Abstract
We analyzed the interaction of NTPs containing modified sugars to develop a better understanding of how DNA primase from herpes simplex virus I catalyzes primer synthesis. During the NTP binding reaction, primase tolerated a large number of modifications to the sugar ring. Altering the 2′ and 3′ carbons, and even converting the furanose sugar into an acyclic sugar, did not prevent binding. Whether or not the base on the NTP could form a correct base-pair with the template base being replicated also had minimal effects on the binding reaction, indicating that primase does not use this process to discriminate between right and wrong NTPs. Rather, the key feature that primase recognizes to bind a NTP is the 5′-γ-phosphate since converting a NTP into a NDP greatly compromised binding. During the polymerization reaction, primase tolerated substantial modification of the 2′-carbon, including the presence of either an ara or ribo hydroxyl, two hydrogens, or two fluorines. However, polymerization absolutely required that the NTP contain a 3′-hydroxyl and an intact sugar ring. Modifications at the 2′-carbon of the nucleotide at the primer 3′-terminus significantly impaired further polymerization events. Compared to a ribonucleotide, incorporation of a 2-deoxyribo- or 2′,2′-difluoro-2′-deoxyribonucleotide resulted in strong chain termination, while incorporation of an aranucleotide resulted in very strong chain termination. The implications of these data with respect to the mechanism of primase and the relationship between human and herpes primase are discussed.
Herpes DNA primase catalyzes a key reaction during viral DNA replication, the synthesis of RNA primers that a replicative DNA polymerase then elongates. Due to the inability of DNA polymerases to initiate DNA synthesis de novo, this enzyme is essential for herpes DNA replication (1-3). The enzymic activity resides within a 3 subunit complex (UL5, UL8, and UL52) that also exhibits helicase activity. Both primase and helicase activity require both the UL5 and UL52 subunits (4-6). While UL8 is dispensable for these activities, it is essential for herpes DNA replication and modulates primase activity under certain conditions (1, 6-11).
After binding a single-stranded template, primase initiates synthesis by binding two purine NTPs and generating a dinucleotide. Primase can initiate primer synthesis on a wide variety of pyrimidine rich templates (12). However, synthesis of primers longer than around 3 nucleotides requires that initiation occurs at the sequence 3′-G-pyrimidine-pyrimidine-5′. Thus, the template requirements of herpes primase are quite similar to a variety of bacterial and phage primases (13-16). The template sequence outside of the 3′-G-pyrmidine-pyrimidine initiation site greatly influences the efficiency with which primase uses that initiation site, however, this phenomenon is not well understood nor is it possible to predict beforehand how these flanking sequences will affect primer synthesis. In contrast, eukaryotic primases do not have strict template requirements and can both initiate primer synthesis and generate long primers on most any pyrimidine rich template (17, 18).
Interactions of herpes primase with NTP analogues have provided important insights into how the enzyme interacts with the base and chooses whether or not to polymerize the NTP. Primase readily misincorporates natural NTPs, and exhibits an overall error frequency of around 1:30 (19). It will also rapidly polymerize NTPs containing base analogues that can form Watson-Crick hydrogen bonds with the template base being replicated. Despite its low fidelity, primase will not readily polymerize NTPs containing unnatural bases that are incapable of forming Watson-Crick hydrogen bonds (20). Primase will, however, bind these NTP analogues incapable of Watson-Crick hydrogen bonding.
In contrast to our knowledge of how primase interacts with the base of a NTP, we know relatively little about how primase interacts with the sugar of the NTP. In these studies, we examined the interaction of primase containing a series of modifications in the sugar of the NTP. Key requirements for NTP polymerization include a cyclic sugar and a 3′-hydroxyl. Incorporation of a nucleotide whose sugar either contains a 2′-OH in the ribo configuration allows further nucleotide polymerization, whereas incorporation of a nucleotide containing a 2′-OH in the ara configuration results in very strong chain termination.
Experimental Methods
Materials
The UL5/UL8/UL52 helicase-primase complex was expressed in baculovirus-infected insect cells grown at the Tissue Culture Core Facility at the University of Colorado Health Sciences Center, and purified as previously described (12). The Tissue Culture Core Facility at the University of Colorado Cancer Center amplified, titered, and maintained the viral stocks.
Radiolabeled NTPs were purchased from Perkin Elmer. GCVTP and ACVTP were generously provided by Glaxo Smith Kline Corp. (Research Triangle Park, NC). Synthetic DNA templates of defined sequence (Table 1) were obtained from either Oligos, etc. or BioSearch Technologies, Inc.. Synthetic RNAs of defined sequence were obtained from Dharmacon (Boulder, CO). Oligonucleotide concentrations were determined spectrally and are reported in terms of 5′-termini. All other reagents were of the highest quality available.
Table 1.
Oligonucleotides Used.
| DNAC | GGGGUAAA(3′) |
| GCCCCATTTCTAATGCATGC(5′) | |
| DNAG | GGGGUAAA(3′) |
| GCCCCATTTGTAATGCATGC(5′) | |
| DNAT | GGGGUAAA(3′) |
| GCCCCATTTTTAATGCATGC(5′) | |
| d(C20GCCCTA15) | CCCCCCCCCCCCCCCCCCCCGCCCTAAAAAAAAAAAAAAA |
| d(GTCT)15 | GTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCT |
| d(GCTC)15 | GCTCGCTCGCTCGCTCGCTCGCTCGCTCGCTCGCTCGCTCGCTCGCTCGCTCGCTCGCTC |
Methods
Assays (10 μL) on single-stranded templates were performed as previously described and typically contained 50 mM Tris, pH 8.0, 10 mM MgCl2, 1 mM DTT, 0.1 mg/mL BSA, 20 μM ssDNA template, 800 μM [α-32P]NTPs, and 100 nM primase (UL5/UL8/UL52) (20). Reactions were initiated by adding enzyme, incubated at 37 °C for 30 minutes, and then quenched by adding 2-4 volumes of gel-loading buffer (90% formamide). Control reactions that lacked enzyme were conducted under identical conditions. Products were separated by denaturing polyacrylamide gel electrophoresis and analyzed by phosphorimagery (Molecular Dynamics). ImageQuant Software (Molecular Dynamics) was utilized for quantitative analysis. Gels typically contained 20% polyacrylamide, 7.5 M urea, although if needed to enhance separation between products, gels contained up to 40% polyacrylamide.
Assays (10 μL) to measure elongation of a pre-existing RNA primer:template were performed similar to those described above, except that they typically contained 50 mM Tris, pH 8.0, 10 mM MgCl2, 1 mM DTT, 0.1 mg/mL BSA, 100 nM 5′-[32P]-primer:template, 800 μM NTPs, and 200 nM primase (UL5/UL8/UL52). Products were analyzed by polyacrylamide gel electrophoresis and phosphorimagery as described above. T4 polynucleotide kinase was used to 5′-[32P] label the RNA primer. Following labeling, the primer was typically purified via gel electrophoresis on a 20% acrylamide gel (non-denaturing), and located and eluted from the gel as previously described (21).
Results
In order to better understand how primase interacts with the sugar of the incoming NTP, we examined a series of NTP analogues containing modified sugars (Chart 1). The modifications primarily focused on changes in the 2′ and 3′ carbons, and were designed to test the importance of each hydroxyl as well as the need for a cyclic sugar. Both the ability of primase to polymerize the analogues and to just bind the analogues was tested.
Chart 1.
Structures and abbreviations of sugars examined.
Since the different analogues contained different bases, we screened for incorporation using different templates that would allow correct base-pairing of each analogue. To control for the possibility that primase might differentially interact with sugar analogues during the primer initiation versus elongation steps, incorporation was measured under conditions where the analogue could be incorporated as the first, second, or a latter nucleotide of a primer.
Sugar requirements for Polymerization as the Second and Latter Nucleotide of the Primer
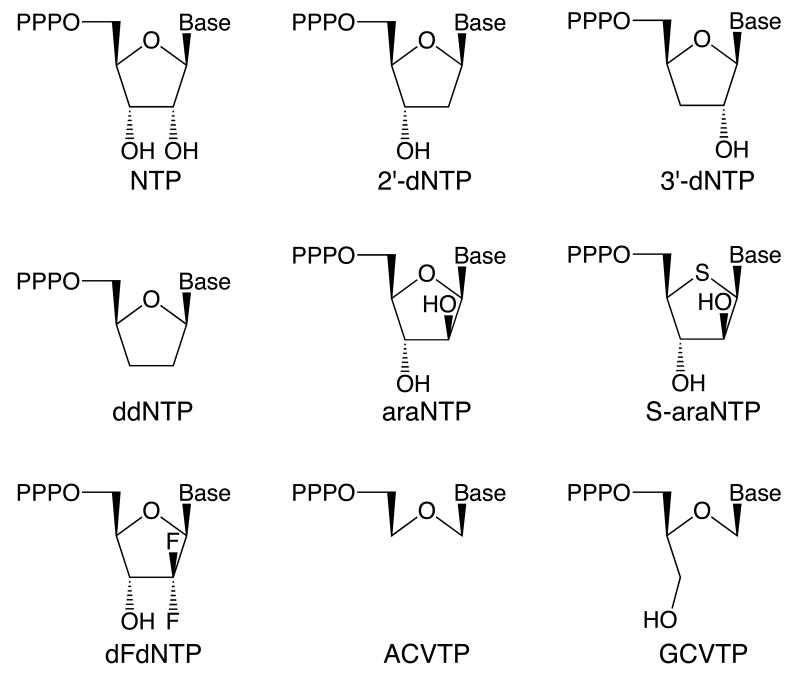
Incorporation of a series of CTP analogues was measured on the template d(GTCT)15 (Table 1) in the presence of ATP and GTP. Under these conditions, primase will synthesize the correctly base-paired trinucleotide pppApGpA, along with smaller amounts of longer products due to misincorporation opposite the template dG (Figure 1A). Adding either CTP or 2′-dCTP resulted in the synthesis of new products of altered mobility, indicating that primase readily incorporates CTP and dCTP opposite the dG. Furthermore, after incorporation of CTP, primase synthesized large amounts of new products with altered electrophoretic mobility > 4 nucleotides long indicating that the enzyme continued polymerizing additional NTPs. Longer products were also synthesized after dCTP polymerization, albeit fewer than after CTP incorporation. Primase also polymerized araCTP, SaraCTP, and dFdCTP as evidenced by the appearance of new products of altered electrophoretic mobility. However, no detectable products > 4 nucleotides long were synthesized upon incorporation of araC or SaraC, indicating that incorporation of these analogues results in significant chain termination. Due to the relatively strong inhibition of primase by 800 μM dFdCTP (Figure 1A), we examined the effects of lower dFdCTP concentrations (Figure 1B). As shown by the products of altered electrophoretic mobility >4 nucleotides long, primase could continue NTP polymerization after dFdCTP polymerization. When primase was incubated with ATP, GTP, and ddCTP, the products synthesized were identical to those generated with just ATP and GTP, albeit in reduced quantities. Increasing the ddCTP concentration to levels that gave >90% inhibition still did not result in new products of altered electrophoretic mobility (data not shown). In combination with previous studies showing that altering the sugar and/or base structure of the nucleotide at the 3′-terminus of a primer significantly alters its electrophoretic mobility (20, 22-25), these data indicate that primase does not readily incorporate ddCTP.
Figure 1.
Incorporation of C analogues. Panel A. Assays were performed as described under Experimental Procedures and contained primase, d(GTCT)20, ATP, [α-32P]GTP, and 800 μM of the indicated NTP (analogue). Panel B. Assays contained d(GTCT)20, ATP, [α-32P]GTP, and either 0, 100, 200, 400, or 800 μM dFdCTP. In both panels, lanes marked ‘C’ lacked enzyme. Primer lengths are noted on the left.
We compared primase's relative ability to polymerize additional NTPs after incorporation of CTP, dFdCTP, and dCTP. In assays containing 800 μM ATP, GTP, and the template d(GTCT)15, primase elongated 75% of the primers after incorporation of CTP. However, the enzyme elongated only 30 and 11 % of the primers after incorporation of dCTP and dFdCTP, respectively. Thus, dCTP and dFdCTP incorporation result in strong chain termination, while as noted above, araCTP and SaraCTP incorporation result in very strong chain termination.
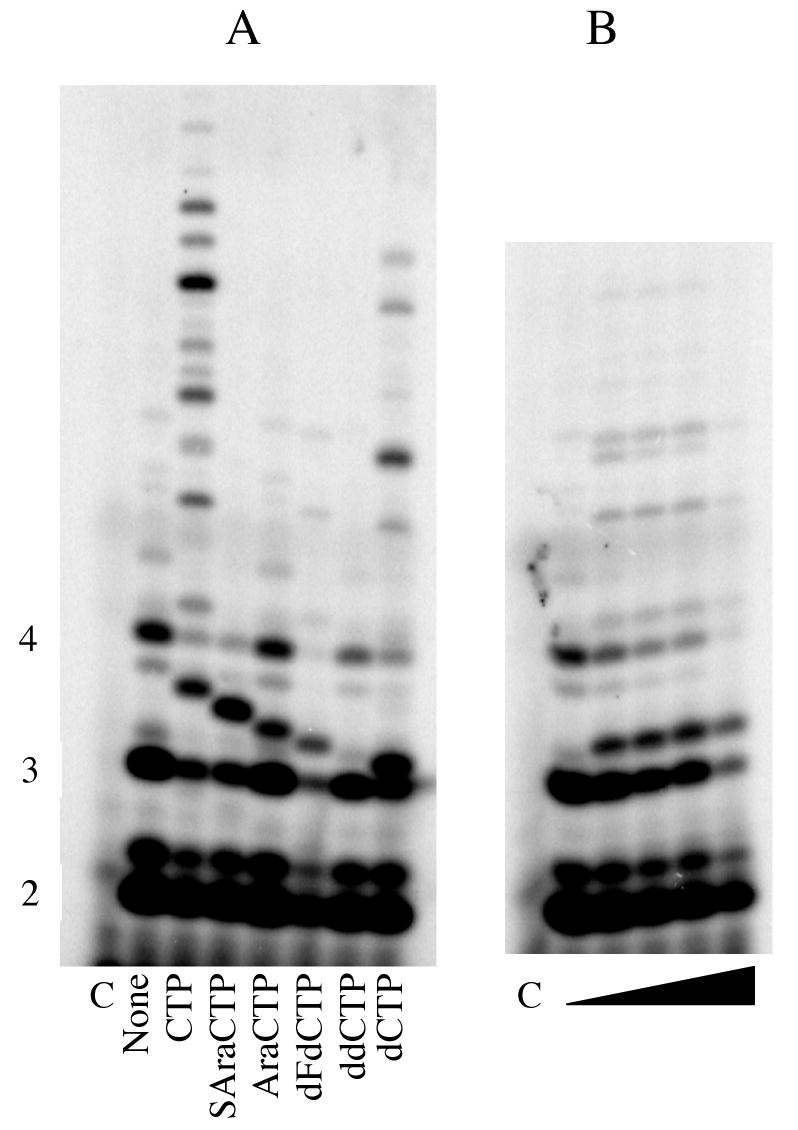
Using a series of adenine and guanine analogues, we extended these studies to examining the effect of removing the 3′-OH (3′-dATP, cordycepin triphosphate) and opening the furan ring of the sugar (ACVTP and GCVTP) on templates that allowed incorporation of either ATP or GTP as the third and later nucleotides of the primer. For example, incorporation of GTP analogues was measured on the template C20GCCCTA15 (Primer initiation site underlined), ATP, [α-32P]GTP, and increasing concentrations of either ddGTP or dGTP (Figure 2). Without any other additions, primase synthesizes products up to 5 nucleotides long due to misincorporation opposite the template T and A. Inclusion of dGTP resulted in new products of altered electrophoretic mobility two and three nucleotides long, thereby demonstrating dGTP polymerization opposite the C's in the template. In contrast, inclusion of ddGTP resulted in no new products, even though primase activity was inhibited up to 90%. Similar results were observed for GCVTP, ACVTP, 3′-dATP and ddATP, indicating that primase strongly discriminates against incorporation of acyclic NTPs and 3′-deoxyNTPs (data not shown).
Figure 2.
Incorporation of ddGTP and dGTP. Assays were performed as described under Experimental Procedures and contained primase, d(C20GCCCTA18). ATP. [α-32P]GTP and either 0, 0.25, 0.5, 0.75, 1, 1.25, 1.5, or 2 mM of ddGTP (left side) or of dGTP. The lane marked ‘C’ lacked enzyme. Primer lengths are noted on the left.
Efficiency of nucleotide analogue polymerization
We extended these initial qualitative studies to determine quantitatively how efficiently primase incorporated the analogues as either the 5′-terminal nucleotide of the primer or the second nucleotide in the primer. Initiation (dinucleotide synthesis) limits the rate of primer synthesis(26), hence we can directly measure the efficiency with which primase polymerized A and G analogues by measuring dinucleotide synthesis under conditions when the product was either pppApG or pppGpA. On the template d(GTCT)15, primase incorporates ATP (or analogue) as the first nucleotide, while on the template d(GCTC)15, primase incorporates ATP (or analogue) as the second nucleotide of the primer. Conversely, primase incorporates GTP (or analogue) as the second nucleotide on the template d(GTCT)15 and as the first nucleotide on the template d(GCTC)15. Since primase only appears to polymerize purine NTPs during initiation (12), we could not measure how efficiently primase incorporated CTP analogues in this reaction.
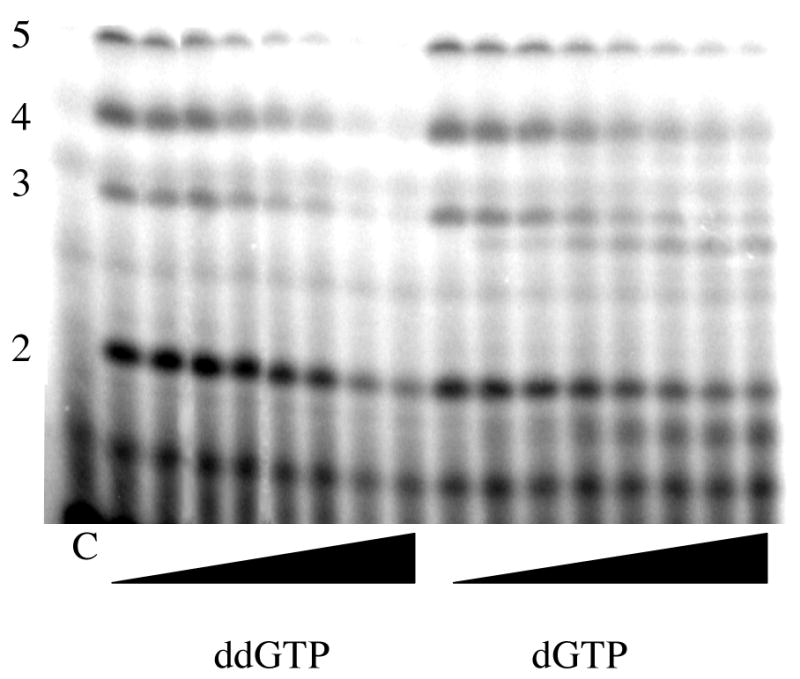
Polymerization of ATP analogues as the second nucleotide was tested in assays containing the template d(CTCG)15 and [α-32P]GTP. In the presence of only [α-32P]GTP, primase does not synthesize significant amounts of di- or trinucleotides since the assays lack the NTP that will become the second nucleotide of the primer (ATP). Including ATP resulted in the production of large amounts of products (Figure 3). The primary product is 3 nucleotides long because the assays lack CTP, the nucleotide required for further elongation of the trinucleotide. Including either 2′-dATP or araATP also resulted in new products, consistent with the studies showing that primase will incorporate either dCTP or araCTP. In contrast, including ddATP did not result in any new products, consistent with the studies showing that primase did not polymerize ddCTP.
Figure 3.
Incorporation of ATP analogues as the second nucleotide of the primer. Assays were performed as described under Experimental Procedures and contained primase, d(GCTC)15, [α-32P]GTP, and the indicated ATP (analogue). The lane marked ‘C’ lacked enzyme, and the length of products is noted on the left.
The efficiency with which primase incorporated ATP (analogues) was obtained by measuring dinucleotide formation in the presence of 800 μM [α-32P]GTP and increasing concentrations of ATP or ATP analogue. Table 2 shows that while primase readily incorporated ATP, araATP, and dATP, it did not detectably incorporate 3′-dATP or ddATP.
Table 2.
Incorporation of Nucleotides as the First or Second Nucleotide of the Primer.
| 5′-Terminal Nucleotide | |||
|---|---|---|---|
| Nucleotide | KM (mM) |
VMax (pmol min-1) |
VMax/KM (pmol min-1 mM-1) |
| ATP | 0.27±0.13 | 14±1.7 | 52 |
| AraATP | 0.34±0.13 | 0.04±0.005 | 1.2 |
| 2′-dATP | 1.8±1.6 | 20±1.1 | 11 |
| GTP | 1.3±0.6 | 1.1±0.27 | 0.82 |
| AraGTP | ND | ND | <0.1 |
| Second Nucleotide | |||
|
KM
(mM) |
VMax
(pmol min-1) |
VMax/KM
(pmol min-1 mM-1) |
|
| ATP | 1.4±0.9 | 5.1±1.8 | 3.6 |
| AraATP | 1.1±0.4 | 1.0±0.16 | 1.0 |
| 2′-dATP | >10 | >10 | 1.0 |
| ddATP | ND | ND | <0.1 |
| 3′-dATP | ND | ND | <0.1 |
| GTP | 0.22±0.10 | 9.5±1.3 | 45 |
| AraGTP | 0.21±0.04 | 4.1±0.3 | 20 |
| 2′-dGTP | 0.98±0.29 | 5.3±0.8 | 5.4 |
| ddGTP | ND | ND | <0.1 |
| ACVTP | ND | ND | <0.1 |
| GCVTP | ND | ND | <0.1 |
Likewise, we examined the polymerization of 2′-dGTP, araGTP, ddGTP, ACVTP, and GCVTP as the second nucleotide using the template d(TCTG)15. Assays contained primase, d(TCTG)15, and [α-32P]ATP, and the effects of adding either GTP or a GTP analogue examined. In the presence of only ATP, primase did not synthesize detectable amounts of di- or trinucleotides. Inclusion of either GTP, 2′-dGTP, or araGTP resulted in production of new products of altered electrophoretic mobility, indicating that primase polymerizes them (data not shown). However, addition of ddGTP, ACVTP, or GCVTP resulted in the production of no new products, indicating that primase does not readily polymerize them. Quantitative analysis of the incorporation of GTP and analogues as a function of concentration show relatively efficient incorporation of GTP, araGTP and 2′-dGTP, but no detectable incorporation of ddGTP, ACVTP or GCVTP (Table 2). The lack of detectable incorporation of 3′-dNTPs, ddNTPs, and acyclic NTPs only allow us to place a limit on the efficiency of incorporation of these analogues. Comparing the data for incorporation of A, C, and G analogues containing identical sugars revealed that qualitatively, primase exhibits similar properties towards NTP analogues containing modified sugars independent of the identity of the base.
Incorporation of nucleotide analogues as the 5′-terminal nucleotide of a primer was likewise quantified. To measure polymerization of GTP (or analogue), assays contained the template d(CTCG)15, [α-32P]ATP, and increasing concentrations of GTP (or analogue). Incorporation of ATP (or analogue) was measured in assays containing the template d(TCTG)15, [α-32P]GTP, and increasing concentrations of ATP (or analogue). In assays containing only [α-32P]ATP and d(CTCG)15, or only [α-32P]GTP and d(TCTG)15, no products were synthesized since the assays lacked the NTP that would become the 5′-terminal nucleotide of the primer. Table 2 shows that while primase used NTPs very efficiently as the 5′-terminal nucleotide, it used 2′-dNTPs and, especially, araNTPs much less efficiently.
Polymerization of nucleotides onto primer:templates
Primase will polymerize NTPs onto a RNA primer hybridized to a DNA template. To a template that will support primase activity in the presence of NTPs, we annealed a RNA primer at the identical position where primase synthesizes primers de novo. As shown in Figure 4, primase elongated this primer when incubated with NTPs. To provide an additional measure of the ability of primase to polymerize nucleotides, we then generated a series of primer:templates where the next correct NTP was either ATP (DNAT), GTP (DNAC), or CTP (DNAG).
Figure 4.
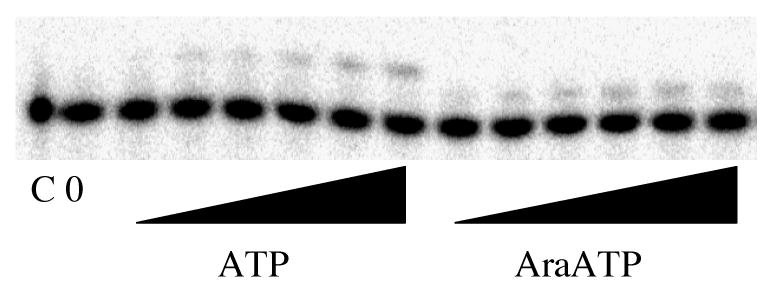
Primer elongation with ATP and araATP. Assays were performed as described under Experimental Procedures and contained primase, DNAT and either 0, 0.1, 0.3, 0.5, 0.75, 1 or 2 mM ATP (left side) or araATP (right side). The lane marked ‘C’ lacked enzyme, and the lane marked ‘0’ contained enzyme but no NTP.
Table 3 shows the efficiency with which primase polymerized various NTP analogues onto primer:templates. Similar to the results for polymerization of NTP analogues as the second nucleotide of the primer, primase incorporated both 2′-dNTPs and araNTPs with efficiencies either slightly less or greater than the efficiency of NTPs. As long as the sugar contained a complete ring and a 3′-OH, primase could incorporate it.
Table 3.
Polymerization of Nucleoside Triphosphates onto a Primer:Template
| KM (mM) |
VMax (pmol h-1) |
VMax/KM (pmol min-1 mM-1) |
|
|---|---|---|---|
| CTP | 1.1±0.28 | 1.2±0.16 | 0.92 |
| AraCTP | 0.360±0.19 | 0.30±0.05 | 0.83 |
| dFdCTP | 0.038±0.026 | 0.078±0.006 | 2.1 |
| 2′-dCTP | 0.64±0.36 | 0.16±0.036 | 0.25 |
| ddCTP | ND | ND | - |
| ATP | 1.700±0.96 | 0.23±0.09 | 0.14 |
| 2′-dATP | 0.77±0.37 | 0.024±0.06 | 0.031 |
| AraATP | 0.19±0.11 | 0.026±0.004 | 0.14 |
| ddATP | ND | ND | - |
| 3′-dATP | ND | ND | - |
| GTP | 0.230±0.17 | 0.060±0.012 | 0.26 |
| AraGTP | 0.30±0.14 | 0.044±0.006 | 0.15 |
| 2′-dGTP | 0.13±0.08 | 0.18±0.02 | 1.4 |
| ACVTP | ND | ND | - |
| GCVTP | ND | ND | - |
| ddGTP | ND | ND | - |
Requirements for binding a nucleotide
The lack of incorporation of a NTP analogue could have resulted from the enzyme lacking the ability either to bind or to polymerize the compound. To differentiate between these two possibilities as well as provide insights into which portions of the nucleotide that primase recognizes during binding, we examined the ability of those nucleotides that primase did not incorporate to inhibit polymerization of a NTP onto a primer:template. In the absence of polymerization, the potency of inhibition reflects the strength of binding of the nucleotide.
Surprisingly, all of the NTP analogues tested inhibited primase with similar efficacy (Table 4). To determine if changing the base on the NTP from one that was complementary to the template base being replicated to a noncomplementary base, we compared the ability of ddATP and ddCTP to inhibit GTP polymerization opposite a template C to the ability of ddGTP to inhibit GTP polymerization. Changing the base from one that is complementary to the template base being copied to a non-complementary base had little effect on binding. Thus, primase does not appear to sense if the base-pair formed between the incoming NTP and the template base is right or wrong during formation of the E-DNA-NTP complex. The only slightly weaker binding of ACVTP than ddGTP, which contains an intact furan ring, indicates that primase does not strongly interact with the 2′ and 3′ carbons.
Table 4.
Inhibition of NTP polymerization onto primer:templates.
| Inhibitor | NTP a | IC50 (mM) |
|---|---|---|
| ACVTP | GTP | 3.2±0.57 |
| GCVTP | GTP | 3.4±0.88 |
| ddGTP | GTP | 1.1±0.18 |
| GDP | GTP (100 μM) | 4.4±0.65 |
| GMP | GTP (100 μM) | 3.8±0.54 |
| ddATP | GTP | 2.4±0.9 |
| ddCTP | GTP | 0.80±0.12 |
| 3′-dATP | ATP | 1.1±0.17 |
| ddATP | ATP | 1.7±0.11 |
| ADP | ATP (100 μM) | >10 |
| AMP | ATP (100 μM) | >10 |
Unless noted, assays contained 800 μM of the noted NTP. Polymerization of GTP and ATP were measured on DNAC and DNAT, respectively.
The fact that all of the modified bases bound with similar efficiency suggested that binding of NTPs to primase was not primarily mediated via interactions with either the sugar or base. Therefore, we tested the roles of the γ and β phosphates by measuring the binding of NDPs and NMPs (Table 4). In contrast to the minimal impacts observed upon modifying either the sugar or base, removing either just the γ or both the γ and β phosphates significantly weakened binding. Thus, binding appears to be largely mediated by interactions with the triphosphate, especially the γ phosphate. For two of the nucleotides, GDP and ddGTP, we determined the type of inhibition (competitive, non-competitive, etc.). In each case, inhibition was competitive with respect to GTP incorporation (data not shown).
Discussion
Herpes primase bound nucleoside triphosphates bearing a remarkably broad range of sugars. Compared to ribose, primase readily tolerated replacement of the 4′-O with S, loss of either or both of the 2′ and 3′ hydroxyls, inversion of the 2′-OH stereochemistry, the presence of 2 fluorines at the 2′ position, and even the complete loss of the 2′ and 3′-carbons. The 2′-hydroxyl does not appear to contribute significantly to binding since both ddATP and 3′-dATP bound primase with similar affinity. The greatest impact occurred upon ablation of the 2′ carbon Both ACVTP and GCVTP bound less tightly than the corresponding ddGTP. Since we cannot measure the KDs for nucleoside triphosphates containing a 3′-hydroxyl due to their polymerization, the effects on binding of removing the 3′-hydroxyl on binding cannot be quantified.
For those NTP analogues that primase did not incorporate, we measured their ability to inhibit polymerization of a NTP onto a primer:template. In contrast to measuring inhibition of total primer synthesis, this assay is advantageous since one is now measuring a specific polymerization event. Inhibition of total primer synthesis is more complicated to interpret since inhibition could reflect effects on one, or more, NTP binding and polymerization events.
Whereas primase tolerated significant modification of the sugar, it was much less tolerant of modification of the triphosphate. Compared to other triphosphates, GDP and GMP bound much less tightly. Indeed, accurate measurement of inhibition of primase activity by GDP and GMP required that we use only 100 μM GTP in the assays, rather than the standard 800 μM. A similar, but larger, effect occurred with ATP – removal of either just the γ-phosphate or both the γ- and β-phosphates dramatically weakened binding to primase. Thus, herpes primase appears to utilize the triphosphate as a primary recognition motif during formation of the E-DNA-NTP complex.
Herpes primase misincorporates NTPs at remarkably high frequencies, varying from 1 in 7 to 1 in 100 depending upon the mismatch (19). In order to begin to understand when in its catalytic cycle primase obtains this admittedly modest selectivity, we compared the binding of ddGTP, ddCTP, and ddATP opposite a template C. All 3 of them bound with similar affinity, suggesting that herpes primase does not discriminate between right and wrong NTPs during the initial binding reaction. In contrast, most higher fidelity polymerases obtain a large amount of their accuracy by specifically discriminating against incorrect (d)NTPs during binding (27-29). This conclusion assumes that the sugar structure does not alter interactions between the enzyme and the base of the NTP during the initial binding reaction. Consistent with this idea, the identity of the base attached to identical sugars had only small effects on either polymerization (Vmax/KM) or binding of the analogue dNTPs.
Primase has two key sugar requirements for polymerization of a nucleoside triphosphate – the presence of a 3′-hydroxyl and a 2′-carbon and/or a cyclic sugar. Testing of a variety of NTP analogues lacking a 3′-hydroxyl (3′-dATP, ACVTP, and ddNTPs) revealed no detectable polymerization, while the lack of detectable polymerization of GCVTP indicates the requirement for a cyclic sugar and/or a 2′-carbon. Primase readily incorporated NTP analogues containing a variety of substituents at the 2′-carbon, including a hydroxyl in either the ara or ribo configuration, two hydrogens, and two fluorines. In light of the very different chemical properties and shapes of these substituents, these data suggest that primase does not interact closely with the 2′-C.
After incorporation of a nucleotide containing a modified 2′-carbon, primase polymerized additional NTPs much less efficiently than after incorporation of a nucleotide containing a 2′-ribo hydroxyl. The most severe chain termination occurred if the nucleotide at the primer 3′-terminus contained the sugar arabinose. The ability of arabinose to inhibit further NTP polymerization was observed both during ongoing polymerization when the ara-nucleotide was incorporated as the fourth nucleotide of the primer and when primase attempted to use an araNTP as the 5′-terminal nucleotide of the primer. Just like ongoing primer sythesis, this latter reaction requires polymerization onto a 3′-hydroxyl with a neighboring trans-2′-hydroxyl.
The chemical requirements of primase during polymerization of the first two nucleotides of the primer (i.e., primer initiation) closely resemble those of ongoing primer synthesis as well as elongation of an existing RNA primer:template. Just as after incorporation of an araNTP primase did not efficiently polymerize additional NTPs, primase did not efficiently elongate an araNTP as the 5′-terminal nucleotide of the primer. Similarly, primase elongated dATP as the 5′-terminal nucleotide less efficiently than it elongated ATP, results that are qualitatively identical to what happens after primase incorporated a dNTP at later positions of the growing primer. As the second nucleotide of the primer, primase only effectively polymerized NTP analogues that contained both a 3′-hydroxyl and a cyclic sugar. Likewise, primase exhibited similar sugar preferences during polymerization of NTP (analogues) onto pre-existing primer:templates as during initiation and/or ongoing primer synthesis. Thus, the interactions of primase with the sugars of the nucleotides do not greatly vary between initiation and ongoing primer synthesis. These similarities also suggest that the 3′-terminus of the growing primer likely resides in the same location as the NTP that will become the 5′-terminal nucleotide of the primer, while NTPs that become the second and later nucleotide of the primer all bind in the same site.
Both herpes primase and human primase exhibit remarkably similar features in terms of how they recognize and polymerize NTPs ((20, 23, 30-33) and vide infra). In order to be polymerized, the incoming NTP must have a 3′-hydroxyl, although it can be substantially modified at the 2′-carbon (Eg., both enzymes polymerize araNTPs, 2′-dNTPs, and dFdCTP). Incorporation of araNTPs by either primase results in very strong chain termination, as does polymerization of 3-deazaATP. Polymerization of 2′-dNTPs results in chain termination by both enzymes, although the termination is more severe with human primase. Initiation of new primers occurs exclusively via the polymerization of two purine NTPs. Both enzymes have very low fidelity – herpes primase discriminates against incorrect NTPs by a factor of 30 while human primase discriminates by a factor of around 100. Even the mechanisms these enzymes use to differentiate between right and wrong NTPs appear similar. In both cases, efficient incorporation requires formation of Watson-Crick hydrogen bonds between the incoming NTP and the template base being replicated. NTPs containing hydrophobic bases such as benzimidazole are polymerized much slower than even an incorrect, natural NTP. Perhaps the only significant mechanistic difference between these enzymes is that synthesis of long primers by herpes primase requires that initiation occurred at the template sequence 3′-G-pyr-pyr while human primase only requires the template sequence pyr-pyr.
These similar “mechanistic fingerprints” indicate that herpes and human primase are closely related evolutionarily. While the actual mechanism of phosphodiester bond formation by either primase has not been probed, the likely conservation of 3 aspartates in the active sites of each enzyme suggests that this aspect of catalysis will also be conserved (34). Many of the conserved properties reflect interactions between the incoming NTP (2′-C, 3′-OH, and base) distal from the site of phosphodiester bond formation (α-phosphate on the 5′-C), hence the enzymes apparently retain conserved pathways of “information transfer” between these distal sites and the catalytic core. Remarkably in light of the almost identical interactions with their substrates, sequence analysis would suggest that these enzymes are distantly related (34), with only a relatively small number of conserved amino acids. Additionally, human primase bears significant homology to the X superfamily of polymerases in terms of both sequence and activity (35), while herpes primase does not exhibit such a relationship. These observations raise the intriguing question of how the primases have retained their mechanistic similarities while diverging in sequence so greatly.
Abbreviations used
- ACVTP
acyclovir triphosphate
- araATP
arabionfuranosyladenine triphosphate
- araCTP
arabinofuranosylcytosine triphosphate
- araGTP
arabinofuranosylguanine triphosphate
- BSA
Bovine Serum Albumin
- ddNTP
2′, 3′-dideoxynucleoside triphosphate
- dFdCTP
2′,2′-Difluoro-2′-deoxycytidine triphosphate
- DTT
Dithiothreitol
- GCVTP
ganciclovir triphosphate
- SaraCTP
4′-thio-arabinofuranosylcytosine triphosphate
- Tris
tris(hydroxymethyl)aminomethane, HCl salt
Footnotes
This work was supported by National Institutes of Health Grant AI059764 (R.D.K.)
References
- 1.Boehmer PE, Lehman IR. Herpes Simplex Virus DNA Replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 2.Carrington-Lawrence SD, Weller SK. Recruitment of Polymerase to Herpes Simplex Virus Type 1 Replication Foci in Cells Expressing Mutant Primase (UL52) Proteins. J Virol. 2003;77:4237–4247. doi: 10.1128/JVI.77.7.4237-4247.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crute JJ, Ctygon CA, Hargrave KD, Simoneau B, Faucher AM, Bolger G, Kibler P, Liuzzi M, Cordingley MG. Herpes Simplex Virus Helicase-primase Inhibitors Are Active in Animal Models of Human Disease. Nature Medicine. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 4.Calder JM, Stow ND. Herpes simplex Virus Helicase-Primase: the UL8 Protein Is not Required for DNA-Dependent ATPase and DNA Helicase Activities. Nuc Ac Res. 1990;18:3573–3578. doi: 10.1093/nar/18.12.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodson MS, Lehman IR. Association of DNA helicase and primase activities with a subassembly of the herpes simplex virus 1 helicase-primase composed of the UL5 and UL52 gene products. Proc Natl Acad Sci USA. 1991;88:1105–1109. doi: 10.1073/pnas.88.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherman G, Gottlieb J, Challberg MD. The UL8 Subunit of the Herpes Simplex Virus Helicase-primase Complex Is Required for Efficient Primer Utilization. J Vir. 1992;66:4884–48892. doi: 10.1128/jvi.66.8.4884-4892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falkenberg M, Bushnell DA, Elias P, Lehman IR. The UL8 Subunit of the Heterotrimeric Herpes Simplex Virus Type 1 Helicase-Primase Is Required for the Unwinding of Single Stand DNA-binding Protein (ICP8)-coated DNA Substrates. J Biol Chem. 1997;272:22766–22770. doi: 10.1074/jbc.272.36.22766. [DOI] [PubMed] [Google Scholar]
- 8.Gac NTL, Villani G, Boehmer PE. Herpes Simplex Virus Type-1 Single-strand DNA-binding Protein (ICP8) Enhances the Ability of the Viral DNA Helicase-primase to Unwind Cisplatin-modified DNA. J Biol Chem. 1998;273:13801–13807. doi: 10.1074/jbc.273.22.13801. [DOI] [PubMed] [Google Scholar]
- 9.Mardsen HS, McLean GW, Barnard EC, Francis GJ, MacEachran K, Murphy M, McVey G, Cross A, Abbotts AP, Stow ND. The Catalytic Subunit of the DNA Polymerase of Herpes Simplex Virus Type 1 Interacts Specifically with the C Terminus of the UL8 Component of the Viral Helicase-Primase Complex. J Vir. 1997;71:6390–6397. doi: 10.1128/jvi.71.9.6390-6397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLean GW, Abbotts AP, Parry ME, Marsden HS, Stow ND. The Herpes Simplex Virus Type 1 Origin-binding Protein Interacts Specifically with the Viral UL8 Protein. J Gen Vir. 1994;75:2699–2706. doi: 10.1099/0022-1317-75-10-2699. [DOI] [PubMed] [Google Scholar]
- 11.Tenney DJ, Hurlburt WW, Micheletti PA, Bifano M, Hamatake RK. The UL8 Component of the Herpes Simplex Virus Helicase-primase Complex Stimulates Primer Synthesis by a Subassembly of the UL5 and UL52 Components. J Biol Chem. 1994;269:5030–5035. [PubMed] [Google Scholar]
- 12.Ramirez-Aguilar KA, Low-Nam NA, Kuchta RD. Key Role of Template Sequence for Primer Synthesis by the Herpes Simplex Virus 1 Helicase-Primase. Biochemistry. 2002;41:4569–4579. doi: 10.1021/bi026680v. [DOI] [PubMed] [Google Scholar]
- 13.Cha TA, Alberts BM. Studies of the DNA Helicase-RNA Primase Unit from Bacteriophage T4. A Trinucleotide Sequence on the DNA Template Starts RNA Primer Synthesis. J Biol Chem. 1986;261:7001–7010. [PubMed] [Google Scholar]
- 14.Hiasa H, Sakai H, Tanaka K, Honda Y, Komano T, Godson GN. Structural Features of the Priming Signal Recognized by Primase: Mutational Analysis of the Phage G4 Origin of Complementary DNA Strand Synthesis. Gene. 1989;84:9–16. doi: 10.1093/nar/18.16.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendelman LV, Richardson CC. Requirements for Primer Synthesis by Bacteriophage T7 63-kDa Gene 4 Protein. Roles of Template Sequence and T7 56-kDa Gene 4 Protein. J Biol Chem. 1991;266:23240–23250. [PubMed] [Google Scholar]
- 16.Yoda K, Okazaki T. Specificity of Recognition Sequence for Eschericia coli Primase. Molecular and General Genetics. 1991;227:1–8. doi: 10.1007/BF00260698. [DOI] [PubMed] [Google Scholar]
- 17.Sheaff R, Kuchta RD. The Mechanism of Calf Thymus DNA Primase: Slow Initiation, Rapid Polymerization and Intelligent Termination. Biochemistry. 1993;32:3027–3037. doi: 10.1021/bi00063a014. [DOI] [PubMed] [Google Scholar]
- 18.Arezi B, Kuchta RD. Eucaryotic DNA Primase. Trends Bioch Sci. 2000;25:572–576. doi: 10.1016/s0968-0004(00)01680-7. [DOI] [PubMed] [Google Scholar]
- 19.Ramirez-Aguilar KA, Kuchta RD. Herpes Simplex Virus I DNA Primase: A Polymerase with Extraordinarily Low Fidelity. Biochemistry. 2004;43:9084–9091. doi: 10.1021/bi049335+. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez-Aguilar KA, Moore CL, Kuchta RD. Herpes Simplex Virus I Primase Employs Watson-Crick Hydrogen Bonding to Identify Cognate NTPs. Biochemistry. 2005;44:15585–15593. doi: 10.1021/bi0513711. [DOI] [PubMed] [Google Scholar]
- 21.Beckman J, Kincaid K, Hocek M, Spratt T, Engels J, Cosstick R, Kuchta RD. Human DNA Polymerase alpha Uses a Combination of Positive and Negative Selectivity to Polymerize Purine dNTPs with High Fidelity. Biochemistry. 2007;46:448–460. doi: 10.1021/bi061243s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilsley DI, Lee SH, Miller W, Kuchta RD. Acyclic Guanosine Analogs Inhibit DNA Polymerases alpha, delta and epsilon with Very Different Potencies and Have Unique Mechanisms of Action. Biochemistry. 1995;34:2504–2510. doi: 10.1021/bi00008a014. [DOI] [PubMed] [Google Scholar]
- 23.Kuchta RD, Willhelm L. Inhibition of DNA Primase by 9-beta-D-Arabinofuranosyladenosine Triphosphate. Biochemistry. 1991;30:797–803. doi: 10.1021/bi00217a033. [DOI] [PubMed] [Google Scholar]
- 24.Kuchta RD, Ilsley D, Kravig KD, Schubert S, Harris B. Inhibition of DNA Primase and Polymerase Alpha by Arabinofuranosylnucleoside Triphosphates and Related Compounds. Biochemistry. 1992;31:4720–4728. doi: 10.1021/bi00134a027. [DOI] [PubMed] [Google Scholar]
- 25.Chiaramonte M, Moore CL, Kincaid K, Kuchta RD. Facile Polymerization of dNTPs Bearing Unatural Base Analogues by DNA Polymerase alpha and Klenow Fragment (DNA Polymerase I) Biochemistry. 2003;42:10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez-Aguilar K, Kuchta RD. Mechanism of Primer Synthesis by the Herpes Simplex Virus 1 Helicase-Primase. Biochemistry. 2004;43:1103–1112. doi: 10.1021/bi035519x. [DOI] [PubMed] [Google Scholar]
- 27.Capson T, Peliska JA, Kaboord BF, West MF, Lively C, Dahlberg M, Benkovic SJ. Kinetic Characterization of the Polymerase and Exonuclease Activities of the Gene 43 Protein of Bacteriophage T4. Biochemistry. 1992;31:10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 28.Yang G, Wang J, Konigsberg W. Base Selectivity Is Impaired by Mutants that Perturb Hydrogen Bonding Networks in the RB69 DNA Polymerase Active Site. Biochemistry. 2005;44:3338–3346. doi: 10.1021/bi047921x. [DOI] [PubMed] [Google Scholar]
- 29.Tsai YC, Johnson KA. A New Paradigm for DNA Polymerase Specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuchta RD, Reid B, Chang LMS. DNA Primase: Processivity and the Primase to Polymerase Alpha Activity Switch. J Biol Chem. 1990;265:16158–16165. [PubMed] [Google Scholar]
- 31.Moore CL, Chiaramonte M, Higgins T, Kuchta RD. Synthesis of Nucleotide Analogues that Potently and Selectively Inhibit Human Primase. Biochemistry. 2002;41:14066–14075. doi: 10.1021/bi026468r. [DOI] [PubMed] [Google Scholar]
- 32.Richardson KA, Vega TP, Moore CL, Rohloff JC, Tomkinson B, Bendele RA, Richardson FC, Kuchta RD. Polymerization of the Triphosphates of AraC, 2′,2′-Difluorodeoxycytidine and ThioAraC by Human DNA Polymerase alpha and DNA Primase. Biochem Pharm. 2004 doi: 10.1016/j.bcp.2004.07.042. In press. [DOI] [PubMed] [Google Scholar]
- 33.Sheaff R, Kuchta RD. Misincorporation of Nucleotides by Calf Thymus DNA Primase and Elongation of Primers Containing Multiple Non-cognate Nucleotides by DNA Polymerase Alpha. J Biol Chem. 1994;269:19225–19231. [PubMed] [Google Scholar]
- 34.Lakshminarayan MI, Koonin EV, Leipe DD, Aravind L. Origin and Evolution of the Archaeo-Eukaryotic Primase Superfamily and Related Palm-Domain Proteins: Structural Insights and New Members. Nuc Ac Res. 2005;33:3875–3896w. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirk BW, Kuchta RD. Arg304 of Human DNA Primase Is a Key Contributor to Catalysis and NTP Binding: Primase and the Family X Polymerases Share Significant Sequence Homology. Biochemistry. 1999;38:7727–7737. doi: 10.1021/bi990247c. [DOI] [PubMed] [Google Scholar]