Abstract
Interleukin 7 (IL-7) plays a critical role in B cell fate decision by regulating early B cell factor (EBF) expression. However, it was not clear when IL-7 stimulation is necessary in hemato/lymphopoiesis in adult mice. Here we show that pre-proB cells derived from IL-7−/− mice have lost B cell potential, despite upregulation of EBF expression following IL-7 stimulation. Pre-proB cells from wild type (WT) mice can give rise to proB cells in the absence of IL-7. In this case, EBF upregulation during the transition from the pre-proB to proB stages occurs normally. In contrast, EBF expression by IL-7−/− pre-proB cells after IL-7 stimulation is ~20 times lower than WT pre-proB cells. In addition, only multipotent progenitors with higher levels of ectopic EBF can give rise to proB cells in the absence of IL-7. Therefore, the primary function of IL-7 prior to the pre-proB stage in B cell development is to maintain the EBF expression level above a certain threshold, which is necessary for pre-proB cells to further transit to the proB stage.
Keywords: B cells, cell differentiation, cytokines, cytokine receptors, transcription factors
Introduction
Interleukin 7 (IL-7) is an indispensable cytokine for T and B lymphocyte development in the mouse (1, 2). IL-7 receptor (IL-7R) is composed of IL-7 receptor α chain (IL-7Rα) and common cytokine receptor γ chain (γc) (3, 4). Lack of IL-7Rα or γc leads to a severe reduction in the number of T and B cells (1, 3, 5–7). Since enforced Bcl-2 expression can rescue impaired T cell development in IL-7Rα deficient (IL-7Rα−/−) or γc deficient (γc−/Y or γc−/−) mice, a main function of IL-7 in T cell development is to support survival of thymocytes (8–12). In contrast, B cell development is not rescued in the same mice (8–11, 13). Although IL-7 plays an important role in rearrangement of the immunoglobulin heavy chain gene (14, 15), the reason why B cell development is significantly impaired in the absence of IL-7R signal has not been clarified.
Recently we and others found that IL-7 critically regulates expression of early B cell factor (EBF) in developing B cells in adult bone marrow (16, 17). EBF is a B cell specific transcription factor and regulates expression of genes that play important roles in B cell development, such as λ5, VpreB, and mb-1 (18, 19). Additionally, EBF in cooperation with E2A positively regulates Pax5 expression (20). These transcription factors, along with PU.1, are indispensable for B cell development and form transcriptional networks that critically regulate B lineage specification and commitment (21). Although there is a hierarchical relationship in the expression of these transcription factors, EBF is especially important in B cell differentiation. For example, PU.1 positively regulates EBF expression, however, enforced EBF expression in hematopoietic progenitors can rescue impaired B cell development in the absence of PU.1 (22). Although the Pax5 gene is a target of EBF, ectopic Pax5 cannot rescue B cell development in the absence of PU.1 (22). In addition, EBF plays an important role in suppression of non-B cell lineages such as T cells and myeloid cells in developing hematopoietic cells (23, 24). Therefore, regulation of EBF expression is critical for the lineage specification at the early stage of B cell development, such as the common lymphoid progenitor (CLP) and pre-proB cell stages.
Previously, it was thought that the phenotype of IL-7Rα−/− mice is more severe than IL-7−/− mice (1, 2). However, we recently found that the phenotype of IL-7Rα−/− and IL-7−/− mice is virtually the same; B cell development is arrested at the pre-proB stage in both mice in adulthood (17). Although EBF expression is severely reduced in pre-proB cells derived from IL-7−/− mice, EBF expression is induced by exogenous IL-7 stimulation in IL-7−/− pre-proB cells (17). This result prompted us to examine whether the developmental arrest of IL-7−/− pre-proB cells can be released if IL-7−/− pre-proB cells are placed in an IL-7-sufficient environment.
Unexpectedly, IL-7−/− pre-proB cells do not give rise to B cells in the presence of IL-7 both in vivo and in vitro, even though IL-7R and its downstream signaling pathways are functional. Further analyses of IL-7−/− pre-proB cells and CLPs demonstrated that B cell potential is irreversibly lost at the pre-proB stage if IL-7 is not available during the transition period from CLP to pre-proB stage. However, ectopic EBF in IL-7Rα−/− pre-proB cells can initiate the stage transition to the proB stage. Therefore, IL-7 stimulation prior to the pre-proB stage is necessary for maintenance of B cell potential, which is ensured by proper EBF expression levels for further maturation to proB cells.
Materials and methods
Mice
The mice used in this study, such as IL-7−/− and RAG2−/− (CD45.1), are described in our previous publication (10). All mice were backcrossed onto C57BL/6 background for more than eight generations. Age-matched C57Bl/6 mice were used as WT control. The age of mice used in this study was between 8 and 12 weeks old. All mice were bred in a specific pathogen-free environment at the mouse facility of Duke University Medical Center. All experimental procedures related to laboratory mice were done according to guidelines specified by the institution.
Plasmids
MSCV-EBF-IRES-GFP was generated previously (17). pPax5-luc reporter construct was provided by Dr. O’Riordan, University of Michigan (20). 1.8 kb 5′ flanking region of the Pax5 gene (Genbank#AF148961) was cloned into the KpnI-XhoI site of pGL3-Basic (Promega).
For generation of EBF/ER cDNA, the coding region of EBF and ER was amplified by PCR with the following primers:
For EBF
5′-ATTGATACCGCGGACCACCATGTTTGGGATCCAGGAAAGCATCC-3′
5′-TATAAGAATTCCATGGGAGGGACAATCATGCCAG-3′
For ER
5′-ATATCAAGCTTCTAGATCGTGTTGGGGAAGCC-3′
5′-ATCGATAAGCTTGATCCACGAAATGAAATGGG-3′
EBF and ER amplicons were digested with EcoRI/SalI and HindIII, and cloned into SacII/EcoRI site and HindIII site of pBluescript SK, respectively (pBS-EBF/ER). Then EBF/ER cDNA was cloned into the SacII-XhoI site of pMSCV-IRES-GFP vector (MSCV-EBF/ER-IRES-GFP). Mouse EBF cDNA and ER mutant (G525R) cDNA (a gift from Dr. Zhuang, Duke University) were used as PCR templates.
Flow Cytometry
Preparation of single cell suspension and antibody staining of cells were done as previously described (10). The purity of doubly sorted cells was more than 99% as shown in Figure 1. Dead cells that were positively stained by propidium iodide (PI) (SIGMA-ALDRICH) were excluded from analysis and sorting. Cell sorting and cell surface phenotyping were performed on a FACSVantage SE with a DiVa option (488 nm argon, 599 nm dye and 408 nm krypton lasers, BD Bioscience Flow Cytometry Systems). Cell cycle analysis was done with a standard protocol. Briefly, FACS sorted cells were incubated with PI buffer containing 0.1% Triton X-100 (EM SCIENCE), 0.5 mg/ml RNaseA, and 50 μg/ml PI for 10 minutes on ice. After washing, PI staining was detected on a FACScan (488 nm argon laser, BD Bioscience Flow Cytometry Systems). Acquired data on FACS machines were analyzed with FlowJo software (Treestar).
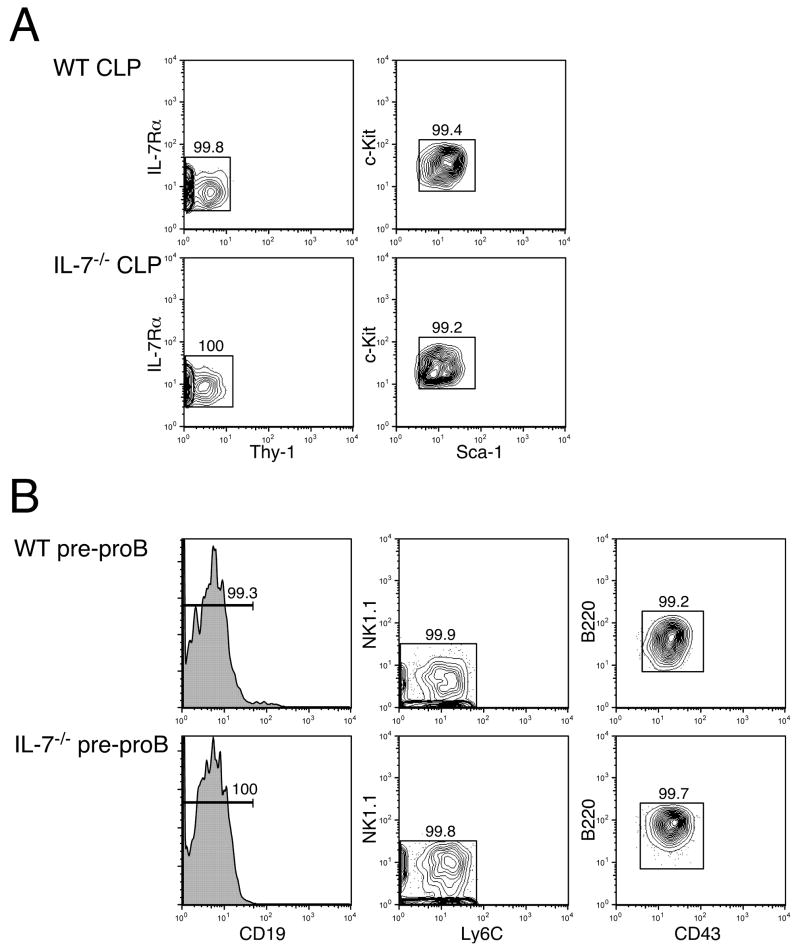
Figure 1. Purity of CLPs and pre-proB cells after sorting.
CLPs (A) and pre-proB cells (B) were doubly sorted as Lin−IL-7Rα+Thy-1−Sca-1loc-Kitlo and B220+CD43+CD19−NK1.1−Ly-6C−, respectively as shown previously (9, 17). After sorting, we examined the purity of the population on FACS. Shown FACS plots are pre-gated on the PI− fraction to exclude dead cells. Since both Lin+ and PI+ cells were gated out in the same PE/Cy5 channel in CLP sorting, Lin expression is not shown in (A). After sorting CLPs and pre-proB cells twice, the purity of the populations is over 99%.
Antibodies
Antibodies (Abs) purchased from BD Biosciences Pharmingen were the following: PE-anti-CD43 (S7), FITC-anti-Ly-6C (AL-21), APC- or PE-Cy7-anti-CD19 (1D3), APC-anti-CD11c (HL3), PE-Cy7-anti-NK1.1 (PK136), and biotin-anti-cytokine common γ chain (TUGm2). Abs purchased from eBioscience were the following: FITC-anti-CD90.1 (Thy-1.1, HIS51), PE-anti-CD19 (6D5), PE-anti-CD40 (1C10), PE-anti-CD86 (B7-2, GL1), APC-anti-CD117 (c-Kit, 2B8), PE-anti-Cy5-CD3ε(145-2C11), FITC- or PE-Cy5-anti-Ly-6G (Gr-1, RB6-8C5), PE-Cy5-anti-TER119, PE-Cy5- or APC-Cy7-anti-CD11b (Mac-1, M1/70), PE-, PE-Cy7-, APC-Cy7- or PE-Cy5-anti-B220 (RA3-6B2), PE-Cy5-anti-CD4 (L3T4), PE-Cy5-anti-CD8a (Ly-2), biotin-anti-CD45.2 (104), biotin-anti-CD80 (B7-1, 16-10A1), biotin-anti-MHC Class II (M5/114.15.2), and biotin-anti-CD127 (IL-7Rα, A7R34). Texas Red (TxR)-goat anti-mouse Igμ Ab (Southern Biotech) was used for surface IgM staining. Anti-Sca-1 (E13-161-7) was purified from culture supernatant and conjugated with Alexa Fluor 594 by a standard procedure in our laboratory. Streptavidin-APC-Cy7 (eBioscience) and Avidin-TxR (BD Biosciences Pharmingen) were used for visualizing biotinylated antibodies. Apoptotic cells were detected by using Annexin V-PE (BD Biosciences Pharmingen).
Reconstitution assay
CLPs or pre-proB cells were purified from either WT (CD45.2+) or IL-7−/− mice (CD45.2+) and injected into the retroorbital venous sinus of sub-lethally irradiated (400 rad) RAG2−/− mice (CD45.1+). Mice were sacrificed 2 weeks after injections and splenocytes were examined by FACS.
Cell culture
Monolayers of OP9 or PA6 cells were prepared in 96-well flat bottom plates (BD Falcon) 1 day before the initiation of the culture. Cells were cultured with Iscove’s Modified Dulbecco’s Medium (IMDM) (GIBCO) supplemented with 5% FCS, 50 μM 2ME, and antibiotics (complete medium). IL-7, Flt3L, and SCF were purchased from R&D Systems. For the limiting dilution assays, the indicated number of cells in the figure was directly sorted into 96-well plates after the first sorting using the ACDU option on the FACSVantage. To stimulate pre-proB cells, FACS sorted cells were incubated in 96-well round bottom plates with the complete medium in the presence or absence of IL-7 for 12 hrs. 293T cells were cultured in DMEM (GIBCO) with 10% FCS and antibiotics.
PCR
RNA purification and first-strand DNA synthesis were done as described previously (17). Briefly, cells were sorted directly into 1.5 ml microcentrifuge tubes with 1 ml TRIzol reagent (Invitrogen). Total RNA was purified based on the manufacture’s instructions. First-strand cDNA was synthesized with Superscript III reverse transcriptase (RT) and oligo-dT primers (Invitrogen). PCR was done with BD Advantage 2 PCR Enzyme System and primer sets described below on GeneAmp PCR System 9700 (Applied Biosystems). Verification of the amount of first strand cDNA was done by amplification of β-actin or GAPDH. The sequences and conditions for the β-actin, GAPDH, EBF and IL-7 primers were described elsewhere (17, 25). The PCR primers for Jak1, Jak3, and Stat5A were as follows. Jak1: forward, 5′-AGAACCTGAGTGTGGCTGCT-3′; reverse, 5′-TGTTGTTGGCTGCTTTTCTG-3′. Jak3: forward, 5′-ATGTGTCTCACCATCCACGA-3′; reverse, 5′-AATTCTGGGCTGCGAGTAGA-3′. Stat5A: forward, 5′-GTGAAGCCACAGATCAAGCA-3′; reverse, 5′-GGAGGTGAAGAGACCAGCAG-3′. The annealing temperature for these primers was 61°C. Forward and reverse primers are not located in the same exon so that bands derived from genomic contamination can be excluded by the size. The primers for CIS were previously described (15). EBF expression level was quantified by using MyiQ™ (BioRad) after first strand DNA synthesis. The amount of first strand DNA applied was normalized by the expression level of GAPDH or β2-microglobulin. The sequence of the primers for β2-microglobulin is as follows: forward, 5′-ACCGGCCTGTATGCTATCCAGAAA-3′; reverse, 5′-GGTGAATTCAGTGTGAGCCAGGAT-3′. The same conditions used for GAPDH were also used for β2-microglobulin. The primer sequences and PCR conditions for EBF and GAPDH were previously described (17).
Reporter gene assay
293T cells were transfected with plasmids indicated in the figures with FuGENE 6 transfection reagent (Roche). After transfection, cells were further cultured for 24 hrs in the presence or absence of 4-HT and harvested. Cell lysates were prepared and the luciferase activity was measured.
Results
Pre-proB cells derived from IL-7−/− mice have lost B cell differentiation potential
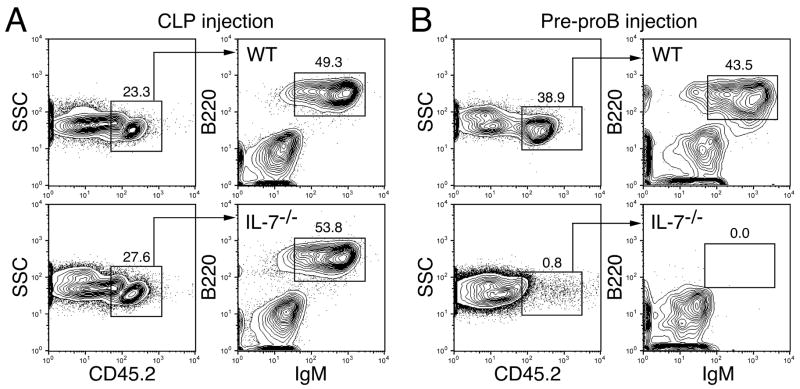
We previously showed that the developmental switch from IL-7-independent fetal type B cell development to IL-7-dependent adult type B cell development occurs at the hematopoietic stem cell (HSC) level between 1 and 2 weeks after birth (26). However, we occasionally observed sporadic B cell development from IL-7Rα−/−HSCs derived from 3–5 week old mice. Therefore, we used IL-7−/− and IL-7Rα−/−mice that were 8 weeks of age or older throughout this study. First, we intravenously injected IL-7−/− pre-proB cells into sub-lethally irradiated RAG2−/−mice in order to test whether pre-proB cells derived from IL-7−/− mice can develop into mature B cells in an IL-7 sufficient condition in vivo. Under these conditions, we could detect donor-derived B cells from both wild type (WT) and IL-7−/− CLPs in the recipient spleens at 2 weeks after injection (Figure 2A). However, no donor-derived B cells were observed in the mice injected with IL-7−/− pre-proB cells (Figure 2B, bottom panels) although WT pre-proB cells gave rise to mature B cells (Figure 2B, top panels). No mature B cells from IL-7−/− pre-proB cells were detected in the host mice even at later time points (data not shown).
Figure 2. In vivo B cell potential of CLPs and pre-proB cells derived from IL-7−/− mice. B cell.
potential of CLPs (A) and pre-proB cells (B) was examined by in vivo reconstitution assay. CLPs (2.0×103) and pre-proB cells (1.4×104) derived from WT or IL-7−/− mice (CD45.2+) were injected into sub-lethally irradiated RAG2−/− mice (CD45.1+). B cell readout was analyzed in the spleen of host mice at 2 weeks post injection. Representative data from three independent experiments were shown. The mean ± S.D. from a total of 5–6 reconstituted mice in each group was calculated and indicated in the FACS plots.
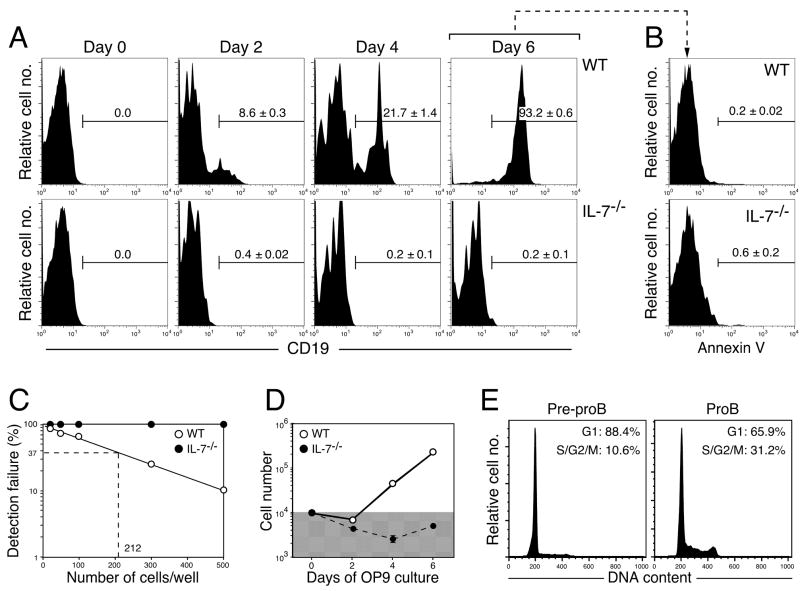
Next, we cultured WT and IL-7−/− pre-proB cells on OP9 stromal cells with IL-7, Flt3L, and SCF in order to examine whether excess amounts of IL-7 can support maturation of IL-7−/− pre-proB cells in vitro. After 6 days of culture, almost all WT pre-proB cells differentiated into CD19+ proB cells (Figure 3A, top panels). However, IL-7−/− pre-proB cells failed to give rise to proB cells throughout the culture period (Figure 3A, bottom panels). The lack of proB cell differentiation is not due to programmed cell death caused by insufficient cytokine receptor signaling since we could not detect significant annexin V+ pro-apoptotic cells in the culture with either WT or IL-7−/− pre-proB cells at day 6 (Figure 3B).
Figure 3. B cell potential of IL-7−/− pre-proB cells in in vitro stromal cell cultures.(A).
Pre-proB cells (1.0×104 cells/well) derived from WT (upper panels) and IL-7−/− mice (lower panels) were cultured on OP9 stromal cells in the presence of IL-7, SCF, and Flt3L for the period shown in the figures. Representative FACS plots from two independent experiments were shown. The mean ± S.D. from more than 6 samples from various time points was indicated in the FACS plots. CD19+ cells in the plots represent proB cells. (B) Apoptotic cells were examined by annexin V staining at 6 days after culture. Stromal cells were excluded by scatter gates. PI exclusion was not done in this assay. (C) The frequency of cells that can give rise to proB cells in the pre-proB population in WT and IL-7−/− pre-proB cells. Indicated numbers of pre-proB cells from either WT (open circle) or IL-7−/− pre-proB cells (closed circle) were cultured in 96 well plates as described in the legend for Figure 2A. Wells containing B220+CD19+ cells were counted as a positive well. We did not observe any CD19+ proB cells from IL-7−/− pre-proB cells even at 1.0×103 cells/well, where all wells seeded with WT pre-proB cells contained proB cells (the limiting number: 1 in 212). (D) Cell numbers in the culture of pre-proB cells. Cell numbers in the culture shown in Figure 2A were counted with a hemocytometer under the microscope. OP9 stromal cells were excluded from the counting based on the difference in cell size. The shaded area in the graph indicates the numbers below the input cell number (1×104). (E) Cell cycle status of pre-proB and proB cells in WT mice was analyzed on FACS.
We performed more comprehensive analysis to determine B cell potential of IL-7−/− pre-proB cells by limiting dilution assay. As shown in Figure 2C, IL-7−/−pre-proB cells did not produce any CD19+ progenies in any of the cell concentrations tested (up to 1×103 cells/well), suggesting that IL-7−/− pre-proB cells have no B cell potential, which is virtually consistence with the results obtained by Dias et al (16). Since cell numbers were not increased in IL-7−/− pre-proB cell cultures (Figure 3D), the cell-intrinsic defect in proliferation might account for the lack of proB cell development from IL-7−/− pre-proB cells. However, the analysis of cell cycle status of WT pre-proB cells showed that pre-proB cells are physiologically quiescent cells compared to proB cells (Figure 3E). These data suggest that although hematopoietic progenitors can give rise to pre-proB cells in the absence of IL-7, IL-7 stimulation prior to the pre-proB stage is necessary for lymphoid progenitors to maintain B cell potential.
IL-7R is functional in IL-7−/− pre-proB cells
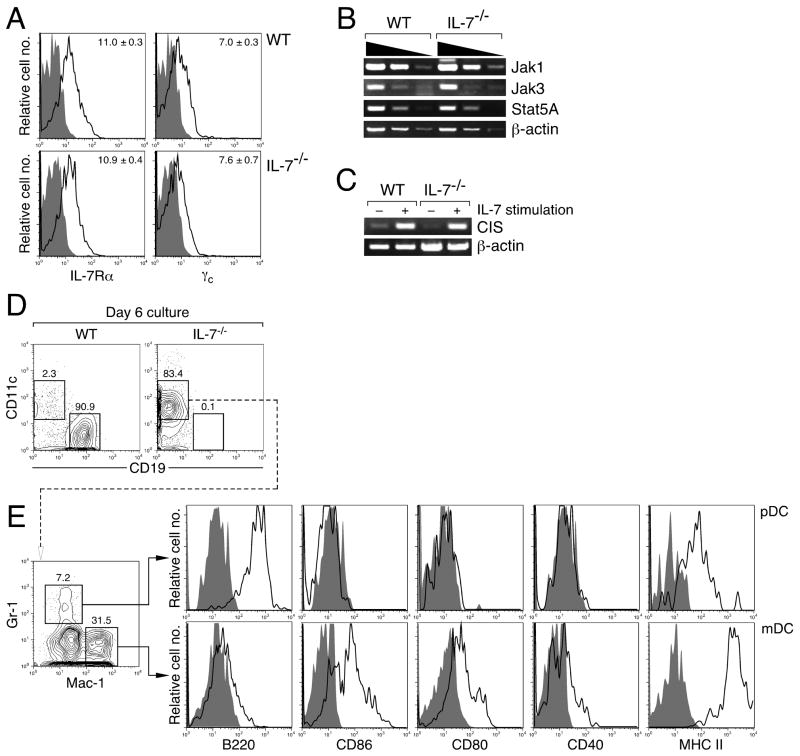
As we previously reported (17), IL-7Rα is normally expressed on IL-7−/− pre-proB cells (Figure 4A, top panels). Expression of γc was also comparable between WT and IL-7−/− pre-proB cells (Figure 4A, bottom panels). Essential components of IL-7R signaling, such as Jak1, Jak3, and Stat5A, were also comparably expressed between WT and IL-7−/− pre-proB cells (Figure 4B). These data demonstrate that the most upstream signaling components in the IL-7R system are intact in IL-7−/− pre-proB cells. Next, we examined whether the IL-7R in IL-7−/−pre-proB cells can actually transmit signals in response to IL-7. For this purpose, we examined expression of CIS, a target of Stat5 (15, 27) in pre-proB cells upon IL-7 stimulation. Prior to IL-7 stimulation, pre-proB cells purified from WT and IL-7−/− mice were cultured without IL-7 for 12 hrs. Two hours after IL-7 stimulation, similar levels of CIS upregulation was observed in both WT and IL-7−/− pre-proB cells (Figure 4C), suggesting that the IL-7R/Jak/Stat5 pathway is fully functional in IL-7−/− pre-proB cells.
Figure 4. IL-7R/Jak/Stat signaling pathway is intact in IL-7−/− pre-proB cells.
(A) IL-7Rα and γc expression were examined in WT and IL-7−/− pre-proB cells. Open histograms represent the expression of IL-7Rα or γc. The shaded histograms represent the negative control stained with isotype-matched irrelevant antibodies. The number in the plot indicates the mean fluorescence intensity (MFI) value. The mean ± S.D. from 4 mice was calculated and indicated in the FACS plots. (B) Expression of essential components of IL-7R signaling was examined by semi-quantitative RT-PCR. WT or IL-7−/− pre-proB cells (1.5×104) were used for RNA purification. Synthesized cDNA was serially diluted by 5-fold and used for PCR amplification of each gene indicated. (C) CIS is normally upregulated in both WT and IL-7−/− pre-proB cells after IL-7 stimulation. Pre-proB cells (2.0×104) were purified from either WT or IL-7−/− mice and pre-cultured in complete medium in the absence of IL-7 for 12 hrs. After this cytokine starvation, cells were further incubated with or without 50 ng/ml IL-7 for 2 hrs. After RNA purification and cDNA synthesis, CIS expression was examined by semi-quantitative PCR. (D) CD11c and CD19 expression after 6 day culture of WT or IL-7−/− pre-proB cells. (E) Expression of various DC markers on the CD11c+ cells derived from IL-7−/− pre-proB cells. Open histograms represent the expression level of various markers and the shaded histograms represent the negative control stained with isotype-matched irrelevant Abs. Representative data from two independent experiments were shown.
Next, we examined whether CD19− cells from IL-7−/− pre-proB cell cultures at day 6 (Figure 3A) remained undifferentiated pre-proB cells or gave rise to other cell types. We found that a majority of cells from IL-7−/− pre-proB cells following culture in the absence of IL-7 were positive for CD11c (Figure 4D), which is a common marker for dendritic cells (28). Our preliminary data suggest that both WT and IL-7−/− pre-proB cells express low levels of CD11c in the same manner (data not shown). Therefore, we investigated the identity of the CD11c+ cells derived from IL-7−/− pre-proB cells after the culture. Further analyses of various markers including B220, Gr-1, Mac-1, MHC class II, and co-stimulatory molecules showed that CD11c+ cells derived from IL-7−/− pre-proB cells contain plasmacytoid DCs (pDCs) and myeloid DCs (mDCs) (29, 30) (Figure 4E). The mDCs in IL-7−/− pre-proB cell culture produced TNFα upon LPS stimulation (data not shown). These data suggest that IL-7−/− pre-proB cells that lack B cell potential still maintain DC potential. Previously, it was reported that DC progenitors are present within the B220+CD43+c-Kit− fraction (31), which overlaps with the pre-proB cell population. These DC progenitors, however, do not express IL-7Rα, which is expressed on pre-proB cells (Figure 4A). In addition, no significant difference in gene expression profiling was found between WT and IL-7−/− pre-proB cells by gene chip assays, except for the lack of B lineage signature genes in IL-7−/− pre-proB cells (data not shown). Therefore, the lack of IL-7 stimulation prior to the pre-proB stage leads to irreversible loss of B cell potential, but retention of DC potential.
B cell development from IL-7−/− CLPs in vitro is reduced but not completely diminished
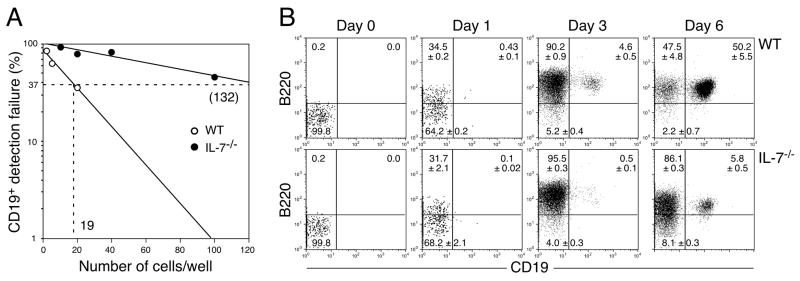
Although we observed normal B cell development from IL-7−/− CLPs in vivo, a dramatic reduction of B cell potential in IL-7−/− CLP was reported using the OP9 stromal cell culture system (16). Thus, we also examined B cell development from CLPs in vitro. We sorted CLPs either from WT or IL-7−/− mice and cultured the cells on OP9 stromal cells in the presence of IL-7, Flt3L, and SCF. Consistent with the previous study (16), the calculated limiting number of IL-7−/−CLPs to produce proB cells were obviously increased (from 1 in 19 to 132, Figure 5A). However, we observed clear proB cell differentiation from IL-7−/− CLPs by 6 days after culture (Figure 5B). These results suggest that B cell potential is still present, albeit at a much lower efficiency, at the CLP stage while B cell potential is completely lost at the subsequent pre-proB stage in the absence of IL-7.
Figure 5. B cell potential of IL-7−/− CLPs.
in vitro. (A) The frequency of cells that give rise to CD19+ proB cells in the CLP population derived from WT and IL- 7−/− mice. Various numbers of CLPs from WT and IL-7−/− mice were plated in wells of 96 well plates and cultured as described in Figure 2A. Wells containing B220+CD19+ cells were counted as a positive well. The limiting numbers in this assay are 1 in 19 for WT pre-proB cells and 1 in 132 for IL-7−/− pre-proB cells. (B) The kinetic analysis of B cell development from WT or IL-7−/− CLPs. CLPs (2×103 cells/well) from either WT or IL-7−/− mice were cultured as described in Figure 3A. In the plots, B220−CD19−, B220+CD19−, and B220+CD19+ cells are CLP, pre-proB, and proB cells, respectively. The period of time in the culture is indicated at the top of the FACS plots. Reanalysis of freshly isolated CLPs before culture was shown as day 0. The mean percentage ± S.D. from more than 6 samples from 2 independent experiments was indicated in the FACS plots.
IL-7 stimulation at the transition from CLPs to pre-proB cells is necessary to maintain sufficient EBF expression levels for further maturation
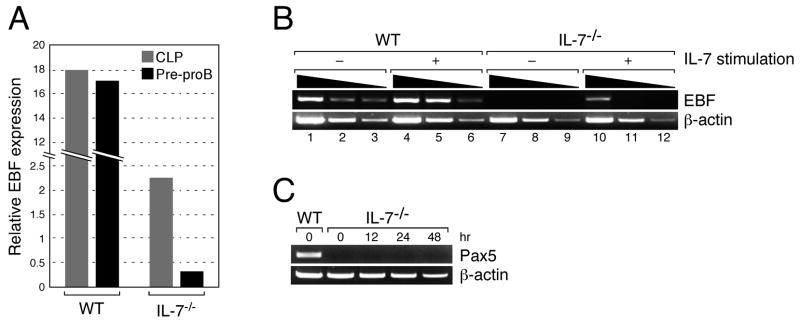
To examine the role of IL-7 during the transition from the CLP to pre-proB stage, we purified CLPs and pre-proB cells from either WT or IL-7−/− mice and examined the EBF expression level in these cells. As previously reported (16), EBF expression was reduced in IL-7−/− CLPs compared to WT CLPs (Figure 6A). When EBF expression was compared between CLPs and pre-proB cells in IL-7−/−mice, the reduction of EBF expression in pre-proB cells was significant. In contrast, the level of EBF expression was comparable between CLPs and pre-proB cells in WT mice (Figure 6A), suggesting that IL-7 stimulation is necessary for maintaining EBF expression during the transition from CLPs to pre-proB cells during B cell development.
Figure 6. EBF expression level in WT and IL-7−/− pre-proB cells.
(A) EBF expression in the CLP (grey bar) and pre-proB (black bar) populations derived from WT or IL-7−/− mice was examined by quantitative PCR. EBF expression in whole BM was arbitrary defined as unit one and the mean value of three independent samples is shown. The same results were obtained by two independent experiments. (B) EBF expression in WT and IL-7−/− pre-proB cells (2.0×104) was examined before and after IL-7 stimulation by semi-quantitative PCR. After RNA purification and first strand synthesis, cDNAs were serially diluted by 5-fold and subjected to PCR. ProB cell markers such as CD19 and BP-1 were not upregulated during the culture (17). (C) Pax5 expression in IL-7−/−pre-proB cells after IL-7 stimulation. IL-7−/− pre-proB cells were stimulated with IL-7 as described above for the period indicated in the figure. Pax5 expression in each sample was measured by RT-PCR. β-actin expression was also examined as a loading control. WT pre-proB cells were used as a positive control for Pax5.
Next, we examined the degree of EBF upregulation in pre-proB cells after IL-7 stimulation. EBF expression was upregulated in IL-7−/− pre-proB cells upon IL-7 stimulation for 12 hrs as we previously reported (Figure 6B, lane 10) (17). However, the level of EBF expression was lower than that of either IL-7 stimulated or unstimulated WT pre-proB cells (Figure 6B, compare lane 10 to lanes 1 and 4). Moreover, expression of Pax5, a gene target of EBF, was not observed in IL-7−/− pre-proB cells upon IL-7 stimulation at any of the time points tested (Figure 6C). These data suggest that the EBF expression level in IL-7−/−pre-proB cells after IL-7 stimulation is insufficient to drive further B cell development.
Required EBF expression levels in pre-proB cells for transition to the proB stage
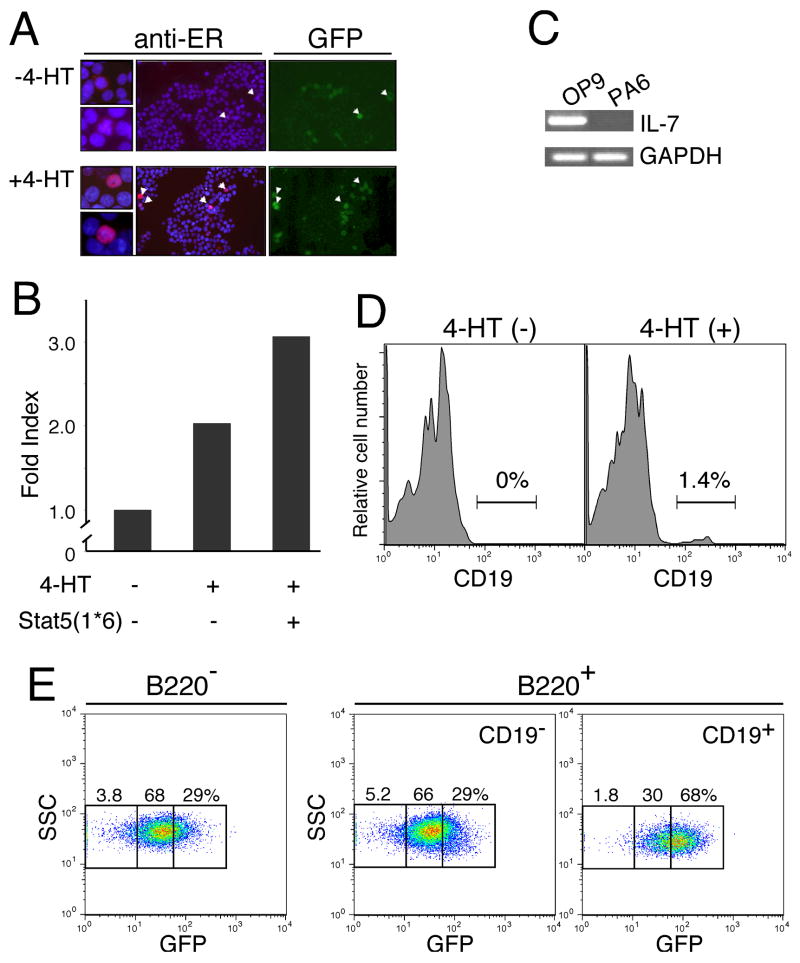
As shown in Figures 1 and 2, IL-7−/− pre-proB cells have lost B cell potential. Based on the fact that the level of EBF expressed in IL-7−/− pre-proB cells after IL-7 stimulation is not sufficient for further maturation, we reasoned that there were two different models that could explain the loss of B cell potential in IL-7−/− pre-proB cells. The first model suggests that the stage transition from pre-proB to proB is blocked by insufficient EBF expression, although other B cell developmental programs in IL-7−/− pre-proB cells are intact. The other model suggests that in addition to EBF, other B cell developmental programs are shut down in IL-7−/− pre-proB cells. To address this issue, we generated an EBF-estrogen receptor (ER) fusion protein (EBF-ER). EBF-ER cDNAs were cloned into a retroviral vector, MSCV-IRES-GFP. EBF-ER was expressed in the cytoplasm of cells after introduction (Figure 7A). EBF-ER proteins translocated to the nucleus in a 4-hydroxytamoxifen (4-HT) dependent manner, similar to the previously reported case for STAT5-ER (32). We confirmed that EBF-ER is functional only when 4-HT is present by measuring the promoter activity of the Pax5 gene (Figure 7B).
Figure 7. EBF plays an indispensable role in the transition from the pre-proB to proB stage.
(A) 293T cells were infected with recombinant viruses with MSCV-EBF-ER-IRES-GFP and cultured in the absence (top) and presence (bottom) of 4-HT (1 μM). Only GFP+ (green) cells were brightly stained with anti-ER (red) as indicated by arrowheads. Therefore, only EBF-ER was detected by anti-ER staining in this figure. EBF-ER was located in the cytoplasm and nucleus in the absence and presence of 4-HT as shown in the left panels. Nuclei were stained with DAPI (blue). (B) EBF activity was measured by the reporter assay with pPax5-luc in 293T cells. EBF can positively regulate the promoter activity of the Pax5 gene (37). This EBF function was observed only when 4-HT was added into the culture. Synergistic effects of EBF-ER with the constitutively active form of Stat5 (Stat5(1*6)) (45) was also observed in the presence of 4-HT. (C) IL-7 expression in OP9 and PA6 cells was examined by RT-PCR. (D) Maturation of pre-proB cells to proB cells in the presence of ectopic EBF and absence of IL-7Rα. After introduction of EBF-ER into MPPs derived from IL-7Rα−/− mice, EBF-ER+ pre-proB cells were purified from EBF-ER+ MPPs that had been cultured for 4 days. EBF-ER+ pre-proB cells were further cultured for 2–3 days in the absence (left) and presence (right) of 4-HT (0.3 μM). A representative result from at least 5 experiments is shown. (E) EBF expression levels required for the transition from the pre-proB to proB stage. VCAM-1+ MPPs (IL-7Rα−) from WT mice were infected with recombinant retrovirus derived from MSCV-EBF-IRES-GFP vectors (17). After infection, GFP+ cells were purified in order to reduce the number of non-infected cells. These GFP+ cells were cultured for 3 days on PA6 stromal cells in the presence of SCF and Flt3L. PA6 stromal cells do not produce any IL-7 (36). GFP expression levels in B220+CD19− pre-proB cells (center panel) and B220+CD19+ proB cells (right panel) as well as B220−CD19−cells (left panel), most of which were Mac-1+ myeloid cells, were examined by FACS.
We found that pre-proB cells are resistant to not only retroviral infection but also lentiviral infection in our experimental system. Therefore, we introduced EBF-ER into multipotent progenitors (MPPs) derived from IL-7Rα−/− mice by using a retroviral system and purified pre-proB cells after culturing EBF-ER+ MPPs in the absence of 4-HT. After an additional 2–3 days of culturing EBF-ER+ pre-proB cells, the appearance of CD19+ proB cells was only observed following the addition of 4-HT to the culture (Figure 7D). Only a small percentage (ranging from 0.5–2.5% in independent cultures) of CD19+ cells were obtained from pre-proB cells in this experimental setting, perhaps in part due to the suboptimal concentration of 4-HT (0.3 μM), which does not induce significant cell death. The optimal dose of 4-HT (1 μM) (33) is toxic to lymphoid progenitors because sex steroids, such as estrogen, induce apoptosis (34). Nevertheless, this result suggests that IL-7Rα−/− pre-proB cells can give rise to proB cells if cells have sufficient amounts of EBF.
We further examined the dosage effect of EBF in the transition from the pre-proB to proB stage. We introduced EBF into VCAM-1+ MPPs (35) from WT mice by using a retroviral system with MSCV-EBF-IRES-GFP vectors (17). Since GFP expression is correlated with the amount of mRNA transcribed in the cells, we can monitor EBF expression levels via levels of GFP expression in this system. After infection with recombinant EBF viruses, we purified GFP+ cells in order to deplete MPPs without ectopic EBF. EBF+ MPPs were further cultured on PA6 stromal cells in the presence of SCF and Flt3L. Since PA6 does not produce any IL-7 (Figure 7C) (36), IL-7 is completely absent in this culture system. Additionally, VCAM-1+ MPPs are negative for IL-7R. Therefore, cells in this experiment were virtually free from the effects of IL-7. After 4–6 days of culture, we detected B220+CD19− (pre-proB) and B220+CD19+ (proB) cells from MPPs (data not shown). Although GFP expression in B220+CD19− pre-proB cells was not significantly different from B220−CD19− cells, which were predominantly myeloid cells, B220+CD19+ proB cells had higher GFP expression than B220+CD19− pre-proB cells (Figure 7E). This result directly demonstrates the presence of a threshold level of EBF expression which determines pre-proB cell fate; whether the cells can mature (higher than the threshold) or not (lower than the threshold).
Dispensability of IL-7 in the transition from the pre-proB to proB stage
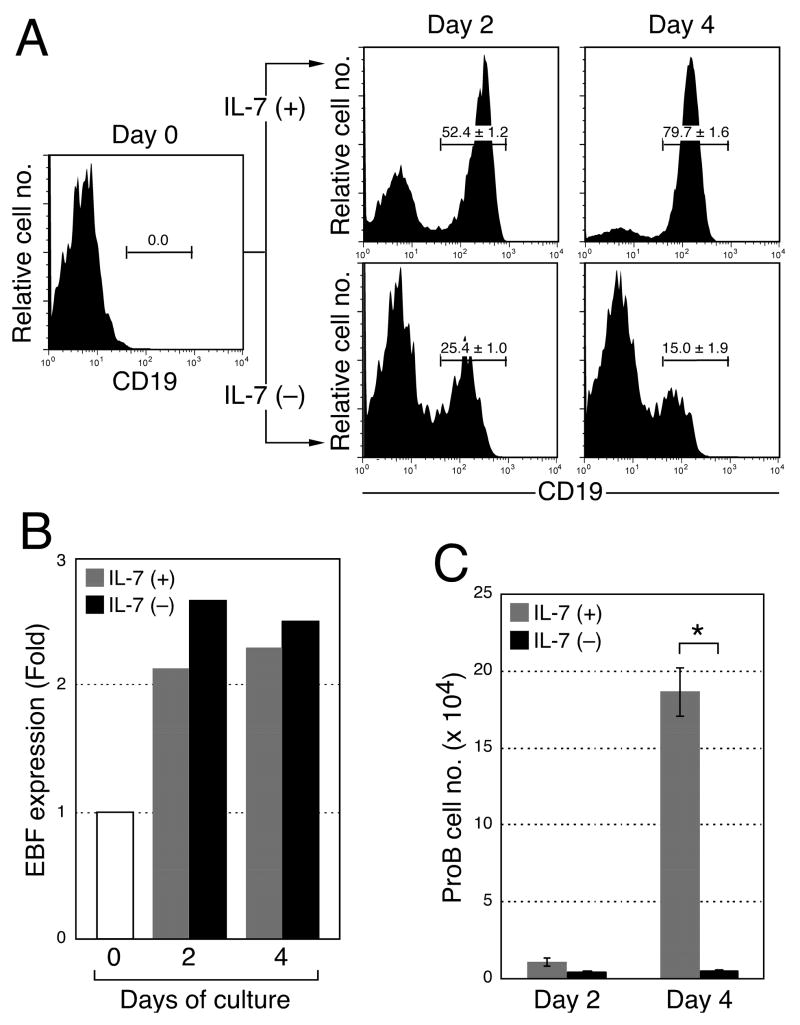
Since we obtained pre-proB and proB cells from in vitro cultures of MPPs without IL-7 by enforced EBF expression, we further examined the requirement of IL-7 in the pre-proB to proB cell transition. We hypothesized if pre-proB cells express EBF higher than the threshold level, IL-7 may be dispensable for the transition from the pre-proB to proB stage. To test our hypothesis, we sorted pre-proB cells from WT mice and cultured the cells on PA6 stromal cells in the presence or absence of IL-7. In both cultures, CD19+ proB cell readout was clearly detected in 2 days, although CD19 expression level was slightly lower in proB cell cultures without IL-7 (Figure 8A). EBF expression in proB cells is higher than pre-proB cells in WT mice as reported previously (17). In fact, EBF expression was upregulated irrespective of IL-7 after the culture (Figure 8B). Therefore, IL-7 is not necessary for the stage transition from pre-proB to proB as long as pre-proB cells express sufficient quantities of EBF. Also these data suggest that the EBF level in WT pre-proB cells is at or above the threshold required for maturation of pre-proB cells. The proB cell numbers after culture in the absence of IL-7 were dramatically reduced compared to the culture in the presence of IL-7 (Figure 8C). Accordingly, if pre-proB cells express enough EBF, the role of IL-7 is not to support the stage transition from pre-proB to proB, but rather to expand the B cell pool at the proB stage.
Figure 8. Dispensability of IL-7 in the transition from the pre-proB to proB stage.
(A) Requirement of IL-7 in the stage transition from the pre-proB to proB stage. Pre-proB cells were purified from WT mice and cultured on PA6 in the presence or absence of IL-7. Although CD19 expression was observed after the culture in the absence of IL-7, CD19 expression levels were constantly lower than the cells cultured with IL-7. The mean ± S.D. from more than 6 samples from two independent analyses were indicated in the FACS plots. (B) EBF expression level was examined in proB cells derived from pre-proB cells after the culture shown in Figure 6C. B220+CD19+ proB cells were sorted from the culture of WT pre-proB cells in the presence (grey) and absence (black) of IL-7. EBF expression was examined by quantitative RT-PCR. The fold expression of EBF in proB cells was calculated against the EBF expression in WT pre-proB cells (day 0). (C) ProB cell numbers after the culture in the presence (grey) and absence (black) of IL-7. The bars are shown as means of triplicate wells ± S.D. *, p < 0.001, calculated by student’s t test.
Discussion
We previously showed that enforced expression of EBF can restore B cell differentiation from IL-7Rα−/− HSCs (17). We demonstrated in this present study that IL-7 stimulation is necessary during the transition from CLP to pre-proB cell stage in order to maintain the necessary EBF expression level that drives further B cell development at the pre-proB stage. B cell development can proceed to the pre-proB stage from HSCs in the absence of IL-7. However, pre-proB cells derived from IL-7−/− mice have completely lost B cell potential (Figures 2 and 3) because IL-7 stimulation cannot upregulate EBF to the level observed in WT pre-proB cells (Figure 6B). MPPs with higher levels of ectopic EBF can preferentially give rise to CD19+ proB cells from CD19− pre-proB cells (Figure 7B). In addition, we recently found that fetal liver pre-proB cells derived from IL-7Rα−/− mice maintain EBF expression at a level comparable to WT pre-proB cells in adult bone marrow (26). Since fetal B cell development is IL-7/IL-7Rα independent, this observation further implicates the importance of EBF expression levels at the pre-proB cell stage for further developmental progression. Based on these results, we conclude that the threshold level of EBF to advance B cell differentiation is higher than the EBF level found in IL-7−/− pre-proB cells after IL-7 stimulation and similar to the level of adult WT pre-proB cells.
Currently, it is not clear why the maintenance of EBF expression by IL-7 at the stage between CLPs and pre-proB cells is so critical. It seems that the EBF gene is regulated by a positive auto-regulatory loop, since EBF can bind and activate its own promoter (37, 38). Thus, the amount of EBF maintained by IL-7 at the transition from CLPs to pre-proB cells might be required to establish the positive regulatory loop of EBF expression. In this case, only pre-proB cells that successfully maintain stabilized basal EBF expression through the activation loop may advance to the proB cell stage, where further EBF upregulation occurs independent of IL-7 (Figure 8).
The inability of IL-7−/− pre-proB cells to upregulate EBF to the level in WT pre-proB cells in response to IL-7 suggests that IL-7 stimulation between CLPs and pre-proB cells may be necessary for protecting the positive regulatory elements of the EBF gene from gene silencing by nucleotide modifications such as methylation (39). In support of these findings, we found a typical CpG island at the 5′ flanking region of the EBF gene by computational analysis (data not shown). Although we demonstrate in this paper that the maintenance of EBF expression is regulated by IL-7, the mechanism of the initiation of EBF expression during lymphopoiesis remains unclear. Since IL-7−/− CLPs express EBF at a low level, initiation of EBF expression in lymphoid-lineage primed VCAM-1− MPPs (40) and/or lymphoid-lineage committed CLPs should occur in an IL-7-independent manner (9). Stat5 is involved in IL-7 mediated EBF expression in pre-proB cells (37). Since a previous study showed that Stat5 is activated by Flt3 in some cell lines (41) and both MPPs and CLPs express Flt3 (35, 42), Flt3 signaling may trigger the expression of EBF. To fully understand the function of IL-7 in the regulation of EBF expression, more careful analysis of the EBF promoter must be carried out.
Involvement of cytokines in cell fate decisions has been observed in many studies (17, 43, 44). In myeloid development, granulocyte colony-stimulating factor signaling has been shown to support the neutrophil cell fate by increasing the relative expression of C/EBPα to PU.1 expression (43). In contrast, the results of this study suggest that the role of IL-7 in B cell differentiation at the transition stage from CLP to pre-proB is to maintain sufficient EBF expression levels for further maturation. The fact that IL-7−/− pre-proB cells cannot express high levels of EBF similar to WT pre-proB cells implies that IL-7 stimulation prior to the pre-proB stage is necessary for maintaining EBF promoter activity intact in pre-proB cells. This novel mode of cytokine function clarified in this study should provide us with new insights into how cytokines regulate cell differentiation during hematopoiesis and lymphopoiesis.
Acknowledgments
This work was supported by the Duke Stem Cell Research Program Annual Award and NIH grants AI056123 and CA098129 to M.K., and NIH grant AI52077 to A.Y.L. M.K. is a scholar of the Leukemia & Lymphoma Society.
We thank Drs. O’Riordan and Grosschedl for the pPax5-luc plasmid and Dr. Zhuang for ER cDNA. We also thank Ms. Martinek and Dr. Cook for great help with FACS sorting and excellent maintenance of the machine; and Ms. Chung for critically reviewing the manuscript.
Abbreviations
- γc
cytokine receptor common γ chain
- EBF
early B cell factor
- WT
wild type
- HSC
hematopoietic stem cell
- ER
estrogen receptor
- 4-HT
4-hydroxytamoxifen
Footnotes
Disclosures
The authors have no conflicting financial interests.
References
- 1.Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohbo K, Suda T, Hashiyama M, Mantani A, Ikebe M, Miyakawa K, Moriyama M, Nakamura M, Katsuki M, Takahashi K, Yamamura K, Sugamura K. Modulation of hematopoiesis in mice with a truncated mutant of the interleukin-2 receptor gamma chain. Blood. 1996;87:956–967. [PubMed] [Google Scholar]
- 4.Leonard WJ, Shores EW, Love PE. Role of the common cytokine receptor gamma chain in cytokine signaling and lymphoid development. Immunol Rev. 1995;148:97–114. doi: 10.1111/j.1600-065x.1995.tb00095.x. [DOI] [PubMed] [Google Scholar]
- 5.Cao X, Shores EW, Hu-Li J, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 6.DiSanto JP, Muller W, Guy-Grand D, Fischer A, Rajewsky K. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc Natl Acad Sci U S A. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugamura K, Asao H, Kondo M, Tanaka N, Ishii N, Ohbo K, Nakamura M, Takeshita T. The interleukin-2 receptor gamma chain: its role in the multiple cytokine receptor complexes and T cell development in XSCID. Annu Rev Immunol. 1996;14:179–205. doi: 10.1146/annurev.immunol.14.1.179. [DOI] [PubMed] [Google Scholar]
- 8.Maraskovsky E, O’Reilly LA, Teepe M, Corcoran LM, Peschon JJ, Strasser A. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1−/− mice. Cell. 1997;89:1011–1019. doi: 10.1016/s0092-8674(00)80289-5. [DOI] [PubMed] [Google Scholar]
- 9.Kondo M, I, Weissman L, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 10.Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 11.Kondo M, Akashi K, Domen J, Sugamura K, Weissman IL. Bcl-2 rescues T lymphopoiesis, but not B or NK cell development, in common gamma chain-deficient mice. Immunity. 1997;7:155–162. doi: 10.1016/s1074-7613(00)80518-x. [DOI] [PubMed] [Google Scholar]
- 12.von Freeden-Jeffry U, Solvason N, Howard M, Murray R. The earliest T lineage-committed cells depend on IL-7 for Bcl-2 expression and normal cell cycle progression. Immunity. 1997;7:147–154. doi: 10.1016/s1074-7613(00)80517-8. [DOI] [PubMed] [Google Scholar]
- 13.Maraskovsky E, Peschon JJ, McKenna H, Teepe M, Strasser A. Overexpression of Bcl-2 does not rescue impaired B lymphopoiesis in IL-7 receptor-deficient mice but can enhance survival of mature B cells. Int Immunol. 1998;10:1367–1375. doi: 10.1093/intimm/10.9.1367. [DOI] [PubMed] [Google Scholar]
- 14.Corcoran AE, Riddell A, Krooshoop D, Venkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- 15.Bertolino E, Reddy K, Medina KL, Parganas E, Ihle J, Singh H. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat Immunol. 2005;6:836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 16.Dias S, Silva H, Jr, Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J Exp Med. 2005;201:971–979. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Annu Rev Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 19.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 20.O’Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 21.Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proc Natl Acad Sci U S A. 2005;102:4949–4953. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Cotta CV, Stephan RP, deGuzman CG, Klug CA. Enforced expression of EBF in hematopoietic stem cells restricts lymphopoiesis to the B cell lineage. Embo J. 2003;22:4759–4769. doi: 10.1093/emboj/cdg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008 doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 25.Ueda Y, Kondo M, Kelsoe G. Inflammation and the reciprocal production of granulocytes and lymphocytes in bone marrow. J Exp Med. 2005;201:1771–1780. doi: 10.1084/jem.20041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikuchi K, Kondo M. Developmental switch of mouse hematopoietic stem cells from fetal to adult type occurs in bone marrow after birth. Proc Natl Acad Sci U S A. 2006;103:17852–17857. doi: 10.1073/pnas.0603368103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubo M, Hanada T, Yoshimura A. Suppressors of cytokine signaling and immunity. Nat Immunol. 2003;4:1169–1176. doi: 10.1038/ni1012. [DOI] [PubMed] [Google Scholar]
- 28.Metlay JP, Witmer-Pack MD, Agger R, Crowley MT, Lawless D, Steinman RM. The distinct leukocyte integrins of mouse spleen dendritic cells as identified with new hamster monoclonal antibodies. J Exp Med. 1990;171:1753–1771. doi: 10.1084/jem.171.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilliet M, Boonstra A, Paturel C, Antonenko S, Xu XL, Trinchieri G, O’Garra A, Liu YJ. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J Exp Med. 2002;195:953–958. doi: 10.1084/jem.20020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano H, Yanagita M, Gunn MD. CD11c(+)B220(+)Gr-1(+) cells in mouse lymph nodes and spleen display characteristics of plasmacytoid dendritic cells. J Exp Med. 2001;194:1171–1178. doi: 10.1084/jem.194.8.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.del Hoyo GM, Martin P, Vargas HH, Ruiz S, Arias CF, Ardavin C. Characterization of a common precursor population for dendritic cells. Nature. 2002;415:1043–1047. doi: 10.1038/4151043a. [DOI] [PubMed] [Google Scholar]
- 32.Scheeren FA, Naspetti M, Diehl S, Schotte R, Nagasawa M, Wijnands E, Gimeno R, Vyth-Dreese FA, Blom B, Spits H. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6:303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 33.Lazorchak AS, Schlissel MS, Zhuang Y. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells. Mol Cell Biol. 2006;26:810–821. doi: 10.1128/MCB.26.3.810-821.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medina KL, Kincade PW. Pregnancy-related steroids are potential negative regulators of B lymphopoiesis. Proc Natl Acad Sci U S A. 1994;91:5382–5386. doi: 10.1073/pnas.91.12.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai AY, Lin SM, Kondo M. Heterogeneity of Flt3-expressing multipotent progenitors in mouse bone marrow. J Immunol. 2005;175:5016–5023. doi: 10.4049/jimmunol.175.8.5016. [DOI] [PubMed] [Google Scholar]
- 36.Sudo T, Ito M, Ogawa Y, Iizuka M, Kodama H, Kunisada T, Hayashi S, Ogawa M, Sakai K, Nishikawa S. Interleukin 7 production and function in stromal cell-dependent B cell development. J Exp Med. 1989;170:333–338. doi: 10.1084/jem.170.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith EM, Gisler R, Sigvardsson M. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J Immunol. 2002;169:261–270. doi: 10.4049/jimmunol.169.1.261. [DOI] [PubMed] [Google Scholar]
- 38.Roessler S, Gyory I, Imhof S, Spivakov M, Williams RR, Busslinger M, Fisher AG, Grosschedl R. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 40.Lai AY, Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J Exp Med. 2006;203:1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S, Fukuda S, Lee Y, Hangoc G, Cooper S, Spolski R, Leonard WJ, Broxmeyer HE. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J Exp Med. 2000;192:719–728. doi: 10.1084/jem.192.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sitnicka E, Bryder D, Theilgaard-Monch K, Buza-Vidas N, Adolfsson J, Jacobsen SE. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 2002;17:463–472. doi: 10.1016/s1074-7613(02)00419-3. [DOI] [PubMed] [Google Scholar]
- 43.Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 44.Kondo M, Scherer DC, Miyamoto T, King AG, Akashi K, Sugamura K, Weissman IL. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407:383–386. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- 45.Hirokawa S, Sato H, Kato I, Kudo A. EBF-regulating Pax5 transcription is enhanced by STAT5 in the early stage of B cells. Eur J Immunol. 2003;33:1824–1829. doi: 10.1002/eji.200323974. [DOI] [PubMed] [Google Scholar]