Abstract
The 18S ribosomal DNA (rDNA) and cytochrome oxidase I region of mitochondrial DNA (mtDNA) were sequenced for 24 Xiphinema americanum-group populations sourced from a number of geographically disparate locations. Sequences were subjected to phylogenetic analysis and compared. 18S rDNA strongly suggested that only X. pachtaicum, X. simile (two populations) and a X. americanum s.l. population from Portugal were different from the other 20 populations studied, whereas mtDNA indicated some heterogeneity between populations. Phylogenetically, based on mtDNA, an apparent dichotomy existed amongst X. americanum-group populations from North America and those from Asia, South America and Oceania. Analyses of 18S rDNA and mtDNA sequences underpin the classical taxonomic issues of the X. americanum-group and cast doubt on the degree of speciation within the X. americanum-group.
Keywords: 18S rDNA, longidorid, Longidoridae, molecular analysis, mtDNA, nematode, taxonomy
The taxonomy of the Xiphinema americanum-group is controversial, comprising either 34 (Luc et al., 1998), 38 (Coomans et al., 2001) or 50 (Barsi and Lamberti, 2004; Lamberti et al., 2004) putative species, depending on the taxonomic authority. For example, Luc et al. (1998) proposed that X. diffusum, X. incognitum and X. taylori were junior synonyms of X. brevicollum. Lamberti et al. (2000) heavily criticized the delineation and definition of the X. americanum-group used by Luc et al. (1998). In turn, Luc and Baujard (2001) heavily criticized Lamberti et al. (2000). However, a hierarchical tree analysis based on morphometrics of 117 X. americanum-group populations indicated that X. brevicollum, X. diffusum, X. incognitum and X. taylori could to varying degrees separate the species (Lamberti et al., 2002). Recently, using molecular techniques, Oliveira et al. (2005) unequivocally demonstrated that X. brevicolle (note, this is the species name used by Oliveira et al., 2005) and X. diffusum were indeed distinct species. In order to try and unravel these taxonomic conundrums, we have taken an expansive view and without a priori prejudice considered the maximum number of potential species notwithstanding synonymies. It is far easier to discover potential species to synonymize by employing an expansive view than to delineate species based on a conservative species list which would arguably be subsequently considered instead as an intra-specific variability.
Species belonging to the X. americanum-group have a cosmopolitan distribution, with several in the Americas and Japan being of particular economic importance, as they are natural virus-vectors of four members of the genus Nepovirus (Taylor and Brown, 1997). Biologically, some species reported from Africa, Europe and the US have been shown to have only three and not the usual four juvenile developmental stages (JDS; Halbrendt and Brown, 1992; Coomans and Heyns, 1997; Barsi and Lamberti, 2002). Recognizing the complex taxonomic issues surrounding the X. americanum-group, Lamberti et al. (2002) recommended the use of molecular tools to discriminate putative X. americanum species and/or populations.
A plethora of recent phylogenetic studies on plant-parasitic nematodes (including the Longidoridae) based on ribosomal DNA (rDNA) have been published (for example, Subbotin et al., 2001; De Ley et al., 2002; Boutsika et al., 2004a, 2004b; De Luca et al., 2004; Ferris et al., 2004; Neilson et al., 2004; Oliveira et al., 2004a, 2004b; Subbotin et al., 2004; Ye et al., 2004), with some authors utilizing rDNA to make definitive taxonomic statements on specific species either with (Subbotin et al., 2002; Oliveira et al., 2004a) or without (Boutsika et al., 2004b) supporting morphological data.
Oliveira et al. (2004b) reported that four putative species (X. brevicolle, X. diffusum, X. peruvianum and X. oxycaudatum) of the X. americanum-group had high 18S rDNA sequence homology. Furthermore, these four species separated from other Xiphinema species with strong statistical support, leading the authors to suggest that a critical taxonomic re-evaluation of the X. americanum-group was required. Similarly, He (2003) noted high sequence homology in the D2/D3 expansion region of rDNA amongst the majority of the 17 putative species of the X. americanum-group studied, although a dichotomy of the X. americanum-group was apparent, with members of the X. pachtaicum sub-group (sensu Lamberti and Ciancio, 1993) separated from the other X. americanum-group species.
Although mitochondrial DNA (mtDNA) evolves very quickly in nematodes (Moritz et al., 1987; Blouin et al., 1998; Denver et al., 2000), studies that have utilized mtDNA as a means of discriminating plant-parasitic species have been primarily restricted to Meloidogyne (Powers et al., 1986; Hugall et al., 1994, 1997; Stanton et al., 1997; Blok et al., 2002). Here we compare the utility of 18S rDNA and mtDNA to discriminate X. americanum-group populations.
Materials and Methods
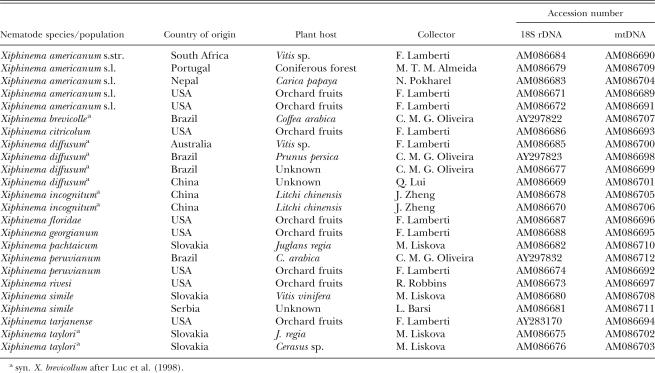
Nematodes: Twenty-four X. americanum-group populations (Table 1) were collected from a number of geographically disparate locations. Specimens were extracted from soil using standard techniques, placed into 1M NaCl, sent to the Scottish Crop Research Institute and frozen at −20°C until DNA extraction.
Table 1.
Populations of Xiphinema americanum-group nematodes used to sequence 18S rDNA and mtDNA.
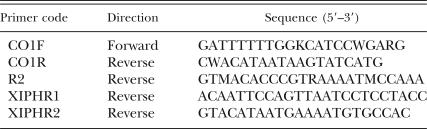
DNA extraction, PCR amplification and sequencing: DNA extraction methodology, 18S rDNA purification, sequencing and the relevant PCR conditions are all described by Oliveira et al. (2004b). A minimum of 3 nematodes from each population were sequenced. Sequences for cytochrome oxidase I (COI) were obtained using the forward CO1F primer in combination with different reverse primers (Table 2). Primers CO1F and CO1R were designed using PRIMER3 software (http://www.genome.wi.mit.edu/genome_software/other/ primer3.html) from nematode mtDNA sequences publicly available from GenBank. Our area of investigation within the COI gene was approximately between nt 9,717 and nt 10,119 based on the mtDNA genome of X. americanum (He et al., 2005; Genbank Accession: AY382608). The remaining primers were designed from X. americanum-group mtDNA sequences obtained during this study. In general, PCR conditions were as follows but were not optimal for all species/populations tested: 1 cycle of 94°C for 1 min, 50°C for a further 1 min and 72°C for 2 min. This was followed by 40 cycles of 94°C for 1 min, 45°C for 1 min and 72°C for 2 min. The PCR was completed with a final extension phase of 94°C for 1 min, 45°C for 1 min and 72°C for 5 min. For mtDNA, the reaction mix comprised a single PCR PureTaq Ready to go Bead (GE Bioscience, UK), 23 μl sterile water, 1 μl template DNA and 0.5 μl of each 10 μM primer. Purified DNA was sequenced directly in both directions using a Big Dye Terminator cycle sequencing kit v3.1 (Applied Biosystems, Warrington, UK), according to the instructions listed by the manufacturer. For each sequencing reaction, the following reagents were added to a 0.5-ml microcentrifuge tube: 4 μl terminator Ready Reaction Mix, 1 μl primer (3.4 μM) and 5 μl template purified DNA. DNA was sequenced in-house using an ABI 377 DNA sequencer. The quality of the sequences produced was checked using Sequence Navigator Software (Applied Bio-systems, Warrington, UK).
Table 2.
Primers used to characterize the cytochrome oxidase I of mitochondrial DNA from populations of Xiphinema americanum-group.
Data analysis: CLUSTAL X v.1.81 (Thompson et al., 1997) was used to generate multiple sequence alignments with default settings for both 18S rDNA and mtDNA sequences. Thereafter, trimming excess nucleotides at the 5′ and 3′ ends to effect a common starting and end point was done using GeneDoc (Nicholas et al., 1997).
MODELTEST (Posada and Crandall, 1998) estimated that a F84 plus gamma + invariant rate heterogeneity model was the most appropriate for the dataset under study.
Phylogenetic analysis was carried out using the Maximum Likelihood (ML) approach. ML was preferred to Maximum Parsimony (MP) because ML uses: i) all columns of the multiple sequence alignment (i.e., not just phylogenetically informative columns) and ii) information on branch lengths when evaluating trees (Swofford et al., 1996). ML trees were estimated (Ts/Tv set at 1.87 and alpha at 0.02 for the 18S rDNA alignment; Ts/Tv set at 3.63 and alpha at 0.28 for the mtDNA alignment) using the PHYLIP v.3.6a DNAML sub-routine (Felsenstein and Churchill, 1996). Furthermore, ML was preferred over Bayesian analysis, as the latter method is recognized to overestimate support for tree topology (Suzuki et al., 2002; Simmons et al., 2004). Sequence data for Californidorus sp. (AY283155) was used as the outgroup for the 18S rDNA analysis, and mtDNA sequences for Caenorhabditis elegans (AY268112), Scutellonema bradys (AY268114) and Meloidogyne hapla (AY268113) were used as the outgroups for the mtDNA analysis.
Results
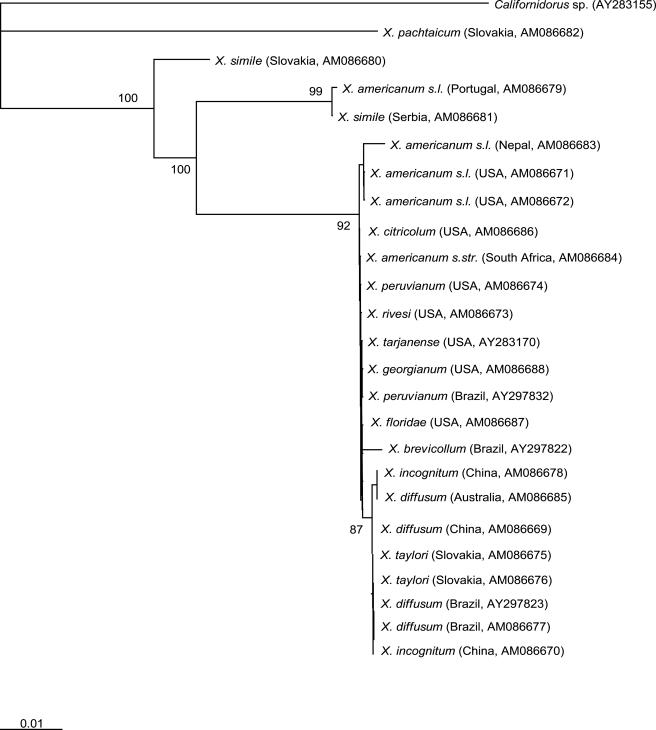
A phylogenetic analysis based on 18S rDNA sequences yielded well resolved groups (Fig. 1). The putative species X. pachtaicum was distinct from all species/populations, and X. simile (two populations) and X. americanum sensu lato (Portugal) were distinct from the remaining populations of X. americanum-group populations studied (Fig. 1). In contrast, the remaining putative X. americanum-group species/populations had sequence homology of >99%, as reflected in their close phylogenetic relationship (Fig. 1). A consensus sequence of 1,752 bp was produced, of which 1,623 bp, i.e., 92.6% of all nucleotides, were constant. The estimated average nucleotide frequencies among the X. americanum-group populations were relatively similar, being 24.8% A, 25.9% C, 21.6% G and 27.7% T. Populations identified using classical morphological techniques as X. diffusum (syn. X. brevicollum after Luc et al., 1998) from China had 18S rDNA sequences identical to those identified as X. diffusum and X. taylori (syn. X. brevicollum) from Brazil and Slovakia, respectively. The majority of X. americanum-group specimens from North America had sequence divergence of <0.5% and were separated from a group comprising the majority of populations from Asia, South America and Oceania.
Fig. 1.
Phylogenetic tree showing relationships between Xiphinema americanum-group populations based on sequences of 18S rDNA. The numbers indicate the bootstrap values higher than 50. Branch lengths are drawn to be proportional to the number of changes inferred.
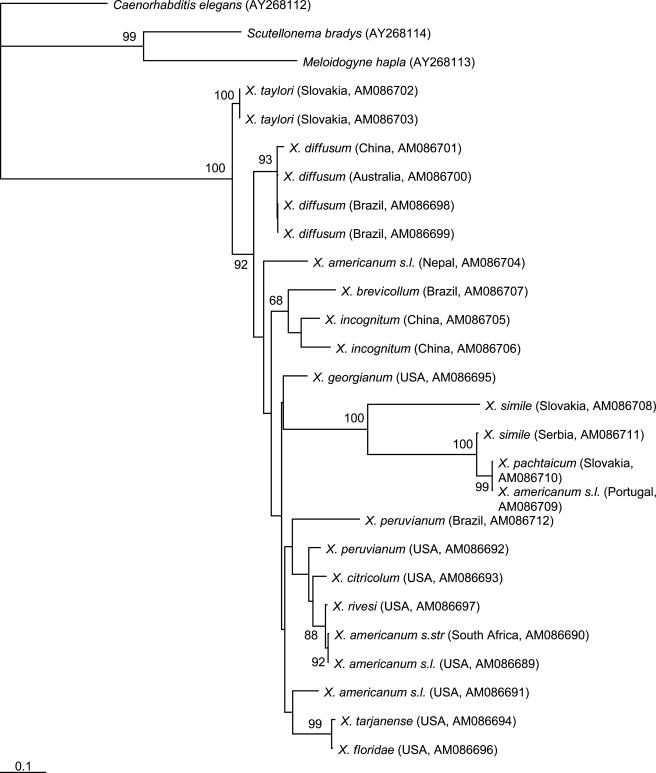
In contrast to 18S rDNA, mtDNA was more heterogeneous, with only 136 bp from a consensus of 405 bp (33.6%) constant. The estimated average nucleotide frequencies were AT-rich, comprising 25.5% A, 14.0% C, 22.4% G and 38.1% T. Furthermore, mtDNA sequences representing X. diffusum (Brazilian populations only) and X. taylori were within each species identical. Xiphinema americanum sensu lato (Portugal), X. pachtaicum and both X. simile populations formed a well resolved group distinct (Fig. 2) from the other species/populations, thus concurring with the 18S rDNA analysis (Fig. 1), with X. simile (Slovakian population) being an outlier. The dichotomy between X. americanum-group populations from North America and those from Asia, South America and Oceania was also apparent for mtDNA, although the groupings were less well resolved than those produced by 18S rDNA (Figs. 1, 2). No intra-population variability in the mtDNA was detected.
Fig. 2.
Phylogenetic tree showing relationships between Xiphinema americanum-group populations based on sequences of cytochrome oxidase I mtDNA. The numbers indicate the bootstrap values higher than 50. Branch lengths are drawn to be proportional to the number of changes inferred.
Discussion
Ever since the inception of the nomenclature “Xiphinema americanum-group” by Tarjan (1969) to encompass morphological and morphometric variability of X. americanum populations, controversy has surrounded their degree of speciation (Luc et al., 1998; Coomans et al., 2001; Lamberti et al., 2004).
The putative species within the X. americanum-group are characterized by minimal inter- and intraspecific variation in both morphological and morphometric characters and, consequently, Loof and Luc (1990) excluded them from their polytomous key of Xiphinema. Subsequently, polytomous keys for the group have been published (Lamberti et al., 2000, 2004), but both keys contain errors that compromise identification of a number of species. Such taxonomic pedantry would normally be irrelevant, but unfortunately a few of the X. americanum-group species are known vectors of Nepovirus (Taylor and Brown, 1997), with the concomitant effect that X. americanum is a quarantined organism in the European Union (http://www.eppo.org/QUARANTINE/listA1.htm); hence correct species identification is imperative within a phytosanitary context.
Consistent with the global tree-of-life project (Stoeckler, 2003), NemATOL (http://nematol.unh.edu) utilizes 18S rDNA as the de facto standard ribosomal region to molecularly characterize nematodes. In this study, with a few exceptions, 18S rDNA was homogeneous for the X. americanum-group populations studied and could not discriminate putative species that were raised according to minute differences in morphology and morphometrics. It is possible that this lack of discriminatory power was due to insufficient evolutionary change occurring in the 18S rDNA to provide useful phylogenetic information. Similarly, with the same exceptions as 18S rDNA, mtDNA did not resolve what is currently perceived to be extant X. americanum-group species, even though mtDNA evolves quickly in nematodes (Moritz et al., 1987; Blouin et al., 1998; Denver et al., 2000). However, analysis of both 18S rDNA and mtDNA indicated that the X. pachtaicum, X. simile and X. americanum s.l. (from Portugal) were different from all other species/populations tested. Both X. pachtaicum and X. simile belong to the “X. pachtaicum group” (Lamberti and Ciancio, 1993), and this separation from other members of the X. americanum-group is consistent with multivariate and cluster analyses of morphological data (Lamberti and Ciancio, 1993; Lamberti et al., 2002). Similarly, the dichotomy of putative species such as X. diffusum and X. taylori from species originating from North America is also consistent with that reported by Lamberti and Ciancio (1993) and Lamberti et al. (2002). Recently, Oliveira et al. (2005) published species-specific diagnostic primers that discriminate between X. brevicolle (syn. X. brevicollum) and X. diffusum, and here our analyses separated the two species into different groups, thus supporting Lamberti et al. (1991, 2002).
In this study, the degree of 18S rDNA sequence divergence (<0.5%) for the X. americanum-group populations originating from North America is similar to intraspecific divergence reported for Longidorus diadecturus (0.6%; Neilson et al., 2004) and X. setariae/vulgare complex (0.4%; Oliveira et al., 2004b). Thus, it could be suggested that extant species such as X. citricolum, X. floridae, X. georgianum, X. peruvianum and X. americanum sensu stricto are merely intraspecific variants of a single species. However, this result should be viewed with caution, as the classical taxonomy of the group is so problematic it is possible that incorrect identifications were made, which in turn would have a concomitant effect on the interpretation of the sequence data; thus, a self-perpetuating scenario ensues. To compound the interpretation further, intraspecific variability of 18S rDNA has to date been largely ignored by nematologists.
Similarly, there are few published data on intraspecific variability of mtDNA in nematodes. Blouin et al. (1998) noted that for the ruminant parasite Ostertagia ostertagi the maximum sequence divergence observed between individuals of the same interbreeding population was 6%. The same authors also stated that mtDNA sequence divergence between closely related species is typically in the range of 10 to 20%. Thus, taking a conservative approach in this study and using 20% mtDNA sequence divergence as a crude measure to determine conspecificity (assuming the classical taxonomy is accurate) would suggest that X. georgianum and X. peruvianum may be valid species, whereas the other populations from North America appear to be examples of intraspecific variability of a single species or perhaps represent cryptic species (Blouin et al. 1998; Blouin, 2002).
In conclusion, analyses of both 18S rDNA and mtDNA sequences have underpinned the taxonomic issues of the X. americanum-group. It is clear that a thorough and combined morphological and molecular investigation is required to unravel the complexities of the X. americanum-group. Such an approach was taken to clarify taxonomic questions surrounding Xiphidorus (Oliveira et al., 2004a). Multiloci sequence typing (MLST), where a number (typically >7) of molecular targets are sequenced (Enright and Spratt, 1998; Maiden et al., 1998), is already used to elucidate taxonomic information of closely related bacterial sequences, and this may be an useful approach for future taxonomic analysis of this complex nematode group.
Footnotes
SSL and CMGO received funding from an UK Royal Society Fellowship and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil), respectively. Research at the Scottish Crop Research Institute is grant-aided by the Scottish Executive Environment and Rural Affairs Department. The authors thank M. T. M. Almeida, L. Barsi, M. Liskova, N. Pokharel, R. T. Robbins, J. Zheng and especially the late Prof. Franco Lamberti for supplying specimens.
This paper was edited by Andrea Skantar.
Literature Cited
- Barsi L, Lamberti F. Morphometrics of three putative species of the Xiphinema americanum group (Nematoda: Dorylaimida) from the territory of the former Yugoslavia. Nematologia Mediterranea. 2002;30:59–72. [Google Scholar]
- Barsi L, Lamberti F. Xiphinema parasimile sp. n. from Serbia and X. simile, first record from Bosnia and Herzegovina (Nematoda, Dorylaimida) Nematologia Mediterranea. 2004;32:101–109. [Google Scholar]
- Blok VC, Wishart J, Fargette M, Berthier K, Phillips MS. Mitochondrial DNA differences distinguishing Meloidogyne mayaguensis from the major species of tropical root-knot nematodes. Nematology. 2002;4:773–781. [Google Scholar]
- Blouin MS. Molecular prospecting for cryptic species of nematodes: Mitochondrial DNA versus internal transcribed spacer. International Journal for Parasitology. 2002;32:527–531. doi: 10.1016/s0020-7519(01)00357-5. [DOI] [PubMed] [Google Scholar]
- Blouin MS, Yowell CA, Courtney CH, Dame JB. Substitution bias, rapid saturation, and use of mtDNA for nematode systematics. Molecular Biology and Evolution. 1998;15:1719–1727. doi: 10.1093/oxfordjournals.molbev.a025898. [DOI] [PubMed] [Google Scholar]
- Boutsika K, Brown DJF, Phillips MS, Blok VC. Molecular characteristics of the ribosomal DNA of Paratrichodorus macrostylus, P. pachydermus, Trichodorus primitivus and T. similis (Nematoda: Trichodoridae) Nematology. 2004a;6:641–654. [Google Scholar]
- Boutsika K, Blok VC, Phillips MS, Lewis SA, Robbins RT, Ferraz LCCB, Brown DJF. Confirmation of the synonymy of Paratrichodorus christiei (Allen, 1957) Siddiqi, 1974 with P. minor (Colbran, 1956) Siddiqi, 1974 (Nematoda: Triplonchida) based on sequence data obtained for the ribosomal DNA 18S gene. Nematology. 2004b;6:145–151. [Google Scholar]
- Coomans A, Heyns J. Three species of the Xiphinema americanum-group (Nematoda: Longidoridae) from Kenya. Nematologica. 1997;43:259–274. [Google Scholar]
- Coomans A, Huys R, Heyns J, Luc M. Character analysis, phylogeny, and biogeography of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae). Annales Musée Royal de L'Afrique Centrale (Zoologie), Tervuren. Belgique. 2001;287:1–289. [Google Scholar]
- De Ley IT, De Ley P, Vierstraete A, Karssen G, Moens M, Vanfleteren J. Phylogenetic analyses of Meloidogyne small subunit rDNA. Journal of Nematology. 2002;34:319–327. [PMC free article] [PubMed] [Google Scholar]
- De Luca F, Reyes A, Grunder J, Kunz P, Agostinelli A, De Giorgo C, Lamberti F. Characterization and sequence variation in the rDNA region of six nematode species of the genus Longidorus (Nematoda) Journal of Nematology. 2004;36:147–152. [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva L, Thomas WK. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans . Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: Identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- Felsenstein J, Churchill GA. A hidden Markov model approach to variation among sites in rate of evolution. Molecular Biology and Evolution. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- Ferris VR, Sabo A, Baldwin JG, Mundo-Ocampo M, Inserra RN, Sharma S. Phylogenetic relationships among selected Heteroderoidea based on 18S and ITS ribosomal DNA. Journal of Nematology. 2004;36:202–206. [PMC free article] [PubMed] [Google Scholar]
- Halbrendt JM, Brown DJF. Morphometric evidence for three juvenile stages in some species of Xiphinema americanum sensu lato . Journal of Nematology. 1992;24:305–309. [PMC free article] [PubMed] [Google Scholar]
- He Y. Gent, Belgium: University of Gent; 2003. Molecular approach to Longidoridae (Nematoda: Dorylaimida): Organelle genomics, phylogeny, population diversity and diagnostics. PhD thesis. [Google Scholar]
- He Y, Jones J, Armstrong M, Lamberti F, Moens M. The mitochondrial genome of Xiphinema americanum sensu stricto (Nematoda: Enoplea): Considerable economization in the length and structural features of encoded genes. Journal of Molecular Evolution. 2005;61:819–833. doi: 10.1007/s00239-005-0102-7. [DOI] [PubMed] [Google Scholar]
- Hugall AC, Moritz C, Stanton J, Wolstenholme DR. Low, but strongly structured mitochondrial DNA diversity in root-knot nematodes (Meloidogyne) Genetics. 1994;136:903–912. doi: 10.1093/genetics/136.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugall AC, Stanton J, Moritz C. Evolution of the AT-rich mitochondrial DNA of the root knot nematode, Meloidogyne hapla . Molecular Biology and Evolution. 1997;14:40–48. doi: 10.1093/oxfordjournals.molbev.a025700. [DOI] [PubMed] [Google Scholar]
- Lamberti F, Ciancio A. Diversity of Xiphinema americanum-group species and hierarchical cluster analysis of morphometrics. Journal of Nematology. 1993;25:332–343. [PMC free article] [PubMed] [Google Scholar]
- Lamberti F, Ciancio A, Agostinelli A, Coiro IM. Relationship between Xiphinema brevicolle and X. diffusum with redescription of X. brevicolle and description of three new species of Xiphinema (Nematoda, Dorylaimida) Nematologia Mediterranea. 1991;19:311–326. [Google Scholar]
- Lamberti F, Hockland S, Agostinelli A, Moens M, Brown DJF. The Xiphinema americanum group. III. Keys to species identification. Nematologia Mediterranea. 2004;32:53–56. [Google Scholar]
- Lamberti F, Molinari S, Moens M, Brown DJF. The Xiphinema americanum-group. I. Putative species, their geographical occurrence and distribution, and regional polytomous identification keys for the group. Russian Journal of Nematology. 2000;8:65–84. [Google Scholar]
- Lamberti F, Molinari S, Moens M, Brown DJF. The Xiphinema americanum-group. II. Morphometric relationships. Russian Journal of Nematology. 2002;10:99–112. [Google Scholar]
- Loof PAA, Luc M. A revised polytomous key for the identification of species of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) with exclusion of the X. americanum-group. Systematic Parasitology. 1990;16:35–66. [Google Scholar]
- Luc M, Baujard P. On specific determination within the Xiphinema americanum group (Nematoda: Longidoridae) Nematology. 2001;3:727–728. [Google Scholar]
- Luc M, Coomans A, Loof PAA, Baujard P. The Xiphinema americanum group (Nematoda: Longidoridae). 2. Observations on Xiphinema brevicollum Lordello & Da Costa, 1961 and comments on the group. Fundamental and Applied Nematology. 1998;21:475–490. [Google Scholar]
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proceedings of the National Academy of Science. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Dowling TE, Brown WM. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Annual Review of Ecology and Systematics. 1987;18:269–292. [Google Scholar]
- Neilson R, Ye W, Oliveira CMG, Hübschen J, Robbins RT, Brown DJF, Szalanski AL. Phylogenetic relationships of selected species of Longidoridae (Nematoda: Longidoridae) from North America inferred from 18S rDNA gene sequence data. Helminthologia. 2004;41:209–215. [Google Scholar]
- Nicholas KB, Nicholas HB, Jr, Deerfield DW., II GeneDoc: Analysis and Visualization of Genetic Variation. EMBNEW.NEWS. 1997;4:14. [Google Scholar]
- Oliveira CMG, Fenton B, Malloch G, Brown DJF, Neilson R. Development of species-specific primers for the ectoparasitic nematode species Xiphinema brevicolle, X. diffusum, X. elongatum, X. ifacolum and X. longicaudatum (Nematoda: Longidoridae) based on ribosomal DNA sequences. Annals of Applied Biology. 2005;146:281–288. [Google Scholar]
- Oliveira CMG, Ferraz LCCB, Monteiro AR, Fenton B, Malloch G, Neilson R. Molecular and morphometric analyses of Xiphidorus species (Nematoda: Longidoridae) Nematology. 2004a;6:715–727. [Google Scholar]
- Oliveira CMG, Hübschen J, Brown DJF, Ferraz LCCB, Wright F, Neilson R. Phylogenetic relationships among Xiphinema and Xiphidorus nematode species from Brazil inferred from 18S rDNA sequences. Journal of Nematology. 2004b;36:153–159. [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Powers TO, Platzer EG, Hyman BC. Species-specific restriction site polymorphism in root-knot nematode mitochondrial DNA. Journal of Nematology. 1986;18:288–293. [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Pickett KM, Miya M. How meaningful are Bayesian support values? Molecular Biology and Evolution. 2004;21:188–199. doi: 10.1093/molbev/msh014. [DOI] [PubMed] [Google Scholar]
- Stanton J, Hugall A, Moritz C. Nucleotide polymorphisms and an improved PCR-based mtDNA diagnostic for parthenogenetic root-knot nematodes (Meloidogyne spp.) Fundamental and Applied Nematology. 1997;20:261–268. [Google Scholar]
- Stoeckler M. Taxonomy, DNA, and the barcode of life. BioScience. 2003;53:2–3. [Google Scholar]
- Subbotin SA, Krall EL, Riley IT, Chizov VN, Staelens A, de Loose M, Moens M. Evolution of the gall-forming plant parasite nematodes (Tylenchida: Anguinidae) and their relationships with hosts as inferred from internal transcribed spacer sequences of nuclear ribosomal DNA. Molecular Phylogenetics and Evolution. 2004;30:226–235. doi: 10.1016/s1055-7903(03)00188-x. [DOI] [PubMed] [Google Scholar]
- Subbotin SA, Sturhan D, Rumpenhorst HJ, Moens M. Description of the Australian cereal cyst nematode Heterodera australis sp.n. (Tylenchida: Heteroderidae) Russian Journal of Nematology. 2002;10:139–148. [Google Scholar]
- Subbotin SA, Vierstraete A, de Ley P, Rowe J, Waeyenberge L, Moens M, Vanfleteren JR. Phylogenetic relationships within the cyst-forming nematodes (Nematoda: Heteroderidae) based on analysis of sequences from the ITS regions of ribosomal DNA. Molecular Phylogenetics and Evolution. 2001;21:1–16. doi: 10.1006/mpev.2001.0998. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Glazko GV, Nei M. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proceedings of the National Academy of Science. 2002;99:16138–16143. doi: 10.1073/pnas.212646199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. Phylogenetic Inference. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. Sunderland, USA: Sinauer Associates; 1996. pp. 407–514. [Google Scholar]
- Tarjan AC. Variation within the Xiphinema americanum group (Nematoda: Longidoridae) Nematologica. 1969;15:241–252. [Google Scholar]
- Taylor CE, Brown DJF. Wallingford, UK: CAB International; 1997. Nematode Vectors of Plant Viruses. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Szalanski AL, Robbins RT. Phylogenetic relationships and genetic variation in Longidorus and Xiphinema species (Nematoda: Longidoridae) using ITS1 sequences of nuclear ribosomal DNA. Journal of Nematology. 2004;36:14–19. [PMC free article] [PubMed] [Google Scholar]