Abstract
Background
Budding yeast Saccharoymyces cerevisiae is a powerful model system for analyzing eukaryotic cell cycle regulation. Yeast cell cycle analysis is typically performed by visual analysis or flow cytometry, and both have limitations in the scope and accuracy of data obtained. This study demonstrates how Multispectral Imaging Flow Cytometry (MIFC) provides precise quantitation of cell cycle distribution and morphological phenotypes of yeast cells in flow.
Methods
Cell cycle analysis of wild-type yeast, nap1Δ, and yeast overexpressing NAP1, was performed visually, by flow cytometry and by MIFC. Quantitative morphological analysis employed measurements of cellular length, thickness and aspect ratio in an algorithm to calculate a novel feature, bud length.
Results
MIFC demonstrated reliable quantification of the yeast cell cycle compared to morphological and flow cytometric analyses. By employing this technique we observed both the G2/M delay and elongated buds previously described in the nap1Δ strain.
Conclusions
Using MIFC, we demonstrate that overexpression of NAP1 causes elongated buds yet only a minor disruption in the cell cycle. The different effects of NAP1 expression level on cell cycle and morphology suggests that these phenotypes are independent. Unlike conventional yeast flow cytometry, MIFC generates complete cell cycle profiles and concurrently offers multiple parameters for morphological analysis.
Keywords: Budding yeast, Cell cycle, MIFC, Multipectral Imaging Flow Cytometry, Nap1
INTRODUCTION
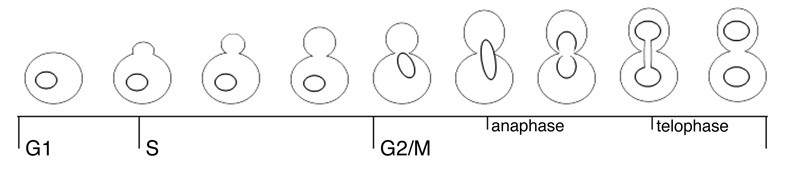
The yeast Saccharoymyces cerevisiae has historically been used as a model system for study of the eukaryotic cell cycle. The main experimental advantage offered by this organism is the ease with which the cell cycle can be manipulated, both through genetic modification and by means of simple culture techniques that alter growth rates physiologically. In other metazoans, cells divide by binary fission and the completion of cytokinesis results in two cells of equal size. In budding yeast, cell division is asymmetrical and the new daughter cell is much smaller than the mother. The smaller daughter cell will then undergo a prolonged growth phase in order to maintain cell size homeostasis, and this requires a strict coordination of cell growth and division. The complexity of cell cycle coordination in budding yeast has made it a focus for recent studies modeling cell population dynamics (1–3). Cell cycle progression in S.cerevisiae is typically monitored using both flow cytometry and visual analysis. Visual analysis requires microscopic observation of key cellular events. Bud emergence is used as a standard marker for entry into S-phase and thus defines the G1/S transition. In large-budded cells, nuclear migration and spindle formation are markers for the G2/M transition, whereas completion of anaphase can be determined by the presence of divided nuclei, Fig.1 (4). In an unsynchronized population with a known doubling time, the number of cells observed in each stage of the cell cycle corresponds to the amount of time spent in that stage and can thus reveal cell cycle delays or checkpoints (5). Though relatively simple, visual analysis is time consuming and is dependent upon a strict correlation between the cytoskeletal events of cell division and the replication of DNA. Under certain growth conditions and in specific mutant strains bud emergence and initiation of DNA replication are uncoupled, and in such cases other methods for monitoring the cell cycle must be employed (6–8).
Figure 1.
Cellular and nuclear morphology of S.cerevisiae during cell cycle progression. See text for details. Adapted from (4,34).
Standard flow cytometric cell cycle analysis of S.cerevisiae involves fluorescently labeling the DNA of fixed cells and analyzing cells on a histogram with peaks at G1 and G2/M corresponding to relative DNA content. The proportion of cells in S-phase can then be extrapolated by calculating the area beneath and between the peaks (9). Though useful for separating populations with 1C and 2C DNA content, cytometric analysis of yeast is imprecise for more complex checkpoint analyses and cannot detect minor perturbations of the cell cycle. Correspondingly, a flow cytometric profile of an unsynchronized population does not provide information about the timing of cell cycle events or delays. In addition, the S-phase estimations attained by these models are often inaccurate (10). Whereas pulse-labeling DNA in order to determine the precise fraction of cells in S phase is possible in other organisms, wild type yeast lack thymidine kinase and cannot incorporate thymidine or BrdU. This means that in the absence of exogenous thymidine kinase, S phase progression in yeast cannot be monitored by incorporation of nucleoside analogs (11). To overcome these limitations, studies of specific cell cycle transitions frequently employ a combination of flow cytometric and visual analysis (1,12). A caveat to this approach is that neither traditional method alone or in combination can accurately distinguish between early and late S-phase and between G2 and M phases.
Multispectral imaging flow cytometry (MIFC) provides flow cytometric analysis of cells while simultaneously acquiring image data from individual cells. Imaging flow cytometers can acquire 6 channels of imagery including brightfield, darkfield and four channels of fluorescent imagery of distinct bandwidth. This allows highly quantitative morphological characterization of cells by a range of criteria, and provides a technique for combining visual analysis and flow cytometric profiles. Here, we demonstrate the use of MIFC to precisely analyze the cell cycle in an asynchronous yeast population. This analysis allowed us to determine how both the cell cycle profile and morphology of yeast are altered by changes in the expression level of the nucleosome assembly protein, NAP1. These data demonstrate both the G2/M delay and elongated bud phenotype previously described in the nap1Δ strain (13). Interestingly, we determined that while both nap1Δ cells and those overexpressing NAP1 exhibited elongated bud morphology in a subset of cells, strains overexpressing NAP1 did not show a delay at G2/M. This suggests that the elongated bud phenotype is independent of the mitotic delay seen in nap1Δ cells. By integrating the analysis of DNA content and budding index, MIFC allows precise quantitation of the cell cycle by both parameters, and measures S-phase directly rather than predicting the proportions by mathematical modeling. This technique therefore represents a significant improvement over existing methods for performing cell cycle analyses in budding yeast, and simultaneously provides a system for sophisticated quantitative morphological analysis.
MATERIALS AND METHODS
Yeast Strains and Manipulations
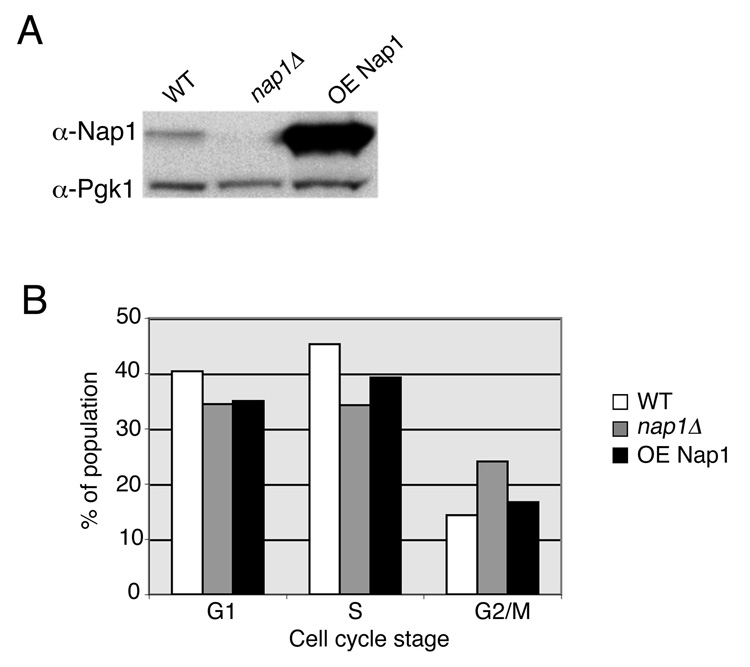
Yeast strains used in this study were derived from DF5, and construction of the nap1Δ strain has been described previously (14). Constitutive overexpression of NAP1 was achieved by cloning NAP1 into pRS426-GPD and transforming this plasmid into wild type DF5 yeast. Exponentially growing cells carrying empty pRS426 or pRS426-GPDNap1 were cultured in SC-ura media. Cells were counted, washed, and fixed in 70% EtOH 30% sorbitol. Samples were then treated with 1µM RNase A (Sigma, St. Louis, Missouri) in 50mM sodium citrate at 37° for 30 min., and stored at 4 C Prior to analysis, cells were stained with 1µM SYTOX Green (504 nanometer excitation and 523 nanometer emission; Molecular Probes, Eugene, OR) and sonicated as previously described (15). For determining the relative expression levels of each strain, whole cell lysates were extracted from equal numbers of cells and the proteins were separated by SDS-PAGE. Following transfer, the membrane was incubated with an anti-Nap1 antibody, followed by anti-Pgk1 antibody and visualized by ECL. Light microscopy was performed as described previously (16) using a Nikon Microphot-SA microscope (Melville, NY) and images were captured using OpenLab software (Improvision, Lexington, MA) with a 100X objective.
ImageStream® Flow Cytometry
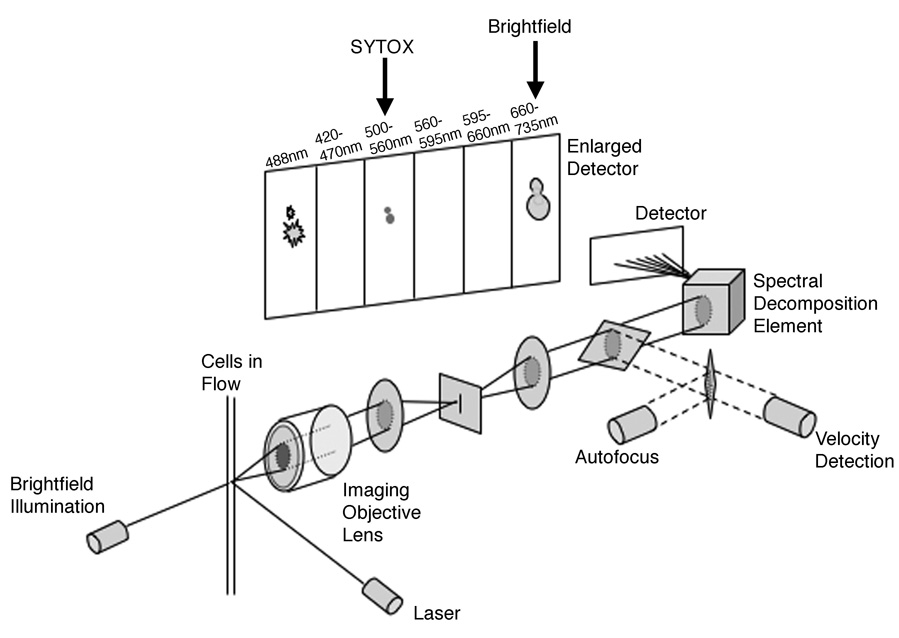
The details of the ImageStream® imaging flow cytometer (Amnis Corporation, Seattle, WA) instrument design and capabilities used here have been previously described and are presented in Figure 2 (17). Briefly, the instrument utilizes traditional hydrodynamic focusing of cells in a core stream that are illuminated by a laser source for both fluorescence and darkfield (analogous to side scatter) image detection. Simultaneously, brightfield images are generated using transillumination from a filtered white light source. All light is collected using an imaging objective lens (0.75 numerical aperture) and projected onto a custom Charge Coupled Device (CCD) consisting of six separate lateral channels. Prior to projection, collected light is passed through a spectral decomposition optical system that separates light by wavelength bands that are passed through a set of filters in an angular array onto to the laterally separated channels of the CCD. This allows the collection of six subimages for each cell based on spectral properties. These six channels generate simultaneous detection of brightfield, darkfield, and four fluorescences according to the spectral filter properties presented in Figure 2. With the use of a filter wheel corresponding to the channel filters, brightfield can be detected in any of the channels except Channel 1, which is dedicated to the detection of scattered blue laser light. Object field size for each channel image is 48µm X 256µm, with a pixel resolution of 0.5µm2 and an approximate image magnification of about 36X. Using the Time Delay Integration mode, a method of electronically tracking the movement of cells in flow, 512 rows of pixels can be accumulated in synchrony with the velocity of the cells in flow measured using the system’s velocity detection system (18). This allows for a large degree of sensitivity without the problem of a streaked image of a moving object. The ImageStream® used in these studies was configured with three lasers (488nm-200mW, 658nm-90mW, and 405nm-350mW) and the Extended Depth of Field (EDF) option, however, only the 488nm laser was employed (19). Images were collected with and without the EDF element in place; however, for the purpose of morphological classification using brightfield images, yeast bud formation was better detected without the EDF element. 10,000 cells per sample were acquired using the INSPIRE® acquisition software. The SYTOX Green signal and corresponding brightfield images were collected in Channel 3 (505–560nm) and Channel 6 (660–730nm) respectively (Fig. 2). Spectral overlap of SYTOX Green emission into the brightfield channel was calculated and subtracted using the IDEAS® v3.0 image analysis software. Out of focus images were excluded by plotting the Gradient RMS feature of the Sytox Green channel. The Gradient RMS feature measures large changes of neighboring pixel values in the image and is a relative indicator of the sharpness quality of the image. Images that are out of focus would have smaller changes in pixel values than those images in focus.
Figure 2.
The ImageStream® multispectral imaging flow cytometer has been previously described (17). Briefly, the instrument uses hydrodynamic focusing of cells in a core stream and illuminates cells in flow for fluorescence, darkfield and brightfield image detection. All light is collected and separated by wavelength bands using a spectral decomposition element, then projected onto a Charge Coupled Device detector operated in time delay integration mode using feedback from a cell velocity measurement sub-system. Six subimages are collected for each cell according to the spectral filter properties described here. The SYTOX Green signal and corresponding brightfield images were collected in Channel 3 (505–560nm) and Channel 6 (660–730nm) as shown.
Comparative Cell Cycle Analysis
Visual cell cycle analysis of images captured by MIFC was performed by assigning cells to one of three morphological categories: single nuclei and no bud (G1), single round nuclei and a visible bud (S-phase) or elongated or divided nuclei and a large bud (G2/M). Standard flow cytometry was performed using a Becton Dickinson FACSCalibur™ dual laser cytometer. For the flow cytometric analysis, 10,000 cells were acquired using a dual laser flow cytometer, collecting the SYTOX Green fluorescence (Area, Height & Width) in the FL1 channel (530/30 BP). Data gated on singlets using the FL1-A and FL1-W parameters were analyzed using two modeling programs, ModFit LT v3.0 (Verity Software House, Inc., Topsham, ME) and FlowJo’s (Treestar Inc., Ashland, OR) Watson Pragmatic v6.4.7. For MIFC cell cycle analysis, intensity of DNA image was plotted against the aspect ratio (minor axis divided by the major axis) of the DNA image and populations were gated as described in the results and quantified. Using this combination of features allows populations to segregate by nuclear shape (aspect ratio) as well as DNA content (intensity).
Morphological Analysis
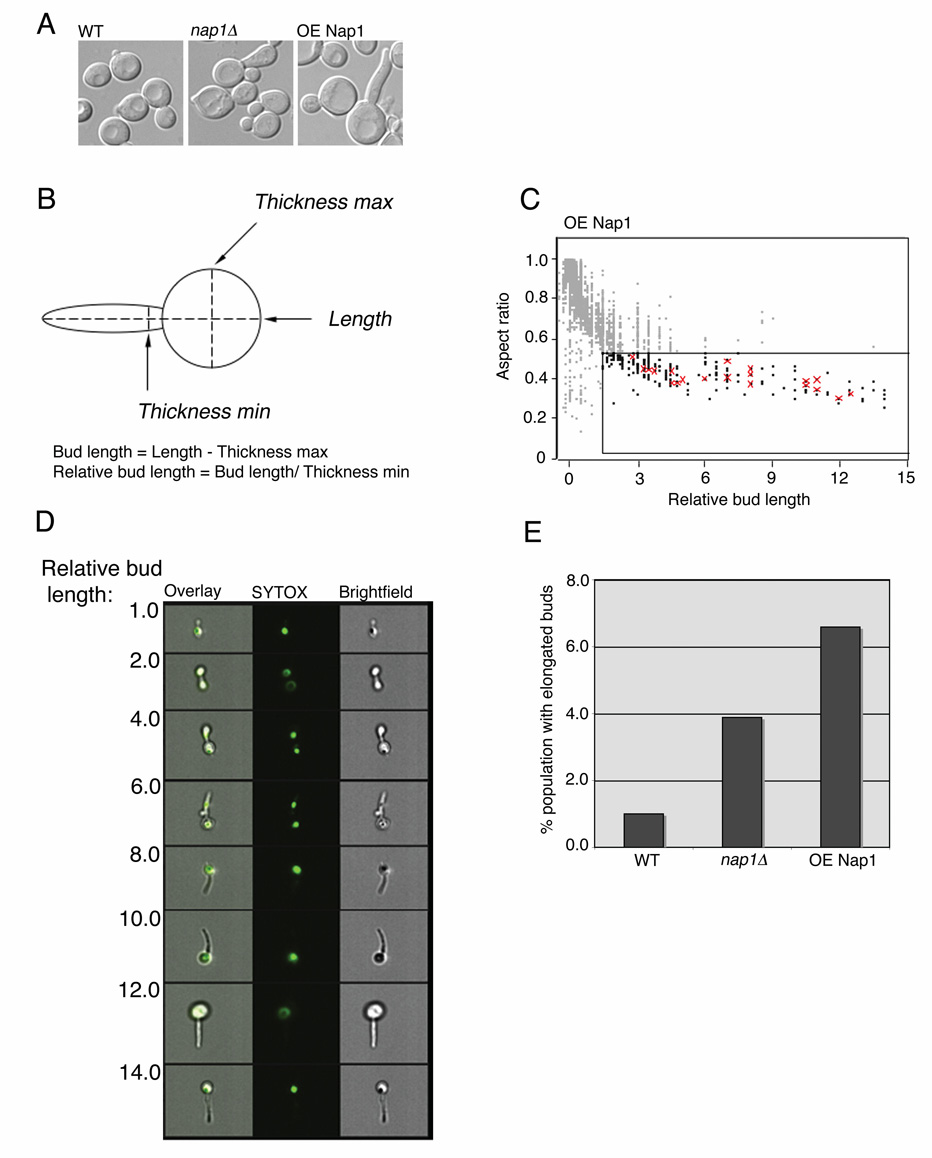
For MIFC analysis, we employed the use of masks that allow a set of pixels to be defined for a specific region of interest and used these values to calculate feature-specific values from the image. For bud morphology, we used a tight object mask (segments the image from background pixel intensity) of the brightfield channel, which was then eroded by 3 pixels ([erode(object(ch.6,tight))3px.]) and the spot count feature was calculated within this mask. In order to remove fields containing more than one cell, only data points with a spot count of one were included in the analysis. In order to evaluate the elongated bud phenotype, the actual bud length was calculated by subtracting the feature thicknessmax of the cell (assumed to be the diameter of the mother cell) from the total length of the cell (object mask described above). The relative bud length was calculated as the ratio between the actual bud length and the feature thicknessmin, assumed to be the width of the bud (Fig. 6 B). The aspect ratio (total cellular length:width) from the brightfield images were then plotted against the relative bud length, and elongated cells, those with an aspect ratio < 0.5 and relative bud length > 1.5, were quantified.
RESULTS
MIFC analysis of the S.cerevisiae cell cycle
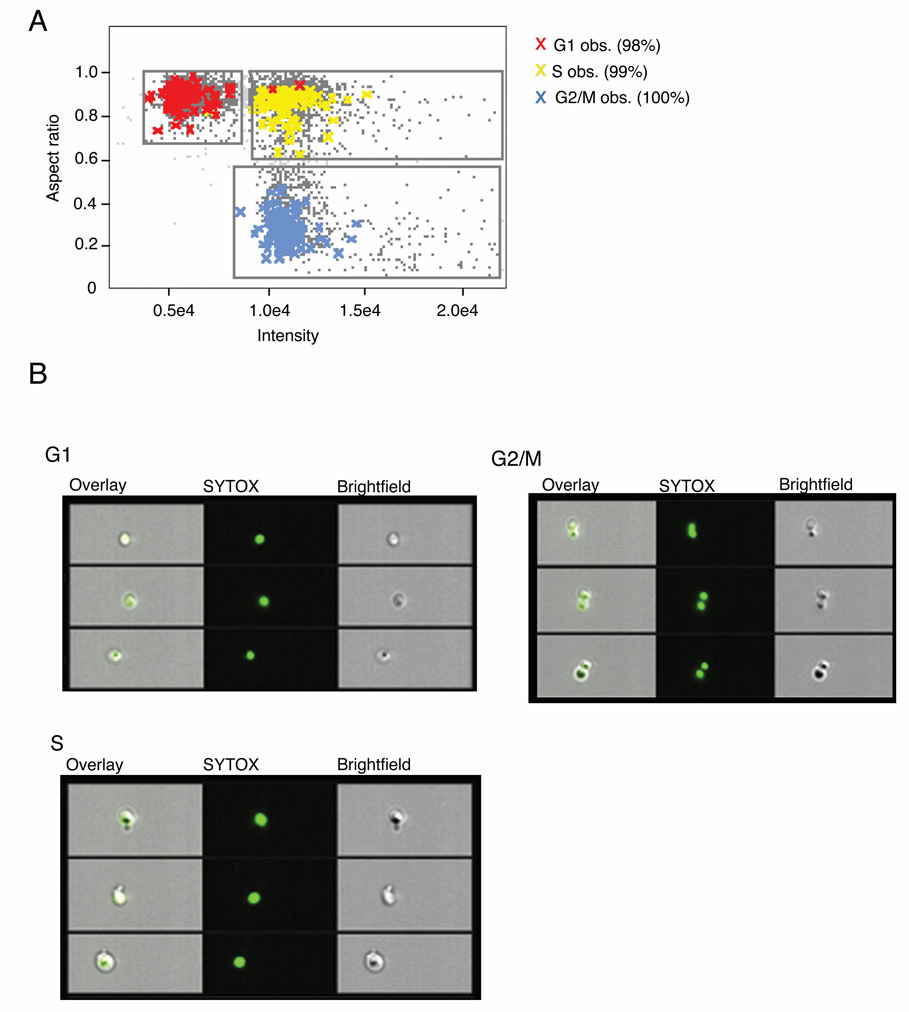
To determine the suitability of MIFC for obtaining a cell cycle profile from budding yeast, yeast from an exponentially growing asynchronous population were fixed and the DNA was stained with SYTOX Green, and 10,000 cells were acquired on an ImageStream® imaging flow cytometer. In order to separate cells at different stages of the cell cycle we decided to analyze the DNA content as determined by the intensity of the DNA stain, and the nuclear morphology using the aspect ratio (the ratio of width to length) of the DNA stain (Fig. 3 A). When these features were applied to a scatter plot, cells spontaneously segregated into three populations: 1N DNA content and round nuclei (aspect ratio ≈ 1), 2N DNA content and round nuclei, and 2N DNA content and elongated or divided nuclei (aspect ratio < 0.6) (Fig. 3 B). We hypothesized that these three populations would consist of cells in G1, S phase and G2/M respectively. In order to test this, images from the unsynchronized population were examined and 100 cells from each of these stages were identified visually and tagged, using the following morphological criteria: G1 cells are unbudded with a single, round nucleus; S phase cells have a small, visible bud and a single, round nucleus; G2/M cells have larger buds and elongated or divided nuclei (Fig. 1). The observed tagged populations were then overlaid on the original scatter plot of the aspect ratio vs. intensity of SYTOX stain (Fig. 3 A). Of the 100 cells visually determined to be in G1, 98 of these were contained within the hypothesized G1 population, whereas 99 of the 100 S phase cells were within the region hypothesized to contain those in S phase. All of the observed G2/M cells were contained within the hypothesized G2/M region (Fig. 3 B). Thus, MIFC analysis utilizes both flow cytometric and morphological analyses in order to accurately quantify the three stages of the cell cycle.
Figure 3.
Cell cycle analysis of S.cerevisiae by Multispectral Imaging Flow Cytometry (MIFC). Unsynchronized wild type yeast cultures were fixed and treated with SYTOX green DNA stain. Images of 10,000 cells were collected simultaneously in brightfield and SYTOX green channels. A: A bivariate plot of the aspect ratio and fluorescence intensity from the SYTOX green channel was generated from the in-focus population. Three spontaneously segregating regions were identified and gated. From the gathered images, 100 G1, S or G2/M cells were visually identified (as indicated by colored crosses) and the tagged populations were overlaid on the original plot. B: Images from within each region were chosen at random. From left to right the overlaid images, SYTOX and brightfield channels are displayed.
Comparison of MIFC cell cycle analysis with flow cytometric and conventional visual analysis
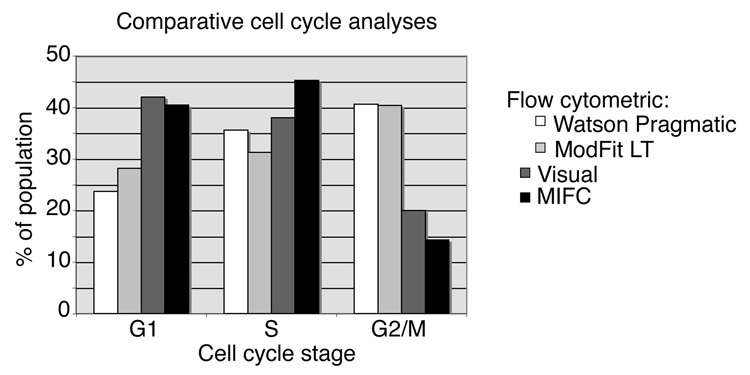
Parallel cell cycle analyses were performed on identical samples of budding yeast by traditional flow cytometry, visual analysis and MIFC analysis. For the flow cytometric analysis, SYTOX stained cells were aquired and the DNA content was quantified and analyzed using two different popular cell cycle modeling programs, FlowJo’s Watson Pragmatic v6.4.7 and ModFit LT v3.0. These two programs use different methods for approximating the proportion of cells in S phase. The Watson Pragmatic model requires that the DNA intensities of the cell population are normally distributed with an identifiable G1 peak, and from these values it approximates the G2/M peak and constructs a probability distribution for S phase. In contrast, ModFit LT requires both the G1 and G2/M peaks to be identified prior to estimating S phase (20). The two analyses yielded similar cell cycle distributions. Both algorithms estimated 40% of the population to be in G2/M, with the remaining cells distributed between G1 and S phases. The Watson Pragmatic algorithm assigned 24% of the population to G1 and 36% to S phase, whereas ModFit LT estimated G1 and S phase at 28% and 31% of the population, respectively (Fig. 4). For the visual analysis, 200 random images captured using the ImageStream® were examined and cell cycle stage was assigned using the morphological criteria described above. By this method, 42% of the cell population was assigned to G1, 38% to S phase and 20% to G2/M. The results from the MIFC analysis using DNA intensity and aspect ratio were similarly distributed, with 40% in G1, 45% in S phase and 14% in G2/M (Fig. 4). As expected, the two flow cytometric models provided relatively similar distributions, whereas results from conventional visual analysis of morphology and MIFC were more similar to each other than to either flow cytometric distribution.
Figure 4.
Comparison of MIFC cell cycle analysis with flow cytometric and conventional visual analysis. Bar graph depicts the results from cell cycle profiles obtained from each of the four indicated analytical methods. Bars indicate the percentage of the population determined to occupy each stage of the cell cycle. For both flow cytometric and MIFC analyses, 10,000 cells were analyzed; for visual observation, 200 cells were analyzed.
Use of MIFC to Discern Changes in Cell Cycle Distribution due to altered Expression of Nap1
To further evaluate the efficacy of MIFC in yeast cell cycle analysis, we decided to examine a mutant strain, nap1Δ, with a published cell cycle phenotype. Nap1 is a histone chaperone involved in the nuclear import of histones H2A and H2B and has chromatin assembly activity in vitro. Deletion of NAP1 has been shown to cause a delayed entry into mitosis and a prolonged passage through mitosis, as determined by assaying the activity of mitotic kinase Clb2/Cdc28 (13). However, we have been previously unable to detect this phenotype using traditional flow cytometry methods (data not shown). We decided to assess the ability of MIFC analysis to demonstrate the phenotype of nap1Δ cells. Additionally, we used this technique to examine the phenotype of a previously undescribed strain, carrying a plasmid with an exogenous promoter from which NAP1 is overexpressed (Fig. 5 A). Exponentially growing populations of wild-type and nap1Δ yeast, and those overexpressing NAP1, were fixed and stained with SYTOX Green. 10,000 cells from each strain were acquired by MIFC and cell cycle analysis was performed by separating cells according to DNA content and nuclear aspect ratio as described previously. By this method, nap1Δ yeast showed an increase in the number of cells in G2/M to 26%, as compared to 14% in the wild-type population (Fig. 5 B). In addition, the nap1Δ cells showed reduced numbers of cells in both G1 and S phase relative to wild-type. We found that overexpression of NAP1 led to a slight increase in the number of cells in G2/M to 18%, and a decrease in the numbers of cells in G1 and S phase, though this difference was less distinct than was seen for the nap1Δ cells (Fig. 5 B). The results from this experiment demonstrate the use of MIFC to successfully discern minor alterations in the cell cycle in budding yeast.
Figure 5.
Budding yeast cell cycle distribution is dependent upon NAP1 expression level. A: To determine the relative expression levels of NAP1, whole cell lysates from the indicated strains were equalized and proteins were visualized by Western blotting with Nap1 or Pgk1 antibodies (WT, wild type; OE, overexpressed NAP1). B: Bar graph depicts the results from cell cycle profiles analyzed by MIFC from each of the three indicated strains. Bars indicate the percentage of the population determined to occupy each stage of the cell cycle. 10,000 cells from each sample were analyzed.
MIFC analysis of morphological phenotypes
In addition to the observed changes in cell cycle distribution, deletion of NAP1 has been demonstrated to cause an elongated bud phenotype (13). This is thought to be due to the role of Nap1 in regulating the switch from polar to isotropic bud growth that occurs at the G2/M transition in S.cerevisiae. Visual analysis showed that overexpression of NAP1 also results in an elongated bud phenotype, and this phenotype appeared to be more severe than that seen in nap1Δ yeast (Fig. 6 A). We wanted to evaluate whether MIFC analysis would be able to recognize and quantify such subtle changes in morphology. Elongated buds are typically defined as those in which the bud length is at least 1.5 times the width of the bud (21,22). Using the IDEAS v.3.0 software, an algorithm was developed to determine the ratio between bud length and width as described in the Materials and Methods (Fig. 6 B). Acquired MIFC data from the wild-type, nap1Δ and NAP1 overexpressing strains were then analyzed for both relative bud length and cellular shape, as described by the aspect ratio of length to width of the cell captured with brightfield imaging. This allows us to distinguish between large-budded mitotic cells and clumps of multiple cells, in which the aspect ratio is close to 1, from elongated cells bearing the long and narrow bud, with aspect ratio less than 0.5. When these features were applied to a scatter plot, the distribution predictably demonstrates an inverse relationship between aspect ratio and relative bud length (Fig. 6 C). We next determined whether this analysis could reliably segregate cells with elongated bud morphology. Images from cells overexpressing NAP1 were examined and 20 cells with elongated buds were identified visually and tagged, using a relative bud length of greater than 1.5 as criteria. The observed tagged populations were then overlaid on the original scatter plot of aspect ratio vs. relative bud length (Fig. 6 C). Of the 20 cells visually identified as having elongated buds, all 20 were contained within the elongated population as determined by the MIFC analysis. In addition, an increase in the calculated relative bud length visually correlates with increasing severity of the elongated bud phenotype (Fig. 6D). Thus, MIFC can accurately recognize and quantify morphological phenotypes in budding yeast. We then quantified the number of cells with elongated buds from each of the strains and determined that whereas in wild-type yeast only 1% of cells exhibit elongated buds, deletion of Nap1 led to an increase to 4.3% and overexpression of NAP1 increased this to almost 7% (Fig. 6 C). Therefore, the elongated bud phenotype was exacerbated in cells overexpressing NAP1 relative to nap1Δ cells, yet the G2/M delay was more pronounced in the deletion strain. This suggests that these are not interdependent phenotypes but may be due to effects of Nap expression levels on different aspects of cellular function. Our results demonstrate that MIFC analysis of budding yeast allows for accurate and quantitative investigation of subtle alterations in the cell cycle, while also providing multiple parameters for characterizing morphological phenotypes.
Figure 6.
Regulation of bud morphology is dependent upon NAP1 expression level. A: Live cells from the indicated strains were examined by DIC microscopy. B: Diagram illustrating the method for calculating relative bud length. C: Images of cells with indicated relative bud length as determined by MIFC. From left to right the overlaid images, SYTOX and brightfield channels are displayed. D: MIFC analysis of bud morphology. A bivariate plot of the aspect ratio and relative bud length from the brightfield channel was generated from the in-focus population. From the gathered images, 20 cells were visually identified as bearing elongated buds and the tagged populations were overlaid on the original plot, as indicated by the red crosses. Boxed region defines cells with elongated buds, where bud length > 1.5 times the bud width and the aspect ratio < 0.5. E: Bar graph indicates the percentage of the population with elongated buds as determined in C, from each of the three indicated strains.
DISCUSSION
This study establishes that MIFC analysis of budding yeast represents a valid method for determining a complete cell cycle profile in budding yeast. Unlike traditional flow cytometry, MIFC makes it possible to detect minor perturbations in the cell cycle without requiring synchronization by cell cycle arrest and release. Gene expression studies have shown that different methods for cell cycle arrest, such as nocodazole treatment, elutriation, α-factor pheromone treatment and temperature-sensitive mutant strains, yield inconsistent results (23–25). Additionally, specific strains are hypersensitive to chemical agents used for synchronization and one arrest technique may not be suitable for analyses with different strains (26,27). Thus, one of the primary advantages of MIFC analysis as presented here is it allows a detailed cell cycle profile to be determined without the additional manipulations required for arrest and release. Visual analysis of the cell cycle does not require synchronization, but is very time consuming; obtaining statistically significant verification of minor defects can be difficult, due to limited sample sizes. This technique is also more prone to subjective errors. MIFC provides the means to objectively evaluate large numbers of cells using both flow cytometry and morphological criteria.
We find that quantification of the cell cycle using standard flow cytometry coupled to standard modeling programs overestimates the proportion of cells in the G2/M phases of the cell cycle and underestimates those in G1, relative to both visual analysis and MIFC. Interestingly, overestimation of the predicted proportion of G2/M cells was a systematic error identified in the original assessment of the Watson Pragmatic model (20). This may be exacerbated in yeast, since a lack of concise separation of late stage G2/M and recently divided cells (doublets) would also lead to an overestimation of G2/M by traditional flow cytometry. Although the analytical parameters differ, both flow cytometry software models fit S-phase to the trough between the G1 and G2/M peaks and attribute some of the area beneath each peak to S-phase. Our findings suggest that both models may assign a proportion of cells to G2/M that are still actively replicating DNA, and should thus be considered in S-phase. Similarly, underestimation of G1 by flow cytometry suggests that a portion of cells beneath the peak corresponding to 1N DNA are being allocated to S phase.
DNA content is measured along a continuum, and thus any algorithm must contain simulated peak boundaries for calculating S phase. This presents a caveat when more complex cell cycle analysis is required, though is not a concern for straightforward analyses in which comparison of relative DNA content (i.e. 1N or 2N) is sufficient. It is important to note that both the Watson Pragmatic model and ModFit LT were designed and evaluated using mammalian cells. Budding yeast exhibit relatively large coefficients of variance (CVs) in fluorescent DNA stain in comparison to mammalian cells (28). The small genome size of yeast renders it more sensitive to variations in cell number and DNA stain concentration between samples, and the asymmetrical cellular morphology of budding yeast can also contribute to the increased variance observed. Another reason for this is that yeast are particularly sensitive to changes in the sheath pressure during flow, and these fluctuations can cause shifts in the fluorescent peak. (28,29). It is therefore feasible that existing cell cycle models based on the mammalian cells could be modified in order to account for these yeast-specific disparities. Other methods for monitoring cell cycle progression by flow cytometry, such as simultaneously tracking protein and DNA content, can also provide better resolution of the cell cycle in heterogeneous yeast populations (30,31).
In comparing the different methods for determining cell cycle, it must be considered that in the MIFC analysis cell populations are not segregated according to DNA content alone, the standard criteria used in traditional flow cytometric cell cycle analysis. Using MIFC we consider multiple criteria, and this provides a more precise demarcation of cell cycle stage. Nuclear elongation is employed here as a marker for the G2/M transition and this is one of a number of morphological events that occur between late S phase and the onset of anaphase. Other events, such as mitotic spindle formation or the migration of the nucleus to the bud neck, could also be easily applied to MIFC analysis.
Using MIFC analysis we confirmed that nap1Δ cells exhibit an almost two-fold increase in the proportion of cells in G2/M of the cycle, indicative of a delayed exit from mitosis, and this is coupled with a shortening of both G1 and S. In cells overexpressing NAP1, this phenotype was less pronounced; the proportion of cells in G2/M increased by about one-third relative to wild-type, and this strain also demonstrated shortened G1 and S phases. These minor perturbations in the cell cycle would not have been detected by traditional flow cytometry, and the small differences would have required very large sample sizes to be statistically significant by purely visual analysis.
The algorithm we developed in order to measure bud length was determined to be a reliable method for evaluating numbers of elongated buds within a population, as tested by previously published criteria. This confirms the utility of MIFC analysis to quantify morphological phenotypes in budding yeast. By categorizing elongated cells as those in which bud length is greater than 1.5 times the width, we determined that nap1Δ cells showed greater than a three-fold increase in elongated buds relative to wild-type yeast whereas in cells overexpressing NAP1, this increase was more than six-fold. This was in agreement with previous unpublished observations that overexpression of NAP1 causes in increase in severity of the elongated bud phenotype relative to wild-type.
It was of particular interest that in cells overexpressing NAP1, the number of cells with elongated buds was higher than in nap1Δ cells, yet the G2/M delay was less pronounced. This implies that the switch from polar to isotropic bud growth is more impaired when excess Nap1 is present than in its absence. Conversely, the delay in G2/M is more severe in the absence of Nap1, and yet fewer of the cells in G2/M exhibit defects in isotropic bud growth. Therefore, the regulation of bud growth and mitotic progression are both affected by expression levels of NAP1, yet the effects are at least somewhat independent. A specific role in mitosis for Nap1 has not been defined, though it interacts with the mitotic cyclin Clb2 (32). Many of the events of mitosis require the activation of the cyclin-dependent kinase Cdc28 by Clb2, including the regulation of bud growth. Nap1 also interacts with the kinase Gin4, though apparently in an independent complex since Gin4 and Clb2 are not detected together (32,33). Interestingly, deletion of the genes for both NAP1 and GIN4 in wild-type cells causes a more severe elongated bud phenotype than either single deletion, whereas in a CLB2-dependent background the phenotypes of the double and single mutants are equivalent (33). Taken together, these results imply that Nap1 and Gin4 share both Clb2-dependent and independent functions in regulating bud growth. It is possible that excess Nap1 impairs the regulation of bud growth by Gin4 without significantly affecting mitotic progression (perhaps by outcompeting other Gin4-interacting proteins), whereas total absence of Nap1 is damaging to both processes. Characterization of other mutants involved in the control of bud growth during mitosis should help elucidate this. In summary, this study validates the use of MIFC in performing tandem cell cycle and morphological analyses on budding yeast. As we have demonstrated, this technique proves particularly useful for characterizing cells in which cell cycle defects have become uncoupled from associated morphological phenotypes.
ACKNOWLEDGEMENTS
We thank Michael Solga in the Flow Cytometry Core Facility and Dr. Shannon Henery of Amnis Corporation for technical help. We thank Dr. Thaddeus George of Amnis Corporation for suggestions and comments. This work was partially supported by NIH Research Grant R01 GM65385.
Footnotes
This work was supported by NIH Research Grant R01 GM65385.
REFERENCES
- 1.Niemisto A, Nykter M, Aho T, Jalovaara H, Marjanen K, Ahdesmaki M, Ruusuvuori P, Tiainen M, Linne ML, Yli-Harja O. Computational methods for estimation of cell cycle phase distributions of yeast cells. EURASIP J Bioinform Syst Biol. 2007:46150. doi: 10.1155/2007/46150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cipollina C, Vai M, Porro D, Hatzis C. Towards understanding of the complex structure of growing yeast populations. J Biotechnol. 2007;128(2):393–402. doi: 10.1016/j.jbiotec.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Hatzis C, Porro D. Morphologically-structured models of growing budding yeast populations. J Biotechnol. 2006;124(2):420–438. doi: 10.1016/j.jbiotec.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Fantes PaB J. The Yeast Nucleus. 2000 [Google Scholar]
- 5.Slater ML, Sharrow SO, Gart JJ. Cell cycle of Saccharomycescerevisiae in populations growing at different rates. Proc Natl Acad Sci U S A. 1977;74(9):3850–3854. doi: 10.1073/pnas.74.9.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. Embo J. 1995;14(15):3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7(7A):1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 8.Rivin CJ, Fangman WL. Cell cycle phase expansion in nitrogen-limited cultures of Saccharomyces cerevisiae. J Cell Biol. 1980;85(1):96–107. doi: 10.1083/jcb.85.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutter KJ, Eipel HE. Flow cytometric determinations of cellular substances in algae, bacteria, moulds and yeasts. Antonie Van Leeuwenhoek. 1978;44(3–4):269–282. doi: 10.1007/BF00394305. [DOI] [PubMed] [Google Scholar]
- 10.Fox MH. A model for the computer analysis of synchronous DNA distributions obtained by flow cytometry. Cytometry. 1980;1(1):71–77. doi: 10.1002/cyto.990010114. [DOI] [PubMed] [Google Scholar]
- 11.Grivell AR, Jackson JF. Thymidine kinase: evidence for its absence from Neurosporacrassa and some other micro-organisms, and the relevance of this to the specific labelling of deoxyribonucleic acid. Journal of General Microbiology. 1968;54:301–317. doi: 10.1099/00221287-54-2-307. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Siede W. Analysis of the budding yeast Saccharomyces cerevisiae cell cycle by morphological criteria and flow cytometry. Methods Mol Biol. 2004;241:77–91. doi: 10.1385/1-59259-646-0:77. [DOI] [PubMed] [Google Scholar]
- 13.Kellogg DR, Murray AW. NAP1 acts with Clb1 to perform mitotic functions and to suppress polar bud growth in budding yeast. J Cell Biol. 1995;130(3):675–685. doi: 10.1083/jcb.130.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosammaparast N, Del Rosario BC, Pemberton LF. Modulation of histone deposition by the karyopherin kap114. Mol Cell Biol. 2005;25(5):1764–1778. doi: 10.1128/MCB.25.5.1764-1778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsberg S. The Forsburg lab pombe pages: Yeast flow cytometry [Google Scholar]
- 16.Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J Cell Biol. 2001;153(2):251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George TC, Basiji DA, Hall BE, Lynch DH, Ortyn WE, Perry DJ, Seo MJ, Zimmerman CA, Morrissey PJ. Distinguishing modes of cell death using the ImageStream multispectral imaging flow cytometer. Cytometry A. 2004;59(2):237–245. doi: 10.1002/cyto.a.20048. [DOI] [PubMed] [Google Scholar]
- 18.Ortyn WE, Hall BE, George TC, Frost K, Basiji DA, Perry DJ, Zimmerman CA, Coder D, Morrissey PJ. Sensitivity measurement and compensation in spectral imaging. Cytometry A. 2006;69(8):852–862. doi: 10.1002/cyto.a.20306. [DOI] [PubMed] [Google Scholar]
- 19.Ortyn WE, Perry DJ, Venkatachalam V, Liang L, Hall BE, Frost K, Basiji DA. Extended depth of field imaging for high speed cell analysis. Cytometry A. 2007;71(4):215–231. doi: 10.1002/cyto.a.20370. [DOI] [PubMed] [Google Scholar]
- 20.Watson JV, Chambers SH, Smith PJ. A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry. 1987;8(1):1–8. doi: 10.1002/cyto.990080101. [DOI] [PubMed] [Google Scholar]
- 21.Iwase M, Toh-e A. Ybr267w is a new cytoplasmic protein belonging to the mitotic signaling network of Saccharomyces cerevisiae. Cell Struct Funct. 2004;29(1):1–15. doi: 10.1247/csf.29.1. [DOI] [PubMed] [Google Scholar]
- 22.Hood-DeGrenier JK, Boulton CN, Lyo V. Cytoplasmic Clb2 is required for timely inactivation of the mitotic inhibitor Swe1 and normal bud morphogenesis in Saccharomyces cerevisiae. Curr Genet. 2007;51(1):1–18. doi: 10.1007/s00294-006-0102-1. [DOI] [PubMed] [Google Scholar]
- 23.Shedden K, Cooper S. Analysis of cell-cycle gene expression in Saccharomyces cerevisiae using microarrays and multiple synchronization methods. Nucleic Acids Res. 2002;30(13):2920–2929. doi: 10.1093/nar/gkf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shedden K, Cooper S. Analysis of cell-cycle-specific gene expression in human cells as determined by microarrays and double-thymidine block synchronization. Proc Natl Acad Sci U S A. 2002;99(7):4379–4384. doi: 10.1073/pnas.062569899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper S, Shedden K. Microarray analysis of gene expression during the cell cycle. Cell Chromosome. 2003;2(1):1. doi: 10.1186/1475-9268-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan RK, Otte CA. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol. 1982;2(1):21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foreman PK, Davis RW. CDP1, a novel Saccharomyces cerevisiae gene required for proper nuclear division and chromosome segregation. Genetics. 1996;144(4):1387–1397. doi: 10.1093/genetics/144.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haase H. Cell Cycle Analysis of Budding Yeast Using SYTOX Green. Current Protocols in Cytometry Volume 7.23. John Wiley & Sons; 2003. [DOI] [PubMed] [Google Scholar]
- 29.Haase SB, Reed SI. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle. 2002;1(2):132–136. [PubMed] [Google Scholar]
- 30.Porro D, Martegani E, Ranzi BM, Alberghina L. Identification of different daughter and parent subpopulations in an asynchronously growing Saccharomyces cerevisiae population. Res Microbiol. 1997;148(3):205–215. doi: 10.1016/S0923-2508(97)85241-2. [DOI] [PubMed] [Google Scholar]
- 31.Porro D, Ranzi BM, Smeraldi C, Martegani E, Alberghina L. A double flow cytometric tag allows tracking of the dynamics of cell cycle progression of newborn Saccharomyces cerevisiae cells during balanced exponential growth. Yeast. 1995;11(12):1157–1169. doi: 10.1002/yea.320111206. [DOI] [PubMed] [Google Scholar]
- 32.Kellogg DR, Kikuchi A, Fujii-Nakata T, Turck CW, Murray AW. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130(3):661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altman R, Kellogg D. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138(1):119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann FT A, Gasser S. Tracking individual chromosomes with integrated arrays of Lacop sites and GFP-Laci repressor (PROT15) The EPIGENOME Network of Excellence Epigenome NoE. 2005 [Google Scholar]