Abstract
Five isolates of M. hapla originating from the Netherlands and California were inbred by sequential transfer of single egg masses to produce six strains. Cytological examination showed that oocytes of these strains underwent meiosis and had n = 16 chromosomes. Strains were tested for ability to infect and to develop on several hosts by in vitro assays. The two strains from California infected tomato roots at a higher rate than those from the Netherlands, but no difference among strains was seen for ability to develop on tomato with or without the resistance gene Mi-1. All strains developed on the common bean cultivar Kentucky Wonder, but strains differed in ability to develop on the nematode-resistant cultivar NemaSnap. Strain-specific differences were also seen in ability to infect and to develop on Solanum bulbocastanum clone SB-22. Strain VW13, derived from nematodes treated with the mutagen EMS, was defective in ability to infect tomato and potato roots in vitro. Comparison of DNA using AFLP markers showed an average of 4% of the bands were polymorphic across the six strains, but no correlation was observed between the geographical origin or virulence and DNA polymorphism pattern.
Keywords: AFLP, Northern root-knot nematode, Meloidogyne hapla, pathogenicity, virulence
Root-knot nematode species can have wide host ranges, but there are within-species differences in host range and in virulence on varieties of a host species (Roberts, 1995). Little is known about the genes in the nematode that account for these differences, in part due to the lack of a tractable genetic system to investigate their inheritance. Three of the most damaging root-knot nematode species, Meloidogyne incognita, M. javanica, and M. arenaria, reproduce by obligate mitotic parthenogenesis and are not amenable to genetic analysis. However, while cytological Race B isolates of M. hapla also reproduce by mitotic parthenogenesis, Race A isolates of this species undergo meiosis and reproduce by facultative meiotic parthenogenesis (Triantaphyllou, 1966, 1985). In this form of reproduction, oocytes undergo meiotic reduction to form a diploid nucleus and diploid polar body. The second meiotic division of the nucleus is arrested at telophase. If the female has been fertilized, nuclear division will be completed, and the sperm will fuse with the haploid egg pronucleus to form a zygote. In the absence of fertilization, the meiosis II products will fuse to restore the somatic chromosome number (Van der Beek et al., 1998). The reproductive mode of M. hapla Race A suggests that it should be possible to carry out genetic crosses and to analyze segregation of pathogenicity and virulence differences.
Although M. hapla reproduces on tomato, potato, carrots, alfalfa, onion, and many other crops, causing considerable economic damage (Mitkowski and Abawi, 2000), isolates of this species differ in host range and pathogenicity (Griffin and McKenry, 1989; Riggs, 1991; Djian-Caporalino et al., 1999). Isolates from the Netherlands have been found to differ in ability to reproduce on the wild potato species Solanum bulbocastanum (Janssen et al., 1997). In another example, isolates of M. hapla were found to differ in virulence on the common bean (Phaseolus vulgaris) cultivar NemaSnap, which carries a single, dominant gene for M. hapla resistance (Chen and Roberts, 2003a). In this case, there is genetic evidence that avirulence in the nematode segregates as a single, dominant locus (Chen and Roberts, 2003b). Although the tomato gene Mi-1 confers resistance to M. incognita, M. arenaria, M. javanica, and potato aphid Macrosiphum euphorbiae (Williamson, 1998), it has been found to be ineffective against M. hapla (Brown et al., 1997; Williamson, 1998).
In this study, we produce and characterize six inbred strains of M. hapla originating from isolates obtained from the Netherlands and from California for cytological race, chromosome number, and DNA polymorphisms. For each strain, we compared ability to infect and develop on specific hosts using an in vitro assay system and investigated the relationship of DNA markers to pathogenicity and geographical origin.
Material and Methods
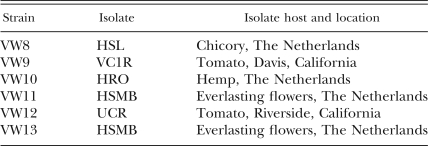
Nematode strains: Six strains of M. hapla derived from five isolates were used in this study (Table 1). Isolate VC1R was obtained from a nematode tank in the Vegetable Crops Department greenhouse at University of California, Davis, that contained a mix of California field isolates of root-knot nematode; UCR was obtained from the Department of Nematology at University of California, Riverside; and HSL, HRO, and HSMB were obtained from G. Janssen in the Netherlands (Janssen et al., 1997). These five isolates were inbred for 18 generations by sequentially transferring a single egg mass on tomato plants to produce strains VW8 to VW12 (Table 1). VW13 was obtained from HSMB eggs that were exposed to the mutagen ethylmethane sulphonate (EMS; Sigma-Aldrich, St. Louis, MO). Eggs were soaked in 25 mM EMS for 20 min with gentle shaking every 4 min (Sulston and Hodgkin, 1988) in a 1.5-ml Eppendorf tube. Eggs were then washed 3–4 times in sterile water and inoculated onto sterile tomato roots in MS medium. Egg masses were harvested 5 wk later and tested for infectivity. One egg mass produced juveniles with decreased infectivity (fewer nematodes in tomato roots) in sterile culture, but no obvious decrease in infectivity was observed in the greenhouse. Strain VW13 was a result of inbreeding of this egg mass.
Table 1.
Meloidogyne hapla strains used in this study.
All M. hapla strains were reared on VFNT tomato (Lycopersicon esculentum) in greenhouses. The original isolates were identified as M. hapla by isozyme analysis (Esbenshade and Triantaphyllou, 1987), perineal patterns (Hirschman, 1985), and a pair of M. hapla species-specific primers (Williamson et al., 1997). The inbred strains were confirmed to be M. hapla with the same species-specific primers.
Specimen preparation for karyotype observations: Four wk after inoculation, soil was rinsed from infected roots, and root pieces were placed into a petri dish with a 0.9% NaCl solution. Young females were dissected from roots and cut gently with a scalpel to release their gonads. The gonads were smeared onto slides according to Triantaphyllou (1985). Chromosomes were stained with Hoechst 33258 (Sigma-Aldrich, St. Louis, MO) using a slight modification of the fixation and staining procedure from Van der Beek et al. (1998) as follows. Smears of gonads were hydrolyzed in 1 M HC1 for 10 min at room temperature on slides, fixed in Carnoy's solution (acetic acid:ethanol, 1:3) for 30 min, and then dried in ambient air. The slides were incubated in PBS (137 mM NaCl, 2.7 mM KC1,10 mM Na2HPO4 and 2 mM KH2PO4) at pH 7.4 for 5 min, immediately stained in 1 μg/ml Hoechst 33258 in PBS for 5 min, and then rinsed in sterile water or PBS for 5–10 min to reduce background. After drying, the smears were embedded in 50% glycerine, and coverslips were sealed with nail polish. Slides were stored in the dark until examination using a Nikon Microphot-SA fluorescent microscope.
Plant materials and pathogenicity assays: Seeds of common bean (Phaseolus vulgaris L.) cultivars NemaSnap and Kentucky Wonder were kindly provided by Dr. Bohac, USDA-ARS at Charleston, SC. Bean and tomato seeds, Motelle (Mi) and Moneymaker (mi), were surface-sterilized by soaking in 10% commercial bleach for 10 min and rinsed with sterile water four times. The seeds then were placed on MS medium to germinate. The roots were excised when they grew to about 2-cm long and placed on MS medium in petri dishes. Solanum bulbocastanum SB-22 is a vegetative clone obtained from Dr. Brown, USDA-ARS at Prosser, WA (Brown et al., 1996). The stems were surface-sterilized (soaked in sterile water with 1–2 drops of Tween 20 for 3 min, 70% ethanol for 5 min, 1% of commercial bleach for 8 min, and 3–4 rinses with sterile water) and placed on MS medium in 1% phytogel with 2% sucrose and supplemented with 0.02 mg/liter of NAA and 0.1 g/liter of myoinositol in Magenta boxes. The roots emerged after 7–10 d in this medium and then were excised when they reached 1–2 cm in length and grown on MS medium with 2% sucrose in petri dishes. The roots were grown at 24°C to 26°C in the dark.
Nematode eggs were isolated from plants and cleaned by sucrose flotation as previously described (Branch et al., 2004). Eggs were then surface-sterilized by shaking vigorously in 10% commercial bleach for 5 min, spun at 900g for 5 min, rinsed with sterile water 1–2 times, pelleted by spinning at 900g, and then treated with 5% commercial bleach followed by four rinses with sterile water. For hatching, eggs were placed on a sterile apparatus consisting of 8–10 layers of Kim-wipe paper tissue placed on a metal screen situated at about 1/3 from the lip of a 500-ml cup filled with sterile water that just touched the bottom of the tissue. Eggs were allowed to hatch for 2–3 d at room temperature. Hatched J2 that had settled to the bottom of the cup were collected and counted.
Approximately 250 J2 (2 J2/μl) were inoculated onto each petri dish containing three excised roots growing on MS medium. The infected roots were stained with acid fuchsin 3 wk after inoculation (Byrd et al., 1983). Roots were examined under a dissecting microscope, and the number of nematodes inside roots and percentage of developed nematodes were assessed. Nematode development was defined as noticeable swelling and change in body shape in comparison to the infective J2 (Ho et al., 1992; Branch et al., 2004). All tests were repeated at least three times, and similar results were obtained. All data were subjected to t-test or analysis of variance (ANOVA) (SAS Institute, Cary, NC).
DNA extraction from nematode eggs: Nematode eggs cleaned by sucrose flotation were rinsed with sterile water three times, pelleted in 1.5-ml Eppendorf tubes, frozen in liquid nitrogen, and stored at −80 °C. Between 200,000 and 1,000,000 frozen eggs were ground with a pre-cooled mortar and pestle for each nucleic acid extraction. One volume of homogenization buffer (50 mM NaCl, 50 mM Tris-Cl pH 7.5, 5 mM EDTA, 0.5% SDS, and 200 μg/ml proteinase K) was added to the ground nematode powder and incubated for 2–3 hr at 50°C. The sample was then purified according to a phenol/chloroform procedure (Sambrook et al., 1989). Following ethanol precipitation, DNA was resuspended in H2O to a final concentration of 50 ng/μl and stored at −20 °C.
AFLP fingerprinting: Eco RI/Mse I AFLP templates were prepared from nematode DNA and amplified with nonselective Eco RI and Mse I primers (Vos et al., 1995). Approximately 150 ng of nematode genomic DNA was digested for each genotype. After PCR with 33P-end-labeled selective Eco RI primer and unlabeled selective Mse I primers, 10 μl of 2× sequencing dye (98% formamide, 10 mM EDTA, 0.25 mg/ml xylene cyanol, and 0.25 mg/ml bromophenol blue) was added to each 10-μl reaction. The samples were denatured at 90 °C for 5 min, rapidly cooled on ice, loaded on a 6% denaturing polyacrylamide gel (National Diagnostics, Atlanta, GA), and electrophoresed for 3 h at 50 watts. The gel was dried using a Bio-Rad Gel Dryer (Model 583) and exposed to Kodak Biomax film for several days at room temperature. Fragment names used in the text designate nonspecific primer sequence (E or M) followed by selective nucleotides and fragment size in nucleotides (Vos et al., 1995).
AFLP fragment isolation, cloning, and sequencing: The area corresponding to the AFLP fragment of interest was excised from the acrylamide gel and incubated for several hours at 37 °C in 0.6 ml elution buffer (0.5 M ammonium acetate, 0.01 M magnesium acetate, 0.1% SDS, and lm M EDTA). The elution buffer containing the fragment was filtered through an Ultrafree-MC filter, 0.45 μM (Millipore, Bedford, MA). Next, 2.5 volumes of 100% ethanol was added to the flow-through, and the mixture was chilled for 10 min in a dry ice/ ethanol bath. After a 10-min centrifugation, the pellet was suspended in 0.4 ml of 0.3 M sodium acetate, then 1 ml ethanol was added and thoroughly mixed. The mixture was incubated overnight at −20 °C. After centrifugation, the pellet was rinsed with 70% ethanol and suspended in 10–20 μl water. Isolated fragments were amplified by PCR using the nonselective primers and then cloned into the vector pGEM-T easy (Promega Inc., Madison, WI). The cloned fragments were sequenced at the UC-Davis Division of Biological Sciences Sequencing Facility.
Phylogenetic analysis of AFLP products: Maximum parsimony and bootstrap parsimony analyses were performed on AFLP data using PAUP* 4.0bl0* (Swofford, 1998). The presence/absence of individual AFLP markers were scored as binary characters, and the branch-and-bound option of PAUP 4.0* was used to find the most parsimonious tree, which was then midpoint rooted. Bootstrap resampling was performed using 1,000 pseudoreplicates.
Results
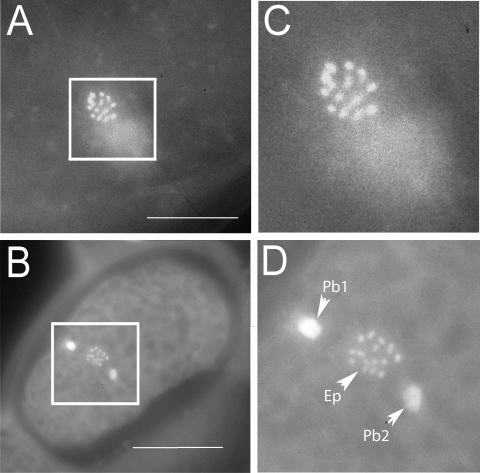
Karyotype observations: Oocytes in the spermatheca and uterus of M. hapla strains VW8, VW9, VW10, VW11, and VW12 were examined after staining. For all strains, two meiotic divisions were seen essentially as described by others for M. hapla isolates that reproduce by facultative meiotic pathenogenesis (Triantaphyllou, 1966; Van der Beek et al., 1998). Before entering the spermatheca, many eggs had nuclei in metaphase I. In most cases, chromosomes were too clustered to count, but occasionally it was possible to distinguish individual chromosomes (Fig. 1A). In other oocytes, the separate chromosomes were visible during anaphase of meiosis II (Fig. 1B). For most spreads, it was necessary to focus through the plane of the oocytes to observe all chromosomes. Counts indicated that strains VW8, VW9, VW10, and VW12 had n = 16 chromosomes. We were not able to identify any oocytes of VW11 in which chromosomes were spread sufficiently to count unambiguously, but we estimate that this strain has approximately 16 chromosomes. VW13 oocytes were not examined; however, because this strain was derived from isolate HSMB as was VW11, it is likely to have the same number of chromosomes as VW11.
Fig. 1.
Meloidogyne hapla oocytes stained with Hoescht 33258. A) Oocyte of VW8 at prometaphase of meiosis I. B) Oocyte of VW10 at anaphase of meiosis II. C, D) Enlarged views of boxed areas of A and B, respectively. In C, 15 chromosome tetrads can be distinguished. By adjusting the focal plane, we identified a total of 16 tetrads in this meiotic figure. In D, 16 chromosomes can be counted in the egg pronucleus (Ep) by adjusting the focal plane. Polar body 1 (Pbl) is highly condensed. Polar body 2 (Pb2) is less condensed but is out of the plane of focus. Bars in A and B represent 4 μm.
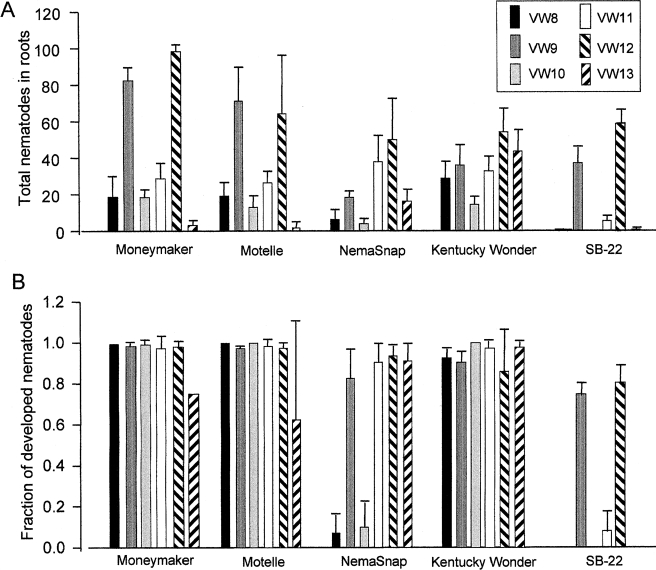
Infection and pathogenicity assays: No difference in ability to infect or to produce galls on tomato with or without the nematode resistance gene Mi-1 was found for any of the strains (Fig. 2A). However, the total number of nematodes of strains VW9 and VW12 inside tomato roots was higher (P < 0.01) than for the other four strains. VW8, VW10, and VW11 infected tomato at an intermediate level. Very few individuals (1.4 ± 2.2 nematodes per plate) of VW13 were detected inside tomato roots. While all six strains infected common bean cultivar Kentucky Wonder, fewer (P < 0.05) VW8 and VW10 juveniles were found inside roots of cultivar NemaSnap, which carries a strain-specific resistance gene to M. hapla (Chen and Roberts, 2003b). Roots of Kentucky Wonder appeared swollen when infected with each of the six strains of M. hapla. Swellings developed on NemaSnap after infection with VW9, VW11, VW12, and VW13, but were not observed on this cultivar after infection with VW8 and VW10. While both VW9 and VW12 were able to infect S. bulbocastanum clone SB22, strains VW8, VW10, VW11, and VW13 infected this host at a lower level (P < 0.01) (Fig. 2A).
Fig. 2.
Virulence differences between Meloidogyne hapla strains. A) The total number of nematodes inside roots 21 d after inoculation of each plate with 250 J2. B) Fraction of nematodes inside roots that had developed at 21 d post inoculation. The standard deviation is based on three or four replicate plates.
Significant differences in the fraction of nematodes that had developed by 3 wk after inoculation were seen between nematode strain/plant combinations by ANOVA analysis (SAS Institute, Cary, NC) (Fig. 2B). For all six strains, most of the nematodes inside tomato roots had developed with no difference due to the presence or absence of Mi-1. Interestingly, even though strain VW13 was present in low numbers inside tomato roots, a high fraction of those that entered were able to develop. Strains VW8 and VW10 failed to develop on NemaSnap (Fig. 2B), whereas for strains VW9, VW11, VW12, and VW13, the majority of the nematodes inside the roots had developed. Strains VW8, VW10, VW11, and VW13, which infected SB-22 poorly, also failed to develop on this host, whereas the majority of nematodes of strains VW9 and VW12 did develop on SB-22.
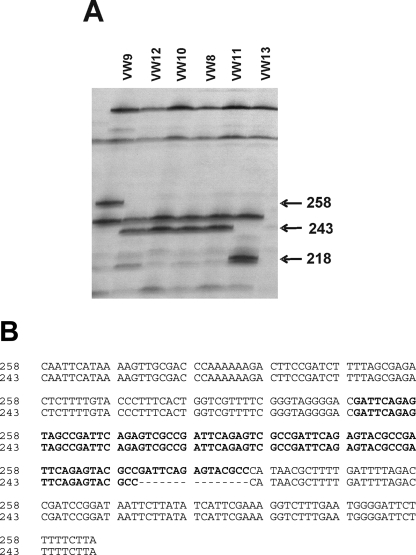
AFLP DNA polymorphisms among strains: Forty-seven pairs of selective AFLP primers were used to generate DNA fingerprints of the six strains of M. hapla. An average of 60 fragments was identified in each lane per primer combination, and between two and 15 polymorphic fragments could be detected. Some polymorphic bands were present only in one strain, but others were present in two or more strains (see Fig. 3 for example). To test the relationship between fragments of the same size in different strains, selected bands were cloned and sequenced. AFLP primers E+AG/M+AG amplified a 359-bp fragment from VW9, VW10, and VW12. Comparison of fragments cloned from VW9 and VW12 revealed that the sequences were identical, and BLAST database searches did not identify notable similarity to other sequences. A 354-bp fragment amplified with primers E+AG/M+GA from only VW12 and VW13 differed by only two nucleotides between these two strains. Primers E+AT and M+GA amplified a 243-bp fragment from VW8, VW10, VW11, and VW12 but a 258-bp fragment from VW9 (Fig. 3A). Sequence analysis showed that the 243-bp fragments cloned from VW11 and VW12 were identical and that the 258-bp fragment differed from the shorter sequences by only a 15-nucleotide insertion. This 15-nucleotide sequence was present in six tandem copies in the 258-bp fragment and in five copies in the 243-bp fragment (Fig. 3B).
Fig. 3.
AFLP pattern of Meloidogyne hapla strains. A) Portion of an AFLP gel showing patterns obtained with primers E+AT and M+GA. The numbers indicate the size in base pairs of polymorphic fragments. Strains are indicated at the top of the lanes. B) Sequence alignment of E+AT/M+GA-258 and E+AT/M+GA-243 shows that these sequences differ by number of tandem copies of a 15-nt sequence. Tandem repeat region is highlighted in bold.
The presence or absence of each of 83 polymorphic bands was scored for the six strains, and this data set was subjected to parsimony analysis. Strains VW12 and VW13 were grouped with a bootstrap value of 93%, and this was the only group receiving reliable (>70%) bootstrap support. Strains VW9 and VW12 from California were not grouped, and VW8 and VW10, the two strains found to be avirulent on SB22, were not sister strains. Interestingly, VW11 and VW13, the strains developed from isolate HSMB, also did not form a group based on AFLP polymorphisms.
Discussion
Based upon karyotype observations, we ascertained that oocytes of the M. hapla strains characterized here undergo meiotic division and therefore belong to cytological race A, as do the majority of M. hapla strains. We found that meiotic prometaphase I and anaphase II are useful stages for counting M. hapla chromosomes. Race A M. hapla isolates have a reduced chromosome number between 13 and 18, depending on the isolate (Triantaphyllou, 1985). Our strains each have approximately 16 chromosomes, indicating that genetic crosses between strains should be possible and that these strains are likely to be amenable to traditional genetic analysis.
Several factors contribute to the pathogenicity of a nematode on a particular host. For root-knot nematodes, these factors include ability to perceive and be attracted to the host, penetrate the host, locate the feeding site, initiate feeding site development, and obtain sufficient nutrients to develop to adulthood and reproduce (Trudgill, 1991; Williamson, 1999). Greenhouse assays that measure reproduction of nematodes and/or galling are commonly used to assess pathogenicity or virulence (Starr et al., 2002). However, greenhouse assays require much time and space. For example, we found that generation of sufficient plant material from S. bulbocastanum clone SB-22 to carry out greenhouse assays was prohibitive. However, for the in vitro assay, we were able to generate roots from stem cuttings in seven days. Number of nematodes inside roots at 21 days after inoculation differed for plant/nematode combinations and likely reflects attraction and ability to penetrate the roots of a particular host. The percentage of nematodes that have developed is an indication of the ability of the nematode to produce a feeding site and to obtain nutrition from the plant. We found reproducible host-specificity differences between our strains in these two traits.
The resistance to M. hapla in common bean cultivar NemaSnap has been previously noted to be strain-specific as assayed by egg masses produced in a seedling growth pouch assay (Chen and Roberts, 2003a). In greenhouse assays, we found VW9, VW11 and VW12, and VW13 were virulent and able to complete their life cycle on NemaSnap, whereas VW8 and VW10 did not reproduce (results not shown). With the in vitro assay, VW9, VW11, VW12, and VW13, but not VW8 and VW10, were able to infect and develop on NemaSnap, consistent with findings of the greenhouse assay.
Strains VW9 and VW12, both derived from California isolates, infected and developed on S. bulbocastanum (SB-22), while strains VW8, VW10, VW11, and VW13, derived from the Netherlands, were not virulent in the in vitro assay. The resistance of SB-22 to these four strains was characterized by both low numbers of nematodes in the roots and by failure of these juveniles to develop. Surprisingly, the isolate HRO, from which strain VW10 was derived, was reported to be virulent on SB-22 (Janssen et al., 1997). The virulence trait may have been lost during the inbreeding process on tomato. Alternatively, the previous study was carried out using whole plants assays, and responses may differ between whole plants and root culture. The two California isolates were found in much higher numbers inside the Solanaceous hosts than were the isolates from the Netherlands. It may be relevant that both California strains originated from M. hapla isolates that were propagated for several years on tomato, whereas the Dutch isolates were isolated originally from non-solanaceous plants (Table 1). We found no difference in infection or development on tomato with or without the resistance gene Mi-1. This supports previous reports that Mi-1 is not effective against M. hapla. It should be noted that we carried out the inbreeding and maintained our stock cultures on tomato cultivar VFNT cherry that does carry Mi-1. Thus, if the original isolates did have variability in compatibility on Mi-1-tomato, we may have selected against this trait.
Strain VW13 can barely infect in vitro tomato roots but does not appear to be impaired in ability to infect common bean in similar assays. Since this strain can infect and reproduce on tomato roots in a greenhouse assay (not shown) and can infect and develop well on bean in vitro, it is unlikely that the failure to develop on tomato in vitro is simply due to a lack of fitness. A possible explanation is that VW13 J2 are unable to perceive an attractant (or are more sensitive to a repellent) from the tomato roots in vitro. It may be that common bean roots and tomato roots under greenhouse conditions produce more or different attractants that are detected by VW13 J2. Those individuals of VW13 that enter roots do appear to develop, supporting the possibility that VW13 is defective in perception. Although this strain was derived from EMS-treated eggs, we do not know whether the observed phenotype is an EMS-induced mutation or was selected by inbreeding. In any case, genetic analysis of the heritability of this trait could be informative.
AFLP markers identified genetic differences between the M. hapla strains. For each primer combination, 3 to 5% of bands were polymorphic, a level of polymorphism that will allow us to obtain sufficient markers to produce a genetic map. Our limited sequence analysis indicated that bands of same size in different strains are identical or almost identical. For the marker band shown in Figure 3, sequences that differed by 15 nucleotides in length were identical except for a 15-nucleotide insertion/deletion, suggesting that the bands are likely to be allelic. Analysis of 83 polymorphic bands showed no obvious correlation of geographic distribution of the isolates with DNA polymorphism pattern. Strains VW11 and VW13 showed many polymorphisms, even though both were derived from Dutch isolate HSMB and did not cluster by parsimony analysis. This suggests that significant sequence polymorphisms exist within isolates.
We have developed assays that distinguish pathogenicity phenotypes of parental lines and should allow investigation of the inheritance of traits involved in parasitism. The potential of a genetic map of pathogenicity traits and molecular markers, coupled with extensive EST resources and a project to sequence the genome of M. hapla, makes this species an excellent candidate for a model system for plant-parasitic nematodes (Liu and Williamson, unpub. data; Mitreva et al., 2005).
Footnotes
Supported by National Science Foundation award 011080 to VMW.
The authors thank C. Brown (USDA-ARS), G. Janssen (CPRO-DLO, Wageningen, the Netherlands) and J. Bohac (USDA-ARS) for generously providing biological material, S. A. Nadler (University of California, Davis) for help with parsimony analysis and for valuable comments on the manuscript, G. E. Bruening for help with graphics, and E. Caswell-Chen for helpful discussions.
This paper was edited by P. A. Roberts.
Literature Cited
- Branch C, Hwang CF, Navarre DA, Williamson VM. Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Molecular Plant-Microbe Interactions. 2004;17:351–356. doi: 10.1094/MPMI.2004.17.4.351. [DOI] [PubMed] [Google Scholar]
- Brown CR, Mojtahedi H, Santo GS. The effect of the Mi gene in tomato on reproductive factors of Meloidogyne chitwoodi and M. hapla . Journal of Nematology. 1997;29:416–419. [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Yang CP, Mojtahedi H, Santo GS. RFLP analysis of resistance to Columbia root-knot nematode derived from Solanum bulbocastanum in a BC2 population. Theoretical and Applied Genetics. 1996;92:572–576. doi: 10.1007/BF00224560. [DOI] [PubMed] [Google Scholar]
- Byrd DW, Jr, Kirkpatrick T, Barker KR. An improved technique for clearing and staining plant tissue for detection of nematodes. Journal of Nematology. 1983;14:142–143. [PMC free article] [PubMed] [Google Scholar]
- Chen P, Roberts PA. Virulence in Meloidogyne hapla differentiated by resistance in common bean (Phaseolus vulgaris) Nematology. 2003a;5:39–47. [Google Scholar]
- Chen P, Roberts PA. Genetic analysis of (a)virulence in Meloidogyne hapla to resistance in bean (Phaseolus vulgaris) Nematology. 2003b;5:687–697. [Google Scholar]
- Djian-Caporalino C, Pijarowski L, Januel A, Lefebvre V, Phally T, Palloix A, Dalmasso A, Abad P. A spectrum of resistance to root-knot nematodes (Meloidogyne spp.) in sweet pepper (Capsicum annuum L.) and inheritance of heat-stable resistance in the PM687 line derived from PI322719. Theoretical and Applied Genetics. 1999;99:496–502. doi: 10.1007/s001220051262. [DOI] [PubMed] [Google Scholar]
- Esbenshade PR, Triantaphyllou AC. Enzymatic relationships and evolution in the genus Meloidogyne (Nematoda: Tylenchida) Journal of Nematology. 1987;19:8–18. [PMC free article] [PubMed] [Google Scholar]
- Griffin GD, McKenry MV. Susceptibility of Nevada synthetic XX germplasm to a California race of Meloidogyne hapla . Journal of Nematology. 1989;21:292–293. [PMC free article] [PubMed] [Google Scholar]
- Hirschman H. The genus Meloidogyne and morphological characters differentiating its species. An advanced treatise on Meloidogyne, In: Sasser J, Carter C, editors. vol. 1. Raleigh, NC: North Carolina State University Graphics; 1985. pp. 79–93. [Google Scholar]
- Ho J-Y, Weide R, Ma HM, van Wordragen MF, Lambert KN, Koornneef M, Zabel P, Williamson VM. The root-knot nematode resistance gene (Mi) in tomato: Construction of a molecular linkage map and identification of dominant cDNA markers in resistant genotypes. Plant Journal. 1992;2:971–982. [PubMed] [Google Scholar]
- Janssen G, van Norel A, Verkerk-Bakker B, Janssen R. Intra- and interspecific variation of root-knot nematodes, Meloidogyne spp., with regard to resistance in wildtuber-bearing Solanum species. Fundamental and Applied Nematology. 1997;20:449–457. [Google Scholar]
- Mitkowski NA, Abawi GS. Pathogenicity and virulence of Meloidogyne hapla populations from vegetables in New York State. Journal of Nematology. 2000;32:446. (Abstr.). [Google Scholar]
- Mitreva M, Blaxter ML, Bird DM, McCarter JP. Comparative genomics of nematodes. Trends in Genetics. 2005;21:573–581. doi: 10.1016/j.tig.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Riggs R. Resistance-breaking races of plant-parasitic nematodes. In: Nickle W, editor. Manual of agricultural nematology. New York: Marcel Dekker; 1991. pp. 827–851. [Google Scholar]
- Roberts PA. Conceptual and practical aspects of variability in root-knot nematode related host plant resistance. Annual Review of Phytopathology. 1995;33:199–221. doi: 10.1146/annurev.py.33.090195.001215. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular cloning: A laboratory manual. [Google Scholar]
- Starr JL, Bridge J, Cook R. Resistance to plant-parasitic nematodes: History, current use, and future potential. In: Starr JL, Bridge J, Cook R, editors. Plant resistance to parasitic nematodes. Wallingford, UK: CAB International; 2002. pp. 1–22. [Google Scholar]
- Swofford DL. Sunderland, MA: Sinauer Associates; 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods) [Google Scholar]
- Sulston J, Hodgkin J. Methods. The nematode Caenorhabditis elegans . In: Wood WB, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- Triantaphyllou A. Polyploidy and reproductive patterns in the root-knot nematode Meloidogyne hapla . Tournal of Morphology. 1966;118:403–414. doi: 10.1002/jmor.1051180308. [DOI] [PubMed] [Google Scholar]
- Triantaphyllou AC. Cytogenetics, cytotaxonomy, and phy-logeny of root-knot nematodes. An advanced treatise on Meloidogyne . In: Sasser J, Carter C, editors. vol. 1. Raleigh, NC: North Carolina State University Graphics; 1985. pp. 113–126. [Google Scholar]
- Trudgill DL. Resistance to and tolerance of plant-parasitic nematodes in plants. Annual Review of Phytopathology. 1991;29:167–192. [Google Scholar]
- Van der Beek J, Los J, Pijanacker L. Cytology of parthenogenesis of five Meloidogyne species. Fundamental and Applied Nematology. 1998;21:393–399. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Homes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson VM. Root-knot nematode resistance genes in tomato and their potential for future use. Annual Review of Phytopathology. 1998;36:277–293. doi: 10.1146/annurev.phyto.36.1.277. [DOI] [PubMed] [Google Scholar]
- Williamson VM. Plant nematode resistance genes. Current Opinion in Plant Biology. 1999;2:327–331. doi: 10.1016/S1369-5266(99)80057-0. [DOI] [PubMed] [Google Scholar]
- Williamson VM, Caswell-Chen EP, Westerdahl BB, Wu FF, Caryl G. A PCR assay to identify and distinguish single juveniles of Meloidoygne hapla and M. chitwoodi . Journal of Nematology. 1997;29:9–15. [PMC free article] [PubMed] [Google Scholar]