol. 1991;112:191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi K, et al. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc. Natl Acad. Sci. USA. 2006;103:14877–14882. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoda K, et al. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl Acad. Sci. USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl Acad. Sci. USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conde e Silva N, et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 62.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 63.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 65.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nature Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 66.Maruyama T, Nakamura T, Hayashi T, Yanagida M. Histone H2B mutations in inner region affect ubiquitination, centromere function, silencing and chromosome segregation. EMBO J. 2006;25:2420–2431. doi: 10.1038/sj.emboj.7601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J. Mol. Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 68.Mello JA, Almouzni G. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 2001;11:136–141. doi: 10.1016/s0959-437x(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 69.Rocha W, Verreault A. Clothing up DNA for all seasons: histone chaperones and nucleosome assembly pathways. FEBS Lett. 2008;582:1938–1949. doi: 10.1016/j.febslet.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 70.Sugasawa K, et al. Nonconservative segregation of parental nucleosomes during simian virus 40 chromosome replication in vitro. Proc. Natl Acad. Sci. USA. 1992;89:1055–1059. doi: 10.1073/pnas.89.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sullivan B, Karpen G. Centromere identity in Drosophila is not determined in vivo by replication timing. J. Cell Biol. 2001;154:683–690. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim SM, Dubey DD, Huberman JA. Early-replicating heterochromatin. Genes Dev. 2003;17:330–335. doi: 10.1101/gad.1046203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pearson CG, et al. Stable kinetochore–microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 75.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takayama Y, et al. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol. Biol. Cell. 2008;19:682–690. doi: 10.1091/mbc.E07-05-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen ES, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003;11:175–187. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 78.Takayama Y, Takahashi K. Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res. 2007;35:3223–3237. doi: 10.1093/nar/gkm213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 80.Carroll CW, Straight AF. Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 81.Keith KC, et al. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol. Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Camahort R, et al. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 85.Stoler S, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl Acad. Sci. USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- 87.Hayashi T, et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 88.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fujita Y, et al. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 90.Dunleavy EM, et al. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol. Cell. 2007;28:1029–1044. doi: 10.1016/j.molcel.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okada M, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nature Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 92.Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell-cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. doi: 10.1083/jcb.200806038. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahmad K, Henikoff S. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 2001;153:101–110. doi: 10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl Acad. Sci. USA. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carlson SR, et al. Meiotic transmission of an in vitro-assembled autonomous maize minichromosome. PLoS Genet. 2007;3:1965–1974. doi: 10.1371/journal.pgen.0030179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nature Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 97.Ikeno M, et al. Construction of YAC-based mammalian artificial chromosomes. Nature Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 98.Hahnenberger KM, Baum MP, Polizzi CM, Carbon J, Clarke L. Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA. 1989;86:577–581. doi: 10.1073/pnas.86.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 100.Okada T, et al. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 101.Nakano M, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENPA chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nature Struct. Mol. Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 104.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grummt I. Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum. Mol. Genet. 2007;16:R21–R27. doi: 10.1093/hmg/ddm020. [DOI] [PubMed] [Google Scholar]
- 106.Rea S, Akhtar A. MSL proteins and the regulation of gene expression. Curr. Top. Microbiol. Immunol. 2006;310:117–140. doi: 10.1007/3-540-31181-5_7. [DOI] [PubMed] [Google Scholar]
- 107.Rieder CL. Ribonucleoprotein staining of centrioles and kinetochores in newt lung cell spindles. J. Cell Biol. 1979;80:1–9. doi: 10.1083/jcb.80.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl Acad. Sci. USA. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005;1:e79. doi: 10.1371/journal.pgen.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Neumann P, Yan H, Jiang J. The centromeric retrotransposons of rice are transcribed and differentially processed by RNA interference. Genetics. 2007;176:749–761. doi: 10.1534/genetics.107.071902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yan H, et al. Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell. 2006;18:2123–2133. doi: 10.1105/tpc.106.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saffery R, et al. Transcription within a functional human centromere. Mol. Cell. 2003;12:509–516. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- 113.Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl Acad. Sci. USA. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wong LH, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 116.Williams SK, Tyler JK. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 117.Reinberg D, Sims RJ. 3rd de FACTo nucleosome dynamics. J. Biol. Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 118.Walfridsson J, et al. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res. 2005;33:2868–2879. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Izuta H, et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 120.Aguilera A. mRNA processing and genomic instability. Nature Struct. Mol. Biol. 2005;12:737–738. doi: 10.1038/nsmb0905-737. [DOI] [PubMed] [Google Scholar]
- 121.Zeitlin SG, Patel S, Kavli B, Slupphaug G. Xenopus CENP-A assembly into chromatin requires base excision repair proteins. DNA Repair (Amst.) 2005;4:760–772. doi: 10.1016/j.dnarep.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 122.Maggert KA, Karpen GH. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 2001;158:1615–1628. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saffery R, et al. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- 124.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]





