Abstract
Comparative studies between Portuguese (T and HF) and Japanese (S10, T4, C14-5 and OKD-1) isolates of the pinewood nematode Bursaphelenchus xylophilus have been made in order to provide information to better understand the possible origin of the Portuguese isolates, recently introduced in the European Union. The main comparative aspects investigated were pathogenicity (seedling mortality ratio), sexual compatibility, and DNA sequences of the rDNA region. Four-year-old Japanese black pine (Pinus thunbergii) seedlings were used as host plants for pathogenicity tests. The Portuguese isolates, and in particular isolate “T,” propagated in higher numbers than the Japanese isolates within pine seedlings. All combinations of crossings produced viable progeny, with higher numbers obtained when crossings were made between Japanese and Portuguese isolates, a possible situation of heterosis and/or inbreeding depression. Reciprocal crossings yielded different values, which may reflect a sex effect (maternal inheritance, mtDNA). Regarding DNA sequencing, both Portuguese isolates displayed nearly identical ITS 1, ITS2, and 5.8S rDNA base sequences as the Japanese isolates. Although biologically very similar, and possibly reflecting a common origin, the Portuguese isolates may present a serious threat to Japanese black pine, due to their higher virulence.
Keywords: Portugal, Japan, pathogenicity, sexual compatibility, DNA sequence
The pinewood nematode (PWN), Bursaphelenchus xylophilus (Steiner and Buhrer, 1934) Nickle, 1970, is a serious pathogen of forest trees, especially of the genus Pinus. Pine wilt disease was first reported in Japan at the beginning of the 20th century (Yano, 1913), although the association between the nematode and the disease was only established in 1971 (Kiyohara and Tokushige, 1971). It has since then spread to other Asian countries such as China (Yang and Wang, 1989), Taiwan (Enda, 1988), and Korea (Enda, 1989). In 1999, PWN was first detected in the European Union (EU) in Portugal (Mota et al., 1999) and immediately prompted several government (national and EU) actions to assess the extent of the nematode's presence and to contain B. xylophilus and its insect vector (Monochamus galloprovincialis, Olivier, 1795) to an area with a 30 km radius in the Setúbal Peninsula, 20 km south of Lisbon. For more complete information on the nematode's global distribution and ecology, see Mota and Vieira (2004) and Ryss et al. (2005).
The origin of PWN found in Portugal remains elusive. Several hypotheses may be considered regarding pathway analysis, basically from two general origins: North America or the Far East. World trade of wood products, such as timber, wooden crates, pallets, etc., plays an important role in the potential dissemination of PWN (Webster, 2004). In fact, human activities involving the movement of wood products are considered the single most important factor in the spread of this nematode.
In Japan during the past 30 years, many PWN isolates have been characterized from the viewpoints of biology, molecular biology, and pathogenicity (Mamiya, 2004). A few isolates have been found to display a very low pathogenic effect on the pine species Pinus densiflora Siebold and Zucc and P. thunbergii Parlatore, Japanese red pine and Japanese black pine, respectively. These isolates have been classified as “avirulent.” Several studies have been carried out regarding the environmental conditions influencing the spread of pine wilt disease in Japan (Akema and Futai, 2004).
In Portugal, no studies have yet been published on the pathogenicity of the isolates collected from the affected area. Twenty-four of these isolates from nearly 100 collected so far are now being studied for their genetic diversity using RAPD-PCR analysis (Paulo Vieira, personal comm.). There is a need to understand the pathogenicity of these isolates and to compare them with isolates from other geographical regions, such as Japan. The purpose of this research was to investigate two Portuguese isolates (T and HF) and compare them with two Japanese isolates (S10 and C14-5) when inoculated onto Japanese black pine. The main comparative aspects investigated were pathogenicity (seedling mortality ratio), sexual compatibility, and DNA sequences of the rDNA region.
Materials and Methods
Nematode isolates and pine species: Bursaphelenchus xylophilus isolates were used from Portugal and Japan; two isolates from Portugal, “Tróia” (T) and “Herdade da Ferraria” (HF), were collected from within the affected zone, an area in the Setúbal Peninsula, of approx. 30 km radius centered in Pegões, 35 km SE of Lisbon. Four PWN isolates from Japan, “S10” and “T4” (virulent) and C14-5 and OKD-1 (avirulent), were used for comparative studies with the Portuguese isolates. One hundred and sixty 4-yr-old Japanese black pine seedlings were used for inoculation and pathogenicity tests.
Pathogenicity experiments: On 8 August 2005, each 4-yr-old pine seedling was inoculated at the base with approx. 5,000 nematodes of one of the four isolates (T, HF, S10, and C14-5) and embedded in cotton to assure nematode survivability during the entry and infection process. Forty-five seedlings were used for each nematode isolate. After inoculation, the bark and cotton were covered with parafilm to prevent them from desiccating. The seedlings were outdoors during the experiment. The maximum and minimum temperatures during the experimental period averaged 31.7°C and 24.2°C, respectively. Symptoms were examined 21, 25, 29, 33, and 37 d after inoculation. Symptoms appearing in each seedling were categorized into three stages: Stage 1—needle necrosis, discoloration, or drooping of some 1-yr-old needles; stage 2—almost all the 1-yr-old needles drooped, displaying necrosis or discoloration; and stage 3—wilting symptoms extending over 50% of current-year needles. Mortality ratio was estimated for each nematode isolate by the number of seedlings reaching stage 3. After 4 wk, the seedlings were cut at the base, and 50-cm shoot segments were weighed (dry weight), cut into small pieces, and extracted on a Baermann funnel. After 24 hr, nematodes were collected and counted. Nematode density (number per gram dry weight) was log-transformed, and the transformed data subjected to analysis of variance (ANOVA) and statistically compared using the Tukey-Kramer post-hoc analysis.
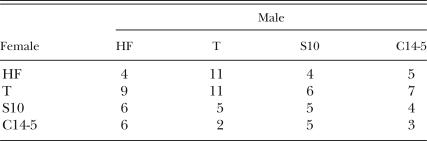
Hybridization experiments: To evaluate the sexual compatibility of the isolates, three males of one isolate and a single female of another isolate were placed in a plastic petri dish (30 mm diam.) together with mycelia of Botrytis cinerea growing on 0.05% malt extract agar. Compatibility of all combinations among the four isolates tested (T, HF, S10, and C14-5) is shown in Table 1. After 5 d, the progeny were extracted with a Baermann funnel and counted. The data, minimum two and maximum 11 replicates (see Table 1), were log-transformed and analyzed.
Table 1.
Replication number of each combination of mating between four nematode isolates (offspring results are shown in Fig. 5).
DNA extraction from nematodes: Six isolates of PWN served for the DNA sequencing: two Portuguese isolates (T, HF), and four Japanese isolates (two virulent, S10, T4, and two avirulent, C14–5, OKD-1). The nematodes were cultured on a fungal mat of B. cinerea grown on autoclaved barley. DNA was extracted using the method described by Iwahori et al. (1998), after the nematodes were isolated with a Baermann funnel.
Briefly, 300 μl of 2× lysis buffer (200 mM NaCl, 200 mM Tris-HCl, pH 8.0, 100 mM EDTA, pH 8.0, 2% SDS, 2% β-mercaptoethanol, 200 μg/ml proteinase K) was added to an equal volume of a nematode suspension in a 2.0 ml test tube. After incubating at 65°C for 30 min, the DNA was extracted twice with an equal volume of chloroform:isoamyl alcohol (24:1), and the aqueous phase was transferred to a new tube each time. The DNA was recovered by ethanol precipitation with 1/10 volume of 3 M sodium acetate and two volumes of 100% ethanol and then rinsed with 70% ethanol. The DNA pellet obtained was dissolved in 200 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and kept at –20°C until used.
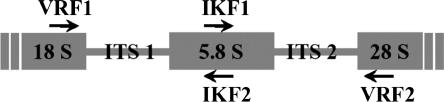
DNA sequencing: The DNA sequencing of the ITS1, 5.8S, and ITS2 regions of the six PWN isolates was performed by PCR direct sequencing (Fig. 1). Firstly, the DNA fragment consisting of these regions was amplified by PCR with a set of primers (VRF1: 5′-CGT AAC AAG GTA GCT GTA G-3′ and VRF2: 5′-TCC TCC GCT AAA TGA TAT G-3′; Iwahori et al., 1998). In addition, another set of primers (IKF1: 5′-GGG TCG ATG AAG AAC GCA G-3′ and IKF2: 5′-CTG CGT TCT TCA TCG ACC-3′; Iwahori et al., 1998) was also used for sequencing.
Fig. 1.
Primer locations for PCR amplification and sequencing of ribosomal DNA (rDNA). To obtain DNA fragment including the 5.8 S rDNA and the regions in between the rDNA genes (18S, 5.8S, and 28 S), i.e., the internal transcribed spacer (ITS) 1 and 2, the primers VRF1 and VRF2 were used for PCR amplification.
The PCR was performed in a final volume of 12.5 μl, containing 1 μl DNA, 1.25 μl 10× Gene Taq Universal Buffer (Nippon Gene, JAPAN), 1.25 μl 2.5 mM dNTP mixture, 0.65 μl each 10 μM primer, and 0.1 μl (corresponding to 1 U) Gene Taq NT (Nippon Gene, Japan) in a thermocycler (TaKaRa PCR Thermal Cycler) under the following conditions: 94°C for 2 min, followed by 35 cycles of 94°C for 45 sec, 48°C for 30 sec, 72°C for 1 min, and 72°C for 10 min. The PCR product was purified by recovering the rDNA fragment after electrophoresis and then used as the template of PCR direct sequencing using the ABI 3130 Genetic Analyzer with a reaction kit (Big Dye Terminator Cycle Sequencing Kit v. 1.0, Applied Biosystems) following the manufacturer's recommendations.
Results
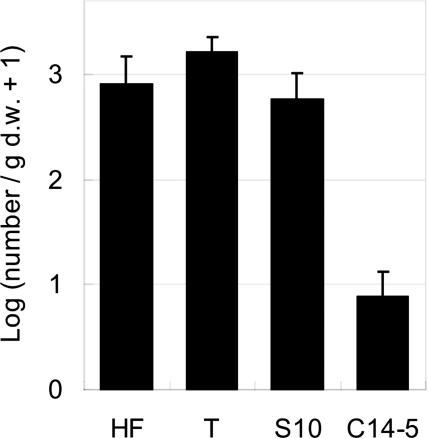
Pathogenicity experiments: In all experiments, the Portuguese isolates, T and HF, produced a higher number of nematodes propagating within the seedlings as compared to the Japanese isolate S10, with a slightly higher value of T (Fig. 2). Isolates HF and S10 generated similar values for nematode propagation.
Fig. 2.
Nematode reproduction for four pinewood nematode (PWN), Bursaphelenchus xylophilus, isolates in Japanese black pine (Pinus thunbergii) seedlings 4 wk after inoculation. Nematode number per gram dry weight was log-transformed and averaged. Bars are standard error.
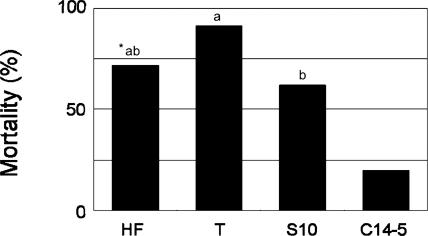
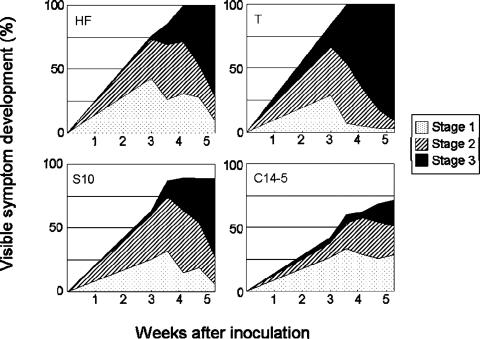
In comparing mortality ratios (rate of seedlings reaching stage-3 symptoms), isolate T was significantly more pathogenic than S10 (ANOVA, F = 21.264, p < 0.001; Tukey's WSD method, α = 0.05), reaching 90% mortality ratio 37 d after inoculation and also reaching a greater speed in causing mortality (Figs. 3, 4). HF and S-10 isolates performed in a very similar fashion, causing 60 to 70% mortality rate at the same time (37 d). The Portuguese isolates (T and HF) were capable of causing a 100% value of visible symptom development (stage 1–3), whereas the Japanese virulent isolate (S10) caused a 90% value (Fig. 4). Regarding nematode propagation in pine seedlings, both Portuguese isolates yielded higher density (800 — 1,700 nematodes/g dry weight (dw) on average) than the Japanese virulent isolate S10 (585 nematodes/g dw). As expected, the avirulent isolate C14-5 yielded significantly lower density (6 nematodes/g dw on average) than the others (p < 0.05, Tukey-Kramer method).
Fig. 3.
Mortality of Japanese black pine seedlings (Pinus thunbergii) 5 wk after inoculation with four isolates of pinewood nematodes, Bursaphelenchus xylophilus (Portuguese isolates HF and T, Japanese virulent isolate S10, and Japanese avirulent isolate C14-5). *There is no difference between two isolates followed by the same letter (Tukey's WSD method, α = 0.05).
Fig. 4.
Development of visible symptoms appeared on Japanese black pine (Pinus thunbergii) seedlings inoculated with pinewood nematodes (Bursaphelenchus xylophilus). Stage 1: Needle necrosis or drooping appeared in some 1-yr-old needles; Stage 2: Almost all the 1-year-old needles drooped and showed necrosis or discoloration; Stage 3: Wilting symptoms extended over 50% of current-year-needles.
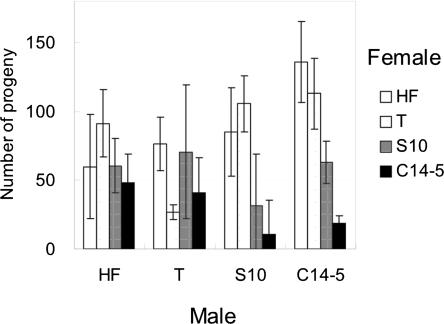
Hybridization experiments: The numbers of progeny that resulted from the mating among isolates are presented in Figure 5; results that yielded fewer than five nematodes were disregarded, thus Figure 5 displays trimmed data. All crossings (male x female, presented in this order) produced viable progeny, with higher numbers from C14-5 × HF, C14-5 × T, S10 × HF, and S10 × T. The lowest values of progeny resulted from C14-5 × C14-5 and S10 × C14-5. A sex effect may be involved, since the numbers of progeny produced from the cross of male C14-5 × female HF were much higher than those produced from the reciprocal crossing of female C14-5 × male HF. The same phenomenon was observed with male S10 × female C14-5 and the reciprocal female S10 × male C14-5 cross.
Fig. 5.
Number of progeny yielded from each combination of mating between two pinewood nematode isolates from Portugal and two from Japan. The resulting offspring were incubated for 5 d on agar cultures and extracted by Baermann funnel (bars represent standard error).
DNA sequencing: The nucleotide sequences of rDNA were determined to compare several isolates of PWN. The accession numbers inscribed in the DDBJ (DNA Data Bank of Japan) are: AB277203 (C14-5), AB277204 (HF), AB 277205 (OKD1), AB277206 (S10), AB277207 (T4), and AB277208 (Tróia). The two Portuguese isolates, HF and T, had the same nucleotide sequence throughout 884 bp. DNA sequencing of ITS regions revealed that both Portuguese isolates were similar to the virulent Japanese isolates, S10 and T4, but different from the avirulent Japanese isolates, C14-5 and OKD-1. The Portuguese isolates, affiliated to B. xylophilus because of the ITS1 and ITS2 sequences, were identical to S10, but had one substitution from T4 in the ITS 1 region and seven substitutions and gaps from C14-5 and OKD-1 in the ITS regions, mainly ITS2.
Discussion
This is the first comparison made between Portuguese and Japanese isolates of PWN and the first record of the pathogenicity of B. xylophilus from Portugal on Japanese black pine. It is unknown at this moment how pathogenicity varies among different isolates of B. xylophilus from Portugal. In particular, isolate T and others originating from Portugal could present a serious threat to Japanese black pine.
As expected, all crosses between Portuguese and Japanese isolates yielded viable F1 progeny (Fig. 5), however, different combinations gave different results. Matings between Japanese and Portuguese isolates, overall, produced more progeny than did isolates from each country, i.e., Japanese × Japanese or Portuguese × Portuguese. This may be the result of inbreeding depression and/or hybrid vigor (heterosis), in which genetically more distant pairs within a species produce larger number of progeny. These results are similar to previous matings between Japanese isolates in which S10 × C14-5 yielded more progeny than S10 × S10 or C14–5 × C14-5 (Takemoto, unpublished data). Certain combinations yielded more progeny than the reciprocal crossings, e.g., male C14-5 × female HF poduced many more progeny than did the reciprocal crossing. Also, the crossing between male S10 × female T produced more progeny than did the reciprocal crossing. In all combinations between Japanese and Portuguese isolates, male × female combinations yielded more progeny than did the reciprocal mating. These results may indicate that maternal inheritance (mtDNA) plays a role in the number of offspring produced.
The molecular analysis of the rDNA region including the 18S and 5.8S coding regions and the noncoding ITS-1 and ITS-2 regions has been thought to be useful for molecular taxonomy and has been used by several authors in phylogenetic analysis of species of Bursaphelenchus (Iwahori et al., 1998; Beckenbach et al., 1999; Irdani, 2000; Kanzaki and Futai, 2002) or simply for identification and diagnostic purposes (Braasch et al., 1995; Hoyer et al., 1998; Iwahori et al., 2000; Liao et al., 2001; Kang et al., 2004; Matsunaga and Togashi, 2004; Cao et al., 2005; Takeuchi et al., 2005). More recently (Metge and Burgermeister, 2005), a technique employing the whole genome has been applied using a multiple displacement amplification technique. In the present study, the number of isolates, two from Portugal and four from Japan, is very small, and major conclusions, e.g. the complete homology between the Portuguese isolates T and HF and the Japanese virulent S10, cannot be extracted from these results. Future work will include a much larger number of isolates which, in combination with analysis of other ribosomal regions and genes for the evaluation of genetic diversity, may help elucidate the relationship between the Portuguese and Japanese isolates.
The presence of PWN in two very distant countries naturally raises questions regarding the possible origin and pathway analysis for each country. Iwahori et al. (1998) reported that three Japanese (S10, T4, S-6), one Chinese (BxC), and one US virulent isolates (MO) were identical to each other in the same rDNA regions as examined in this study; however, the route for their worldwide distribution and the mechanisms involved in variation including speciation remain unsolved. These two Portuguese isolates may reflect natural variability in pathogenicity, or they may represent two separate introductions into the country, which is difficult to explain considering the very short time of probable entry (5–10 years). More studies are needed using a wider range of PWN isolates from Portugal, as well as other isolates, including avirulent ones from Japan.
Footnotes
We thank Drs. Jon Eisenback, Rita Costa and Paulo Vieira for critical review of the manuscript. The first author acknowledges partial support from EU programs QLK5-CT-2002-00672 “PHRAME” (“Development of improved Pest Risk Analysis techniques for quarantine pests, using pinewood nematode, Bursaphelenchus xylophilus, in Portugal as a model system”) and EU-SSPE-RTD-502348, “PortCheck” (“Development of generic ‘on site’ molecular diagnostics for EU quarantine pests and pathogens”).
This paper was edited by Ed Lewis.
Literature Cited
- Akema T, Futai K. Ectomycorrhizal development in a Pinus thunbergii stand in relation to location on a slope and effect on tree mortality from pine wilt disease. Journal of Forestry Research. 2004;10:227–238. [Google Scholar]
- Beckenbach K, Blaxter M, Webster JM. Phylogeny of Bursaphelenchus species derived from analysis of ribosomal internal transcribed spacer DNA sequences. Nematology. 1999;1:539–548. [Google Scholar]
- Braasch H, Burgermeister W, Pastrik K-H. Differentiation of three Bursaphelenchus species by means of RAPD-PCR. Nach-richtenbl. Deut. Pflantzenschutzd. 1995;47:310–314. [Google Scholar]
- Cao AX, Liu XZ, Zhu SF, Lu BS. Detection of the pinewood nematode, Bursaphelenchus xylophilus, using a real-time polymerase chain reaction assay. Phytopathology. 2005;95:566–571. doi: 10.1094/PHYTO-95-0566. [DOI] [PubMed] [Google Scholar]
- Enda N. Current status of pine wilt disease in Taiwan. Forest Pests. 1988;37:161–166. (in Japanese). [Google Scholar]
- Enda N. Current status of pine wilt disease in Korea (tentative translation by authors) Forest Pests. 1989;38:148–152. (in Japanese). [Google Scholar]
- Hoyer U, Burgermeister W, Braasch H. Identification of Bursaphelenchus species (Nematoda: Aphelenchoididae) on the basis of amplified ribosomal DNA (ITS-RFLP) Nachrichtenblatt des Deutschen Pflanzenschutzdienstes. 1998;50:273–277. [Google Scholar]
- Irdani T. Genetic analysis of three Bursaphelenchus species by random amplified polymorphic DNA. Nematologia Mediterranea. 2000;28:11–120. [Google Scholar]
- Iwahori H, Kanzaki N, Futai K. A simple, polymerase chain reaction-restriction fragment length polymorphism-aided diagnosis method for pine wilt disease. Forest Pathology. 2000;30:157–164. [Google Scholar]
- Iwahori H, Tsuda K, Kanzaki N, Izui K, Futai K. PCR-RFLP and sequencing analysis of ribosomal DNA of Bursaphelenchus nematodes related to pine wilt disease. Fundamental and Applied Nematology. 1998;21:655–666. [Google Scholar]
- Kang JS, Choi KS, Chin SC, Moon IS, Lee SG, Lee SH. Development of an efficient PCR-based diagnosis protocol for the identification of the pinewood nematode, Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae) Nematology. 2004;6:279–285. [Google Scholar]
- Kanzaki N, Futai K. A PCR primer set for determination of the phylogenetic relationship of Bursaphelenchus species within the xylophilus group. Nematology. 2002;4:35–41. [Google Scholar]
- Kiyohara T, Tokushige Y. Inoculation experiments of a nematode, Bursaphelenchus sp., onto pine trees. Journal of Japanese Forest Society. 1971;53:210–218. (In Japanese; English summary.) [Google Scholar]
- Liao JL, Zhang LH, Eng ZX. Reliable identification of Bursaphelenchus xylophilus by rDNA amplification. Nematologia Mediterranea. 2001;29:131–135. [Google Scholar]
- Mamiya Y. Pine wilt disease in Japan. The pinewood nematode, Bursaphelenchus xylophilus. Nematology Monographs & Perspectives, vol. 1. In: Mota M, Vieira P, editors. Leiden-Boston: Brill Academic Publishers; 2004. pp. 9–20. [Google Scholar]
- Matsunaga K, Togashi K. Among-tree difference in the inhibition of systemic dispersal of Bursaphelenchus xylophilus (Nema-toda: Aphelenchoididae) by Pinus densiflora . Applied Entomology and Zoolology. 2004;39:271–277. [Google Scholar]
- Metge K, Burgermeister W. Multiple displacement amplification of DNA for ITS-RFLP analysis of individual juveniles of Bursaphelenchus . Nematology. 2005;7:253–257. [Google Scholar]
- Mota MM, Braasch H, Bravo MA, Penas AC, Burgermeister W, Metge K, Sousa E. First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology. 1999;1:727–734. [Google Scholar]
- Mota M, Vieira P. Leiden-Boston: Brill Academic Publishers; 2004. The pinewood nematode, Bursaphe-lenchus xylophilus. Nematology Monographs & Perspectives, vol. 1. [Google Scholar]
- Ryss A, Vieira P, Mota M, Kulinich O. A synopsis of the genus Bursaphelenchus Fuchs, 1937 (Aphelenchida: Parasitaphe-lenchidae) with keys to species. Nematology. 2005;7:393–458. [Google Scholar]
- Takeuchi Y, Kanzaki N, Futai K. A nested PCR-based method for detecting the pine wood nematode, Bursaphelenchys xylophilus, from pine wood. Nematology. 2005;7:775–782. [Google Scholar]
- Webster J. The pine wood nematode: Implications of factors past and present for pine wilt disease. The pinewood nematode, Bursaphelenchus xylophilus. Nematology Monographs & Perspectives, vol. 1. In: Mota M, Vieira P, editors. Leiden-Boston: Brill Academic Publishers; 2004. pp. 55–64. [Google Scholar]
- Yang B, Wang Q. Distribution of the pine wood nematode in China and susceptibility of some Chinese and exotic pine to the nematode. Canadian Journal of Forest Research. 1989;19:1527–1530. [Google Scholar]
- Yano S. Investigation on pine death in Nagasaki prefecture. Sanrin-Kouhou. 1913;4:1–14. (in Japanese). [Google Scholar]