Abstract
We report the results of an investigation of the inhibition of the ATP-mediated HIV-1 reverse transcriptase catalyzed phosphorolysis in vitro of AZT from AZT-terminated DNA primers by a series of 42 bisphosphonates. The four most active compounds possess neutral, halogen-substituted phenyl or biphenyl sidechains and have IC50 values < 1 μM in excision inhibition assays. Use of two comparative molecular similarity analysis methods to analyze these inhibition results yielded a classification model with an overall accuracy of 94%, and a regression model having good accord with experiment (q2 = 0.63, r2 = 0.91), with the experimental activities being predicted within, on average, a factor of 2. The most active species had little or no toxicity against three human cell lines (IC50avg >200 μM). These results are of general interest since they suggest that it may be possible to develop potent bisphosphonate-based AZT excision inhibitors with little cellular toxicity, opening up a new route to restoring AZT sensitivity in otherwise resistant HIV-1 strains.
Keywords: Reverse transcriptase, Nucleoside reverse transcriptase inhibitor (NRTI), Bisphosphonate, Quantitative structure activity relationship (QSAR)
1. Introduction
HIV-1 is a major cause of AIDS with ~40,000,000 individuals infected1. The first drug approved for the clinical treatment of HIV-1 infection was 3′-azido, 3′-deoxythymidine (AZT, Retrovir®, Retrovis® and Zidovudine®). When taken up into cells AZT is converted to AZT-triphosphate, which can then be incorporated into nascent viral DNA2, resulting in a so-called “blocked DNA primer” that contains a terminal azide (-N3) rather than an -OH group. The terminal OH →-N3 substitution prevents DNA elongation and, consequently, viral DNA replication is terminated. However, shortly after the introduction of AZT, cases of resistance were found. This resistance originates in the phosphorolytic excision of AZT by ATP (and, to a lesser extent, PPi) catalyzed by viral reverse transcriptase3–5 (RT), as illustrated below:

In most cases, resistance originates from specific RT mutations called thymidine analog mutations (TAMs)6–10. Specific inhibitors of AZT excision, and in principle the excision of other nucleoside reverse transcriptase inhibitors (NRTIs), could have therapeutic utility, since they might restore the activity of the NRTI against TAM-containing HIV-1. Optimally, such inhibitors should not reduce incorporation of an NRTI, since this would clearly antagonize their anti-retroviral activity. We demonstrate here that selective inhibitors of RT-catalyzed phosphorolytic excision of NRTIs can be identified.
While screening large libraries of compounds for activity in inhibiting AZT excision might be one approach to discovering NRTI-excision inhibitors, current assay methods are not well suited for high-throughput screening. An alternative approach might, however, be to screen smaller libraries of compounds containing hydrolytically stable analogs of diphosphate or triphosphate, which could block AZT (or PPi) binding to RT. Conceptually, the simplest analogs of PPi to consider are the so-called bisphosphonate class of drugs (used to treat bone resorption diseases) in which the central, bridging oxygen atom is replaced by a methylene (2) or substituted methylene group, such as 3, 4:

and in previous work11 we reported that the bisphosphonate 3 had some activity in this context. More recently, Cruchaga et al.12 have followed up on this work, finding that the bisphosphonate pamidronate (4) competitively inhibited the phosphorolytic activity of RT, and synergistically inhibited HIV-1 RT in combination with AZT-triphosphate, in the presence of either ATP or PPi. And in more recent work, this group found similar results with the anti-viral compound 2-O-galloylpunicalin (2GP13) Here, we report the results of an investigation of the activity of 3 and 41 other bisphosphonates in inhibiting RT-catalyzed AZT excision by ATP. The most active compounds are far more potent than are 3 or pamidronate (4), and we use here a variety of structure-activity relationship methods to relate their structure with function. We also investigate the direct inhibition of RT catalyzed DNA synthesis by these bisphosphonates, finding in most cases that, unlike 2GP, they have little or no activity, an observation of potential importance for their further development as inhibitors of NRTI excision during HIV-1 replication.
2. Results and Discussion
2.1. Structure-activity relationships (SAR)
We investigated 42 bisphosphonates for activity in RT-catalyzed ATP-mediated AZT excision reactions in vitro. The structures of 25 compounds (3, 5–28) having activity in the range 0.5–50 μM are shown (rank ordered in terms of decreasing activity) in Figure 1A and the structures of 17 additional compounds (29–45) having low (≥ 100 μM) activity are shown in Figure 1B. The synthesis and characterization of all compounds except 29 and 30 have been reported elsewhere14 and the samples used here were from the same batches. The synthesis of 29 and 30 were shown in the Experimental Section below, using our previous methods.
Figure 1.
Structures of bisphosphonate inhibitors. A, 25 active species investigated, rank ordered by decreasing activity. B, Additional, inactive species (IC50 ≥ 100 μM).
We show in Figure 1A the structures of 25 bisphosphonates having high-to-moderate activity in inhibiting RT-catalyzed AZT excision by ATP, ordered in terms of decreasing activity. The IC50 values are in the range 500 nM–51.3 μM (Table 1). The structures of a second set of compounds having low activity (IC50 values ≥ 100 μM) are shown in Figure 1B. What is immediately apparent from these results is that none of the conventional, nitrogen containing bisphosphonates, such as incadronate (35), zoledronate (36) or minodronate (44), Figure 1B, used to treat bone-resorption diseases, have appreciable activity in inhibiting AZT-excision, since their IC50 values are all ≥100 μM. Several of the other inactive compounds (31, 38 and 39) are also known to be active in inhibiting bone resorption15, 16, and in each case their target is the enzyme farnesyl diphosphate synthase (FPPS). However, targeting FPPS is an undesirable feature for an AZT-excision inhibitor since it is likely to lead to toxicity in host cells.
Table 1.
Inhibition of ATP-mediated AZT excision from HIV-1 reverse transcriptase
| Compound a | Excision Inhibition IC50 (μM) | MCF-7 IC50 (μM) | SF-268 IC50 (μM) | NCI-H460 IC50 (μM) |
|---|---|---|---|---|
| 5 | 0.5 | 213 | 208 | 241 |
| 6 | 0.9 | 41 | 48 | 39 |
| 7 | 1.0 | >300 | >300 | >300 |
| 8 | 1.0 | 225 | 236 | 253 |
| 9 | 1.3 | 259 | 225 | 248 |
| 10 | 1.5 | 165 | 122 | 141 |
| 11 | 1.9 | 173 | 224 | 175 |
| 12 | 2.1 | >300 | >300 | >300 |
| 13 | 2.2 | >100 b | >100 b | >100 b |
| 14 | 2.4 | 128 | 139 | 118 |
| 15 | 2.9 | >300 | >300 | >300 |
| 3 | 3.2 | 289 | 269 | 253 |
| 16 | 3.3 | 188 | 176 | 212 |
| 17 | 3.5 | >100 b | >100 b | >100 b |
| 18 | 3.9 | >100 b | >100 b | >100 b |
| 19 | 3.9 | >300 | >300 | >300 |
| 20 | 4.6 | 84 b | 95 b | 95 b |
| 21 | 5.0 | 296 | 297 | 301 |
| 22 | 5.2 | n.d. | n.d. | n.d. |
| 23 | 5.4 | >300 | >300 | >300 |
| 24 | 7.9 | >300 | >300 | >300 |
| 25 | 9.5 | >300 | >300 | >300 |
| 26 | 11.2 | >300 | >300 | >300 |
| 27 | 38.0 | 205 | 186 | 187 |
| 28 | 51.3 | >300 | >300 | >300 |
Structures shown in Figure 1.
Compounds dissolved in dimethyl sulfoxide (DMSO)
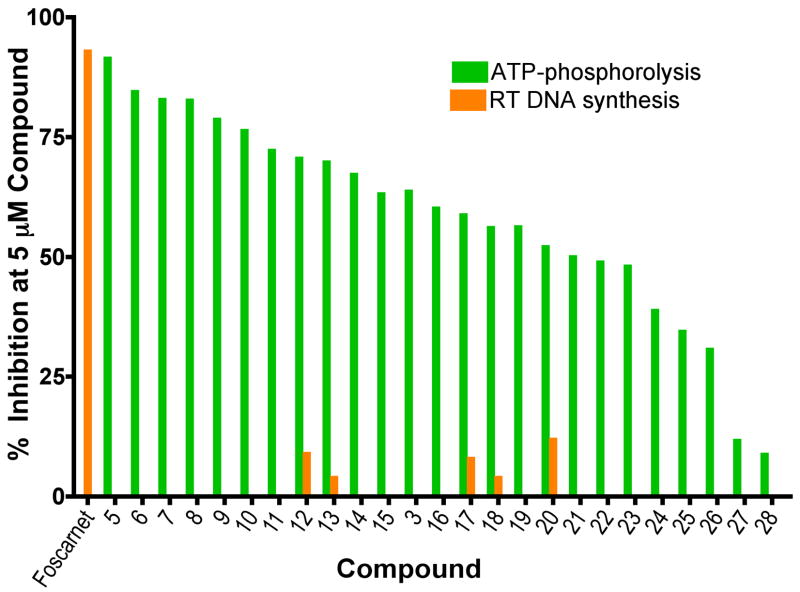
In the case of the bisphosphonates that are far more active in inhibiting AZT excision, we find a clearly different pattern of sidechain substitution to that seen in the nitrogen containing bisphosphonates. All of the nitrogen-containing bisphosphonates active in bone resorption have a basic nitrogen atom that is protonated when bound to the FPPS enzyme17, whereas in the case the potent excision inhibitors, the sidechain is expected (based on pKa values) to be neutral. The four most active compounds have IC50 values ≤ 1 μM and in each case, these compounds contain halogen substituents on a phenyl or biphenyl ring. In contrast, the two least “active” compounds (27 and 28, with IC50 values of 38, 51.3 μM, respectively), each contain electron-donating ring substituents. Of all of the active compounds, none were potent inhibitors of RT-catalyzed DNA synthesis, as shown in Figure 2, a desirable feature for an excision inhibitor. We also found that there was little or no activity of these bisphosphonates against the human cell lines tested: IC50 values are given in Table 1. For example, 3 had an IC50 (averaged over the three cell lines investigated, MCF-7, SF-268, NCI-H460) of ~229 μM, while the most active species (5) had an IC50 of ~234 μM. In contrast, species such as zoledronate and minodronate (36, 44) had IC50 values of ~15, 5 μM, due to potent inhibition of the farnesyl diphosphate synthase enzyme. As expected, 3 and 5 had very weak activity against this target (44, 8.5 μM vs 0.14, 0.12 μM for minodronate, zoledronate: data not shown). The presence of both phosphonate groups is critical for activity, as can be seen in the series 5, 29, 30. Both of the latter compounds have P-C (phosphinate) groups, replacing 1 or 2 –OH groups with CH3 groups, and neither compound has any appreciable activity in inhibiting AZT excision.
Figure 2.
Inhibition of RT catalyzed ATP-mediated phosphorolysis (green) and DNA synthesis (orange) at 5 μM inhibitor. Foscarnet, used as a control.
2.2. Quantitative structure-activity relationships (QSAR)
We next sought to see to what extent it might be possible to make a more detailed analysis of the activity results shown in Table 1. We first carried out a pharmacophore modeling investigation and obtained the common feature hypothesis shown in Figure 3A. However, this approach did not yield any highly predictive models. We thus next moved to using CoMSIA modeling methods, which, in previous work on bisphosphonate inhibitors18–20, have been shown to be highly predictive. These methods require an alignment of all molecules within a common frame of reference (Figure 3B), which in this case was constructed using the flexible alignment algorithm in the program MOE21. We then used the default settings in Sybyl 7.3 to generate steric, hydrophobic, electrostatic, donor and acceptor CoMSIA descriptors at grid points surrounding the alignment. Given the discrete distribution of the RT-enzyme inhibition data, we first investigated the possible utility of a classification approach, rather than regression methods, to analyze the CoMSIA results. For such classification approaches, compound activity can be assigned using an ordered scale (i.e. active, moderately active, inactive) rather than specific numeric values. The soft independent modeling of class analogy22 (SIMCA) module in Sybyl 7.3 provides one approach with which to create such classification models, based on the CoMSIA fields of the aligned structures. Using this approach we find that the resulting cross-validated model has an overall classification accuracy of 94%, with 100% accuracy among inactive and moderately active groups and 90% accuracy among actives, Figure 4A (see the Supplementary Material, Table S1, for more details).
Figure 3.
Structure-activity schematics. A, Common feature pharmacophore: Yellow spheres, combined feature of acceptor and metal ligator representing a phosphonate group; purple sphere, donor or hydrophobic; green sphere, aromatic; cyan sphere, aromatic or hydrophobic. B–E, CoMSIA alignment and results; B, Flexible common feature alignment used for CoMSIA, showing heavy atoms only; C, Steric fields; green favored, yellow disfavored; D, Electrostatic fields; blue positive charge favored, red, positive charge-disfavored; E, Hydrophobic fields; yellow favorable, grey disfavored.
Figure 4.
CoMSIA modeling results; A, SIMCA results, the overall classification accuracy is 94%. Actual and predicted classes shown, yellow box indicating classification error; B, PLS regression results, q2 = 0.63, r2 = 0.90, F = 73, n = 35. Test and training set results shown: training =
 , test = ○; C, Activity-electron affinity correlations. ●= EA, r2 0.55, F = 28;
, test = ○; C, Activity-electron affinity correlations. ●= EA, r2 0.55, F = 28;
 = VEA, r2 = 0.37, F =14.
= VEA, r2 = 0.37, F =14.
Given confidence in the results of the classification approach, which can be applied to all of the compounds investigated, we then investigated use of a standard regression method. Due to the limited range of the numeric data, we assigned arbitrarily large (300 μM) activities to the inactive compounds, in order to increase the range, basically the same approach we used in a previous study20. We then performed a CoMSIA analysis on all compounds, using a partial least squares (PLS) regression approach. The optimal number of components in the final PLS model was determined to be 4 by using q2 values, as obtained from the SAMPLS23 leave-one-out cross-validation technique implemented in Sybyl. This four component CoMSIA model (n=35) gave q2 = 0.63, r2 = 0.91 and F= 73, as shown graphically in Figure 4B (full details shown in Supplementary Material Table S2). Five iterations of a leave-two-out analysis were performed and 10 iterations of activity data randomization were then applied to confirm the predictive ability of the model, and that the final model was not the result of a chance correlation. The hydrophobic, electrostatic and steric fields generated from the model are shown in Figures 3C–E. The (green) favored steric features (Figure 3C) are coincident with the meta position of the phenyl ring, while the unfavorable steric feature (yellow, Figure 3C) correlate with the occurrence of para-biphenyl substituted and inactive n-alkyl species. The (yellow) favored hydrophobic feature (Figure 3E) is basically coincident with the (red) negative charge-favorable field feature (Figure 3D), and can be attributed to the importance of having either a halogen or aromatic ring feature in the meta position of the phenyl ring. Experimental and predicted pIC50’s including statistical parameters for the final model are given in Table 2, with training and test set predictions shown graphically in Figure 4B, there being on average a factor of 2x error in the IC50 predictions.
Table 2.
CoMSIA Results for inhibition of AZT excision by bisphosphonates
| CoMSIA pIC50 Predictions
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Experimental
|
Test Sets b |
|||||||||
| Compound a | IC50 (μM) | pIC50 (M) | Training Set | 1 | 2 | 3 | 4 | 5 | Pred | Residual |
| 5 | 0.5 | 6.3 | 6.1 | 6.1 | 6.1 | 6.1 | 6.2 | 6.1 | 6.1 | 0.2 |
| 6 | 0.9 | 6.1 | 5.9 | 5.9 | 5.9 | 5.8 | 5.9 | 5.9 | 5.8 | 0.2 |
| 7 | 1.0 | 6.0 | 5.6 | 5.8 | 5.8 | 5.8 | 5.8 | 5.8 | 5.8 | 0.2 |
| 8 | 1.0 | 6.0 | 5.9 | 5.8 | 5.8 | 5.8 | 5.9 | 5.8 | 5.8 | 0.1 |
| 9 | 1.3 | 5.9 | 5.9 | 5.8 | 5.8 | 5.7 | 5.8 | 5.6 | 5.6 | 0.3 |
| 10 | 1.5 | 5.8 | 5.9 | 5.9 | 5.8 | 5.8 | 5.8 | 5.8 | 5.8 | 0.0 |
| 11 | 1.9 | 5.7 | 5.9 | 5.8 | 5.7 | 5.7 | 5.8 | 5.7 | 5.8 | 0.0 |
| 12 | 2.1 | 5.7 | 5.5 | 5.6 | 5.5 | 5.6 | 5.6 | 5.6 | 5.5 | 0.1 |
| 13 | 2.2 | 5.7 | 5.5 | 5.7 | 5.7 | 5.7 | 5.6 | 5.7 | 5.7 | 0.0 |
| 14 | 2.4 | 5.6 | 5.8 | 5.6 | 5.6 | 5.6 | 5.9 | 5.6 | 5.9 | −0.3 |
| 15 | 2.9 | 5.5 | 5.4 | 5.4 | 5.4 | 5.5 | 5.7 | 5.5 | 5.5 | 0.0 |
| 3 | 3.2 | 5.5 | 5.5 | 5.7 | 5.7 | 5.6 | 5.6 | 5.6 | 5.7 | −0.2 |
| 16 | 3.3 | 5.5 | 5.6 | 5.5 | 5.6 | 5.6 | 5.6 | 5.6 | 5.6 | −0.1 |
| 17 | 3.5 | 5.5 | 5.0 | 5.1 | 5.1 | 5.2 | 5.2 | 5.2 | 5.2 | 0.3 |
| 18 | 3.9 | 5.4 | 5.4 | 5.5 | 5.4 | 5.4 | 5.4 | 5.4 | 5.4 | 0.0 |
| 19 | 3.9 | 5.4 | 5.2 | 5.2 | 5.2 | 5.2 | 5.5 | 5.2 | 5.2 | 0.2 |
| 20 | 4.6 | 5.3 | 4.7 | 4.4 | 5.5 | 5.3 | 5.3 | 5.3 | 4.4 | 0.9 |
| 21 | 5.0 | 5.3 | 5.5 | 5.6 | 5.6 | 5.5 | 5.5 | 5.5 | 5.5 | −0.2 |
| 22 | 5.2 | 5.3 | 5.8 | 5.8 | 5.8 | 5.8 | 5.8 | 5.8 | 5.8 | −0.5 |
| 23 | 5.4 | 5.3 | 5.4 | 5.3 | 5.2 | 5.1 | 5.2 | 5.2 | 5.1 | 0.2 |
| 24 | 7.9 | 5.1 | 4.9 | 5.1 | 5.1 | 5.1 | 5.1 | 5.0 | 5.1 | 0.0 |
| 25 | 9.5 | 5.0 | 4.9 | 5.0 | 5.0 | 5.0 | 4.9 | 4.7 | 4.7 | 0.3 |
| 26 | 11 | 5.0 | 5.0 | 4.8 | 5.8 | 5.2 | 5.1 | 5.2 | 5.8 | −0.9 |
| 27 | 38 | 4.4 | 4.4 | 4.2 | 4.2 | 4.2 | 4.2 | 4.2 | 4.2 | 0.2 |
| 28 | 51 | 4.3 | 4.9 | 4.4 | 4.6 | 4.5 | 4.5 | 4.5 | 4.5 | −0.2 |
|
| ||||||||||
| 32 | 302 | 3.5 | 3.8 | 3.3 | 3.4 | 3.3 | 3.2 | 3.3 | 3.3 | 0.2 |
| 38 | 302 | 3.5 | 3.1 | 3.3 | 3.3 | 3.3 | 3.4 | 3.3 | 3.3 | 0.2 |
| 41 | 302 | 3.5 | 3.9 | 3.5 | 3.5 | 3.5 | 3.6 | 3.5 | 3.5 | 0.0 |
| 42 | 302 | 3.5 | 3.7 | 3.3 | 3.3 | 3.3 | 3.4 | 3.3 | 3.3 | 0.2 |
| 40 | 302 | 3.5 | 4.2 | 4.1 | 4.0 | 4.1 | 4.1 | 4.0 | 4.1 | −0.5 |
| 33 | 302 | 3.5 | 3.2 | 3.4 | 3.3 | 3.3 | 3.2 | 3.3 | 3.3 | 0.2 |
| 43 | 302 | 3.5 | 3.9 | 3.9 | 3.9 | 3.9 | 3.9 | 3.9 | 3.9 | −0.4 |
| 34 | 302 | 3.5 | 3.6 | 3.6 | 3.6 | 3.7 | 3.6 | 3.6 | 3.6 | −0.1 |
| 31 | 302 | 3.5 | 3.3 | 3.6 | 3.6 | 3.6 | 3.6 | 3.6 | 3.6 | −0.1 |
| 45 | 302 | 3.5 | 3.4 | 3.7 | 3.6 | 3.6 | 3.4 | 3.6 | 3.6 | 0.0 |
|
| ||||||||||
| q2 | 0.63 | 0.70 | 0.68 | 0.66 | 0.67 | 0.61 | ||||
| r2 | 0.91 | 0.96 | 0.95 | 0.95 | 0.97 | 0.95 | ||||
| F test | 73 | 115 | 108 | 107 | 151 | 110 | ||||
| N | 35 | 33 | 33 | 33 | 33 | 33 | ||||
| n | 4 | 5 | 5 | 5 | 5 | 5 | ||||
| % Steric | 0.19 | 0.19 | 0.19 | 0.19 | 0.18 | 0.18 | ||||
| % Electrostatic | 0.32 | 0.33 | 0.35 | 0.35 | 0.34 | 0.35 | ||||
| % Hydrophobic | 0.28 | 0.27 | 0.27 | 0.27 | 0.29 | 0.27 | ||||
| % Donor | 0.20 | 0.20 | 0.20 | 0.20 | 0.21 | 0.20 | ||||
|
|
||||||||||
Structures shown in Figure 1.
Bold values represent predicted activities of compounds that were not included in the training set.
We next considered what features might be missing from the CoMSIA fields, or equivalently, why is the dichlorophenyl species 5 so active? One possibility is that these halogenated species might be involved in charge-transfer interactions with an electron-rich amino-acid sidechain in the protein, such as a tyrosine or phenylalanine. Indeed, a TAM mutation found in many HIV-1 mutants and present in our HIV-1 involves T215, and in previous work24 it has been proposed that a major contact between ATP and RT involves aromatic ring stacking interactions between the ATP adenine base and an aromatic ring at position 21524. In the case of a highly electron deficient bisphosphonate sidechain, this interaction could be quite substantial. Of course, in the absence of concrete structural information, we cannot rule out other possibilities, such as a v-v interaction with an electron rich DNA base, although this would probably be expected to have an effect on DNA polymerase activity, which is generally not seen. In either case, however, the presence of halogen substituents in the most potent inhibitors suggests the possibility that v/v or charge transfer interactions might be involved. To test this hypothesis, we computed semi-empirical electron affinities (EA) as well as the ab initio vertical electron affinities (VEA) for each of the bisphosphonate sidechains to see: 1) if there was any correlation between electron affinity and excision activity and 2), whether electron affinity might be a useful additional descriptor. The computed EA and VEA values are shown in Table S3 in the Supplementary Material, and we observe good agreement between ab initio and semi-empirical calculations, r2 = 0.81, F = 99, p < 0.0001 (Table S3 and Figure S2, Supplementary Material). When the experimental pIC50 = (−log10 [IC50 (M)]) values for AZT excision by the active compounds are plotted against the EA and VEA values, we obtain the result shown in Figure 4C, in which we see a modest correlation: r2 = 0.55, F = 28 and p < 0.0001 for EA and r2 = 0.37, F = 14, p = 0.0012 for the VEA. These results are of interest since they suggest that it might be possible to improve upon the CoMSIA results by incorporating the EA or VEA values as additional descriptors. This indeed turns out to be the case (Supplemental Information Tables S4-6) with incorporation of the VEA term resulting in significant improvements in r2 (0.91→0.97), F (73→144) and the rms error (0.32→0.10). Clearly then, using the CoMSIA and VEA results lead to good activity predictions, and should be of help in the future design of improved excision inhibitors.
The bisphosphonates we have investigated here have IC50 values as low as 500nM in AZT excision. This is ~6x higher than that found with 2GP, however, unlike 2GP, the bisphosphonates tested here have essentially no activity on DNA polymerase activity. This is of importance since our purpose is to block, exclusively, AZT excision (not AZT incorporation). We also find that the most potent inhibitors, in general, have IC50 values for the inhibition of human cell growth >250μM, since they do not inhibit the human FPPS (or GGPPS) enzymes, again making them of interest for further development.
3. Conclusion
The results we have presented above are of interest for a number of reasons. First, we have tested a library of 42 bisphosphonates for their ability to inhibit the HIV-1 reverse transcriptase catalyzed phosphorolysis of AZT from AZT-terminated primers by ATP. The four most active compounds are all halogen containing aromatic species having IC50 values of ≤1 μM. Second, we have used 3D-QSAR methods to investigate structure-activity relationships. A classification method was found to have an accuracy of 94% in categorizing compounds into three discrete classes: active, moderately active and inactive. In addition, a CoMSIA partial least square regression model was found to yield a predictive model (q2 = 0.63, r2 = 0.90, F = 73, n=35) for AZT excision. Interpretation of the resulting fields showed a preference for large, hydrophobic/steric substituents at the meta- rather than para- position of the first ring. Additionally, electron-withdrawing ring substituents (negative charge favored) enhanced activity. Third, we find that there is no significant correlation (r2 = 0.10) between RT-catalyzed AZT excision inhibition and the growth inhibition of three human cell lines, with the most potent excision inhibitors having essentially no activity (>250 μM) in cell growth inhibition. Fourth, we find that there is no inhibition of RT catalyzed DNA synthesis by these potent bisphosphonate excision inhibitors. When taken together, these results strongly support the idea that bisphosphonates may have utility as inhibitors of AZT-excision catalyzed by HIV-1 RT, provided suitable formulations or delivery vehicles are available to facilitate uptake of the charged bisphosphonates into HIV-infectable target cells such as T-lymphocytes. Nonetheless, given the low cytotoxicity of the novel bisphosphomnates described in the present work, because they do not target the FPPS enzyme, such compounds are of interest in the context of the future development of novel anti-infectives for HIV-1.
4. Experimental Section
4.1. General
All reagents used were purchased from Aldrich (Milwaukee, WI). The purities of all compounds were routinely monitored by using 1H and 31P NMR spectroscopy at 400 or 500 MHz on Varian (Palo Alto, CA) Unity spectrometers using, in some instances, absolute spin-count quantitative analyses. TLC was performed on silica gel F254 plates (Merck) and UV light (254 nm) and 1% KMnO4 solution were used for visualization.
4.2. Synthesis
The synthesis and characterization of all compounds except 29 and 30 have been reported elsewhere14 and the samples used here were from the same batches. The synthesis of 29 and 30 were shown below:
4.2.1. 2-(3,4-Dichlorophenyl)ethyl-1,1-bis(methylphosphinic acid) (29)
NaH (44 mg, 1.1 mmol, 60% in oil) was added at 0°C to diethyl methylenebis(methylphosphinate)25 (228 mg, 1 mmol) in dry THF. 3,4-Dichlorobenzyl bromide (240 mg, 1 mmol) was added to the resulting solution and after 1 h, the reaction was quenched with an NH4Cl solution and extracted with diethyl ether (3 × 20 mL). The organic layer was dried and evaporated and the residue purified using column chromatography (silica gel, ethyl acetate: MeOH/10: 1). The diethyl ester obtained was treated with 3 equiv. of TMSBr in CH2Cl2 for 6h. The mixture was then evaporated to dryness, dissolved in water/MeOH, and neutralized to pH ~6. Compound 29 was obtained by filtration as a white powder. 1H NMR (400 MHz, D2O): δ 1.21 (d, J = 14.4 Hz, 6H, CH3), 2.3–2.45 (m, 1H, CH), 2.96 (td, J = 15.2Hz, 6.8 Hz, 2H, CH2), 7.1–7.3 (m, 3H, aromatic); 31P NMR (162 MHz, D2O): δ 47.4; Anal. Calcd for C10H13Cl2O4P2·1.5H2O·0.6NaBr: C, 27.19; H, 3.65. Found: C, 27.14; H, 3.47.
4.2.2. 2-(3,4-Dichlorophenyl)-1-(methylphosphino) ethylphosphonic acid (30)
NaH (44 mg, 1.1 mmol, 60% in oil) was added at 0°C to triethyl methylphosphinomethylenephosphonate26 (258 mg, 1 mmol) in dry THF. 3,4-Dichlorobenzyl bromide (240 mg, 1 mmol) was added to the resulting solution and after 1 h, the reaction was quenched with an NH4Cl solution and extracted with diethyl ether (3 × 20 mL). The organic layer was dried and evaporated and the residue purified using column chromatography (silica gel, ethyl acetate: MeOH/10: 1). The diethyl ester obtained was treated with 4.5 equiv. of TMSBr in CH2Cl2 for 6h. The mixture was then evaporated to dryness, dissolved in water/MeOH, and evaporated again to give compound 30 as a white powder. 1H NMR (400 MHz, D2O): δ 1.23 (d, J = 14.4 Hz, 3H, CH3), 2.2–2.35 (m, 1H, CH), 2.85–3.05 (m, 2H, CH2), 7.1–7.3 (m, 3H, aromatic); 31P NMR (162 MHz, D2O): δ 18.3 (s, 1P), 49.6 (s, 1P). Anal. Calcd for C9H12Cl2O5P2: C, 26.75; H, 3.24. Found: C, 26.90; H, 3.00.
4.3. Biological activity testing
4.3.1. Protein Expression
TAM-RT with the mutations D67N/K70R/T215F/K219Q was expressed and purified as previously described27. The protein concentration of the purified enzyme was determined by spectrophotometry at 280 nm using an extinction coefficient of 260,450 M−1 cm−1. [3H]-TTP and [γ-32P]-ATP were products of PerkinElmer Life Sciences (Boston, MA). The homopolymeric template-primer (T/P) poly(rA)-oligo(dT)12–18 used for steady state kinetic studies was obtained from GE Healthcare (Piscataway NJ). AZTTP as well as DNA oligonucleotides used to prepare the heteropolymeric T/P for excision assays were obtained from Trilink Biotechnologies (San Diego, CA).
4.3.2. Steady-state analysis of bisphosphonate inhibition of RT DNA polymerase activity
TAM-RT RNA-dependent DNA polymerase activity in the absence or presence of varying concentrations of bisphosphonate inhibitor was determined using 0.2 units/mL of poly(rA)-oligo(dT)12–18, 2 nM RT, 5 μM [3H] TTP in a final volume of 50 μL of 50 mM Tris-HCl, pH 8.0 containing 60 mM KCl and 10 mM MgCl2. Samples were incubated for 20 min at 37 °C and then quenched by adding 200 μL of ice-cold 10% trichloroacetic acid containing 20 mM sodium pyrophosphate. After a 20 minute incubation on ice, samples were filtered through 1.2 μm glass fiber type-C filters in 96-well microplates (Millipore, Bedford MA). The filters were washed sequentially with 10% TCA and ethanol, and the extent of [3H]-TMP incorporation into the precipitated nucleic acid was quantified by liquid scintillation spectrometry.
4.3.3. Assay of RT-catalyzed ATP-mediated phosphorolysis of chain-terminating AZT
In vitro ATP-dependent AZTMP excision reactions were carried out using a direct assay, essentially as previously described (4). Briefly, a 19-nt DNA primer (3′-CAG GGG GGA AAA GAA AAT T -5′) was annealed to a 57-nt DNA template (5′-CGT TGT CAG TGA ATC AGC CCT TCC AGT CCC CCC TTT TCT TTT AAA AAG TGG CTA AGA-3′) to prepare the heteropolymeric template/primer (T/P). The 19 nt DNA primer was 5′-radiolabeled with [γ-32P]-ATP and T4 polynucleotide kinase (New England BioLabs, Ipswich, MA) according to the manufacturer’s instructions. The labeled primer was purified using denaturing polyacrylamide gel electrophoresis on 7M urea-16% polyacrylamide gels, followed by elution from the gel. The purified 5′-32P-labeled primer was then annealed to the 57 nt DNA template using a 1:1.2 molar ratio of template to primer, by heating to 90°C followed by slow cooling to ambient temperature to form the 57 nt/19 nt template-primer (57/19 T/P) substrate. AZTMP was added to the primer 3′-terminus of the 57 nt/5′-[32P]-19 nt T/P by incubating with wt RT and 100 μM AZTTP for 30 min at 37°C. The resulting [32P]-labeled, chain-terminated 20 nt primer was purified by elution of the appropriate band following electrophoretic resolution on 7M urea-16% acrylamide denaturing gels. The purified chain-terminated primer was then re-annealed to the 57 nt DNA template DNA primer and stored at −20°C for use as T/P in the phosphorolysis experiments.
The phosphorolytic removal of chain-terminating 3′-AZTMP in the absence or the presence of varying concentrations of bisphosphonates was assayed by incubating TAM-RT with the AZTMP-terminated T/P (4:1 ratio of RT:T/P) in 50 mM Tris-HCl pH 8.0, 50 mM KCl. Reactions were initiated by addition of 10 mM MgCl2 and 3.0 mM ATP. Inorganic pyrophosphatase (0.01 U; Sigma) was included in phosphorolysis reactions using ATP, to ensure removal of any contaminating PPi in the ATP preparation. Aliquots were removed at defined times and quenched with an equal volume of gel loading dye (98% deionized formamide, 10 mM EDTA and 1 mg/mL each of bromophenol blue and xylene cyanol). The 20 nt 3′-AZTMP terminated substrate and 19 nt excision products were separated by denaturing gel electrophoresis and quantified by phosphorimaging.
4.3.4. Human Cell Growth Inhibition Assay
Human tumor cell lines MCF-7 (breast adenocarcinoma), NCI-H460 (lung large cell), and SF-268 (central nervous system glioblastoma) were obtained from the National Cancer Institute and maintained at 100% humidity and 5% CO2 at 37°C. Cell lines were cultured in RPMI-1640 medium supplemented with 2mM L-glutamine and 10% fetal bovine serum (Gibco, Grand Island, NY) at 37°C in a 5% CO2 atmosphere with 100% humidity. 0.01 M compound stock solutions were prepared in water or DMSO. A broth microdilution method was used to determine bisphosphonate growth inhibition IC50 values. Compounds were half-log serial diluted from 316μM - 10nM into 96-well TC-treated flat bottom plates (Corning Inc., Corning, NY) using cell culture media. In cases where DMSO was used, the range of dilution was 100μM - 10nM. Cells were plated at a density of 5,000 cells/well and contained 10 μL of the compound of interest in a final volume of 100 μL. Cells were then incubated for 4 days, at which time an MTT ((3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide) cell proliferation assay (ATCC, Manassas, VA) was performed, in order to obtain growth inhibition dose response curves. Briefly, cells were incubated for 2 hours with the tetrazolium salt, lysed with detergent and incubated O/N, protected from light and at room temperature. The absorbance at 600nm was read the following day using a SpectraMax Plus 384 spectrophotometer (Molecular Devices, Sunnyvale, CA). The compound concentrations for 50% growth inhibition (IC50 values) were obtained by fitting the absorbance data to a rectangular hyperbolic function: , where I is the percent inhibition, Imax = 100% inhibition and C is the concentration of the inhibitor, using GraphPad PRISM 3.0 software for Windows (Graphpad Software Inc., San Diego, CA).
4.3.5. FPPS Inhibition
Human FPPS was expressed and purified as described previously28. Human FPPS assays were carried out using 96 well plates with a 200 μL reaction mixture in each well. The condensation of geranyl diphosphate with isopentenyl diphosphate was monitored by a continuous spectrophotometric assay for phosphate releasing enzymes29. The reaction buffer contained 50 mM Tris-HCl, 1 mM MgCl2, at pH 7.4. The compounds investigated were pre-incubated with the enzyme for 30 minutes at 20°C. The IC50 values were obtained by fitting the inhibition data to a normal dose-response curve using Origin 6.1 (OriginLab Corporation, Northampton, MA).
4.4. Computational Methods
4.4.1. Pharmacophore Model
A common feature pharmacophore was constructed in MOE21 using the consensus pharmacophore module and the polarity-charge-hydrophobicity (PCH) scheme. The pharmacophore was perceived from energy minimized structures of the most active compounds using tolerances of 1.2 and a threshold of 25%. The final model was simplified by combining the features of each individual P-O group into two master functions, each describing the phosphonate group as an anionic metal ligator. Tolerances (sphere sizes) encompassing each feature were enlarged uniformly in order to relax the mapping requirements.
4.4.2. SIMCA analysis
To carry out classification analysis we used the Soft Independent Modeling of Class Analogy22 (SIMCA) module in Sybyl 7.3. The dataset was divided into three classes: active (IC50 < 10 μM), moderately active (10–100 μM), and inactive (IC50 > 100 μM). SIMCA analysis was then performed by using the assigned classes and CoMSIA fields (see below), with default settings (CoMFA standard scaling and column filtering at 2.0 kcal/mol). Models were automatically cross-validated by the algorithm, with classification rates, sum of squared residuals and other details being reported.
4.4.3. CoMSIA Alignment
To carry out our comparative molecular similarity indices analysis (CoMSIA)30 calculations, we began by building and geometry optimizing the bisphosphonate structures in the Molecular Operating Environment program (MOE)21. Minimized structures were generated using the MMFF94x forcefield 31 and default minimization parameters, which apply three successive non-linear optimization methods: steepest descent, conjugate gradient and truncated-Newton. Minimized structures were then manually aligned to the lowest energy conformation of the most active compound (5). The manual alignment was subsequently refined using the flexible alignment module in MOE, which perceives common features between molecules (e.g. aromaticity and volume) and attempts to improve the alignment of such features. The aligned compounds were then exported to Sybyl 7.332 for analysis: all CoMSIA fields were computed on a rectangular grid, using default grid spacing and probe atoms.
4.4.4. Quantum Chemical Calculations
Vertical electron affinities (VEA) were calculated for the sidechains of aromatic bisphosphonates using the Gaussian 03 program33 following an approach described previously.34 Basically, compounds were geometry optimized using the B3LYP functional and a 6-31G* basis set, and a frequency calculation was used to confirm that the geometries did not represent saddle points. Single point energy calculations were then carried out for the neutral, optimized geometries, using B3LYP, a 6-311+G(2df,p) basis set with tight convergence, no symmetry constraints, and an ultrafine grid. UB3LYP was used to calculate the single point energy structures, after adding an additional electron, again using the 6-311+G(2df,p) basis set and the options specified for the neutral molecules. The stability of the wavefunction for the charged species was verified with the Stable=Opt keyword. The VEA was taken to be the difference between the total eigen-energies of the neutral and charged species. For purposes of comparison, sidechain electron affinities were also computed semi-empirically within Maestro using the PM3 Hamiltonian35. Structures of the sidechains were prepared using LigPrep36 with default options. Electron affinities (EA) were computed using QikProp37 specifying full, rather than fast mode.
Acknowledgments
This work was supported by the United States Public Health Service (NIH grants AI-060452 and AI-052010 to M.A.P., GM-073216 to E.O.). Y.S. was supported by a Leukemia and Lymphoma Society Special Fellowship. J.M.W.C. was a Jean Dreyfus Boissevein Fellow.
References
- 1.UNAIDS/WHO AIDS Epidemic Update. United Nations Programme on HIV/AIDS; December, 2006, [Google Scholar]
- 2.Mitsuya H, Weinhold KJ, Furman PA, St Clair MH, Lehrman SN, Gallo RC, Bolognesi D, Barry DW, Broder S. Proc Natl Acad Sci U S A. 1985;82:7096. doi: 10.1073/pnas.82.20.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer PR, Matsuura SE, So AG, Scott WA. Proc Natl Acad Sci U S A. 1998;95:13471. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dharmasena S, Pongracz Z, Arnold E, Sarafianos SG, Parniak MA. Biochemistry. 2007;46:828. doi: 10.1021/bi061364s. [DOI] [PubMed] [Google Scholar]
- 5.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. Mol Cell. 1999;4:35. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 6.Larder BA, Kemp SD. Science. 1989;246:1155. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 7.Kellam P, Boucher CA, Larder BA. Proc Natl Acad Sci U S A. 1992;89:1934. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrigan PR, Kinghorn I, Bloor S, Kemp SD, Najera I, Kohli A, Larder BA. J Virol. 1996;70:5930. doi: 10.1128/jvi.70.9.5930-5934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooker DJ, Tachedjian G, Solomon AE, Gurusinghe AD, Land S, Birch C, Anderson JL, Roy BM, Arnold E, Deacon NJ. J Virol. 1996;70:8010. doi: 10.1128/jvi.70.11.8010-8018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemp SD, Shi C, Bloor S, Harrigan PR, Mellors JW, Larder BA. J Virol. 1998;72:5093. doi: 10.1128/jvi.72.6.5093-5098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parniak MA, McBurney S, Oldfield E, Mellors JW. Antiviral Therapy. Tenerife Sur-Costa Adeje; Canary Islands, Spain: 2004. Bisphosphonate inhibitors of nucleoside reverse transcriptase inhibitor excision. [Google Scholar]
- 12.Cruchaga C, Anso E, Rouzaut A, Martinez-Irujo JJ. J Biol Chem. 2006;281:27744. doi: 10.1074/jbc.M603360200. [DOI] [PubMed] [Google Scholar]
- 13.Cruchaga CAE, Font M, Martino VS, Rouzaut A, Martinez-Irujo JJ. Biochem J. 2007;405:165. doi: 10.1042/BJ20061831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotsikorou E, Song Y, Chan JM, Faelens S, Tovian Z, Broderick E, Bakalara N, Docampo R, Oldfield E. J Med Chem. 2005;48:6128. doi: 10.1021/jm058220g. [DOI] [PubMed] [Google Scholar]
- 15.McCloskey EV, Yates AJ, Beneton MN, Galloway J, Harris S, Kanis JA. Bone. 1987;8(Suppl 1):S35. [PubMed] [Google Scholar]
- 16.Widler L, Jaeggi KA, Glatt M, Muller K, Bachmann R, Bisping M, Born AR, Cortesi R, Guiglia G, Jeker H, Klein R, Ramseier U, Schmid J, Schreiber G, Seltenmeyer Y, Green JR. J Med Chem. 2002;45:3721. doi: 10.1021/jm020819i. [DOI] [PubMed] [Google Scholar]
- 17.Mao J, Mukherjee S, Zhang Y, Cao R, Sanders JM, Song Y, Zhang Y, Meints GA, Gao YG, Mukkamala D, Hudock MP, Oldfield E. J Am Chem Soc. 2006;128:14485. doi: 10.1021/ja061737c. [DOI] [PubMed] [Google Scholar]
- 18.Sanders JM, Gomez AO, Mao J, Meints GA, Van Brussel EM, Burzynska A, Kafarski P, Gonzalez-Pacanowska D, Oldfield E. J Med Chem. 2003;46:5171. doi: 10.1021/jm0302344. [DOI] [PubMed] [Google Scholar]
- 19.Sanders JM, Song Y, Chan JM, Zhang Y, Jennings S, Kosztowski T, Odeh S, Flessner R, Schwerdtfeger C, Kotsikorou E, Meints GA, Gomez AO, Gonzalez-Pacanowska D, Raker AM, Wang H, van Beek ER, Papapoulos SE, Morita CT, Oldfield E. J Med Chem. 2005;48:2957. doi: 10.1021/jm040209d. [DOI] [PubMed] [Google Scholar]
- 20.Hudock MP, Sanz-Rodriguez CE, Song Y, Chan JM, Zhang Y, Odeh S, Kosztowski T, Leon-Rossell A, Concepcion JL, Yardley V, Croft SL, Urbina JA, Oldfield E. J Med Chem. 2006;49:215. doi: 10.1021/jm0582625. [DOI] [PubMed] [Google Scholar]
- 21.MOE. Chemical Computing Group, Inc; Montreal, Quebec: 2006. 2006.08. [Google Scholar]
- 22.Wold S, Sjostrom M. Chemometrics: theory and application. American Chemical Society; 1977. [Google Scholar]
- 23.Bush BL, Nachbar RB. J Comput Aided Mol Des. 1993;7:587. doi: 10.1007/BF00124364. [DOI] [PubMed] [Google Scholar]
- 24.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. J Virol. 2001;75:4832. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prishchenko AA, Novikova ZS, Lutsenko IF. Zhurnal Obshchei Khimii. 1977;47:2689. [Google Scholar]
- 26.Teulade MP, Savignac P, Aboujaoude EE, Collignon N. J Organomet Chem. 1986;312:283. [Google Scholar]
- 27.Fletcher RS, Holleschak G, Nagy E, Arion D, Borkow G, Gu Z, Wainberg MA, Parniak MA. Protein Expr Purif. 1996;7:27. doi: 10.1006/prep.1996.0004. [DOI] [PubMed] [Google Scholar]
- 28.Kavanagh KL, Guo K, Dunford JE, Wu X, Knapp S, Ebetino FH, Rogers MJ, Russell RG, Oppermann U. Proc Natl Acad Sci U S A. 2006;103:7829. doi: 10.1073/pnas.0601643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webb MR. Proc Natl Acad Sci U S A. 1992;89:4884. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klebe G, Abraham U, Mietzner T. J Med Chem. 1994;37:4130. doi: 10.1021/jm00050a010. [DOI] [PubMed] [Google Scholar]
- 31.Halgren TA. J Comput Chem. 1996;17:490. [Google Scholar]
- 32.Sybyl 7.3. Tripos Inc; St. Louis: 2006. [Google Scholar]
- 33.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, et al. Gaussian, Inc; Wallingford, CT: 2004. [Google Scholar]
- 34.Mariano D, Vera A, Pierini AB. Phys Chem Chem Phys. 2004;6:2899. [Google Scholar]
- 35.Maestro 8.0. Schrodinger Inc; Portland, OR: [Google Scholar]
- 36.LigPrep 2.1. Schrodinger Inc; Portland, OR: [Google Scholar]
- 37.QikProp 3.0. Schrodinger Inc; Portland, OR: [Google Scholar]