Abstract
Diagnosis of an Argentinean population of Nacobbus sp. infecting sweet pepper (lamuyo) was carried out including morphology, scanning electron microscopy, and molecular studies. In light of our morphometric, molecular and host-range results, we consider the studied population to belong to N. aberrans s. l., and by host range tests the population is assigned to the "sugar beet group." ITS-PCR analysis on individual male and immature female specimens of this population yielded amplification products of approximately 922 bp. RFLP profiles and sequencing of the ITS region revealed that, in addition to the host group, the present population can be assigned to the "Argentina 2" group. Disease development and histopathology were investigated with glasshouse observations using tomato, pepper, sugar beet and potato seedlings exposed to nematode infection for 45 days at 28 ± 2°C. Histopathology of tomato roots confirmed that all immature stages and young females and males are migratory, whereas mature females are obligate sedentary endoparasites. Rather than syncytia, large regions of cortical necrosis and cavities were detected in tomato swellings infected by juveniles. However, syncytia were associated only with adult females. Large root galls, hyperplasia, abnormal proliferation of lateral roots and asymmetry of root structure were common anatomical changes induced by the nematode feeding in tomato roots.
Keywords: Argentina, false root-knot nematode, histopathology, host-parasite relationships, morphology, PCR-RFLP profiles, quarantine, tomato
The false root-knot nematode genus Nacobbus Thorne and Allen, 1944 comprises endoparasitic, gall-inducing nematodes indigenous to and distributed mainly in North and South America (Sher, 1970; Manzanilla-López et al., 2002; Reid et al., 2003; Anthonie and Mugnièry, 2005). According to Sher (1970) it contains only two valid species: Nacobbus dorsalis Thorne and Allen, 1944 and N. aberrans (Thorne, 1935) Thorne and Allen, 1944. Nacobbus aberrans is an important parasite (with annual estimated agricultural losses of 65%) in the Americas, and it is subject to strict quarantine pest regulations in various parts of the world where it is not yet established, although sporadically detected (CABI/EPPO, 1997). Several records and quarantine interceptions of the genus (some of which have not been confirmed) include England (Franklin, 1959), The Netherlands (de Bruijn and Stemerding, 1968), Finland, the former USSR (Kirjanova and Lovanova, 1975), India (Prasad et al., 1965), and China (Yin and Feng, 1981).
The morphology and host range of N. aberrans show great variability among populations from different geographical areas (Zamudio, 1983). Intraspecific physiologic and molecular diversity have been shown to be particularly great among isolates from different geographical origins, including Mexico, Peru and Argentina (Baldwin and Cap, 1992; Ibrahim et al., 1997; Manzanilla-López et al., 1999, 2002; Reid et al., 2003; Manzanilla-López et al., 2004). Based on the great importance of N. aberrans for Argentinean agriculture, its extraordinary adaptation to a wide range of climatic conditions and its variability in morphology and host range, the objectives of the present study were: (i) to carry out a detailed characterization of all nematode stages of this Argentinean population; (ii) to study the molecular variability of this population by using ITS-RFLP analysis and sequencing of two ribosomal regions, D3 expansion segment of the 26S rDNA and the ITS region; and (iii) to study the parasitism and histopathology of this on tomato.
Materials and Methods
Nematode isolation and culture: The isolate of N. aberrans used in our studies was originally collected from infected roots of sweet pepper (lamuyo), Capsicum annuum L. cv. Zafiro, in the locality Colonia Urquiza, La Plata, in the Buenos Aires province, by the second author (A.I. Nico). The sample was collected from a six-year-old plastic greenhouse which had been cropped with tomato (Lycopersicum esculentum Mill.) since its establishment. Sampling was carried out in early February in coincidence with full reproductive stage of sweet pepper and highest summer temperatures. One individual plant showing aboveground symptoms of nematode infection (incipient wilting and stunting) was selected. To establish and increase inoculum of N. aberrans for experiments, a single egg mass was removed from an individual gall from infected Zafiro roots, containing a single female nematode. Egg mass was surface-disinfected with 1% NaOCl for 4 min, rinsed thoroughly in sterilized water and placed onto roots of tomato (L. esculentum Mill. cv. Rutgers) under strict quarantine conditions in a glasshouse at 28 ± 2°C. The inoculated plants were incubated for at least 55 d and renewed three times to increase nematode population (for a total of 6 mon). Thereafter, nematode populations were extracted for host-range, morphological, molecular and histopathological studies (Coolen, 1979).
Light and Scanning Electron Microscopy observations: Juveniles, immature females, males and adult females were observed in water-agar mounts (Esser, 1986) under light microscopy (LM). At least 25 specimens of each life-stage were examined and measured, and the main diagnostic features were photographed.
Host range studies: The group of the N. aberrans isolate from Colonia Urquiza, La Plata, in the Buenos Aires province (Argentina) was determined according to the definition of Manzanilla-López et al. (2002) by the ability, or not, to develop on bean (Phaseolus vulgaris L. cv. Nectar), lamuyo pepper (C. annum cv. Zafiro), potato (Solanum tuberosum L. cv. Spunta), sugar beet (Beta vulgaris L. cv. Abril) and tomato (L. esculentum cv. Platense). For that determination, seeds of tomato, lamuyo pepper, sugar beet and potato were allowed to germinate and develop until the roots were 3 to 5 cm long. Then the seedlings were transplanted to an artificially infested autoclaved (121°C for 1 hr, twice on 2 consecutive days) soil mixture (sand: clay loam, 2: 1, v/v) with the Argentinean N. aberrans population. Three months later when the nematode completed the life cycle, the galls and individuals (mature females) in infected roots were observed to define the nematode/host combination.
DNA extraction, amplification and sequencing: For molecular studies, A3 was the population code assigned to this isolate of N. aberrans from Colonia Urquiza, La Plata, in the Buenos Aires province (Argentina), after Reid et al. (2003). Twenty-four individual specimens (12 males, 8 larvae and 4 females) derived from the original population were handpicked, and each one was placed in 3 ul of lysis buffer (10 mM Tris-HCl pH 8.4, 50 mM KC1, 15 mM MgCl2, 0.045% NP40, 0.045% Tween 100, 0.1% gelatin with 90 μg/ml proteinase K) and cut into 3 to 5 small pieces by using a sterilized syringe needle under a dissecting microscope (De Luca et al., 2004). The suspension was recovered from the glass slide, which was washed with a further 7 μl of lysis buffer. The suspension and washing buffers were transferred into a cold 0.5 ml microcentrifuge tube. Each 10 ul sample was incubated at 60°C for 1 hr and then at 95°C for 10 min. The crude DNA extracted from each individual nematode was directly amplified by diluting the sample to 100 ul, such that it contained 0.2 mM of each dNTP, 20 pmols of each primer and 2.5 units of Taq DNA polymerase (Roche, Germany). Oligonucleotides used were the universal 18S-26S primers spanning the 3′ end of the 18S rDNA to the 5′ end of the 26S rDNA and including the ITS1 and ITS2 regions and the 5.8S rDNA (Vrain et al., 1992). In addition, the D3 expansion segment of the 26S rDNA was amplified using the forward primer D3A (5′-GACCCGTCTTGAAACACGGA-3′) and the reverse primer D3B (5′-TCGGAAGGAACCAGCTACTA-3′) (Al-Banna et al., 1997) in four individual nematodes. The PCR conditions used for amplification of both segments were identical: an initial denaturation at 94°C for 5 min and then 35 cycles of 95°C for 50 sec, 55°C for 50 sec and 72°C for 90 sec, followed by a final step at 72°C for 7 min. The size of amplification products were determined by comparison with the molecular marker, Ladder 100 (Fermentas, St. Leon-Rot, Germany), following electrophoresis of 10 μl of each sample on a 1.5% agarose gel.
The PCR products containing the ITS region from 12 individual nematodes were digested overnight with the following restriction enzymes: Alu I, Dde I, Hae III, Hinf I, Hpa II and Rsa I at 37°C. The digested DNA fragments were loaded onto a 2.5% agarose gel and visualized by ethidium bromide staining. All gel images were stored digitally.
The PCR products obtained with the 18S–26S primers from three individual nematodes were purified from agarose gel using the High Pure PCR elution kit (Machinery-Nagel, Düren, Germany) and cloned into PCR 2.1-TOPO plasmid and TOP10 competent cells transformed using the TOPO-TA cloning kit (Invitrogen, Cergy-Pontoise, France), according to the manufacturer's instructions. Two clones from each individual were sequenced using an ABI Prism 377 sequencer (PE Applied Biosystems, Foster City, CA). Both strands of each clone were sequenced using Ml3 forward and Ml3 reverse primers. The sequences obtained in this study were also aligned with the ITS sequences for N. aberrans populations deposited in the database (Reid et al., 2003; Anthoine and Mugnièry, 2005).
Alignments were performed using ClustalW (Thompson et al., 1994). Formatting of alignments was performed with GeneDoc (Nicholas et al., 1997).
The D3 amplified products from three individual nematodes were isolated from gels and directly sequenced using an ABI Prism 377 sequencer and dye terminator sequencing reagents at CNR, Bari.
The sequences have been submitted to GenBank with the following accession numbers D3 region: AM412741, AM412742, and AM412743. Also, the sequences obtained from the ITS region of the A3 population were coded with a clone number and submitted to GenBank with the following accession numbers: A315 (AM412748), A318 (AM412749), A321 (AM412746), A322 (AM412747), A326 (AM412745) and A328 (AM412744).
The availability of several ITS sequences for A3 population allowed us to identify the consensus sequence. Phylogenetic trees were generated by the maximum parsimony (MP) method with UPGMA cluster analysis using Bionumerics 4.5 software (Applied Maths, Kortrijk, Belgium). The phylograms were bootstrapped 1,000 times to assess the degree of support for the phylogenetic branching indicated by the optimal trees.
Histopathology: To study the development of parasitism and histopathological changes induced by the nematode, galled roots from artificially inoculated N. aberrans-miected tomato (cv. Platense) plants were selected. Infected roots were gently washed free of adhering soil and debris, and individual galls were selected together with healthy roots (non-inoculated). These root tissues were fixed in formaldehyde chromo-acetic solution for 48 hr, dehydrated in a tertiary butyl alcohol series (40–70–85–90–100%) and embedded in 58°C-melting point paraffin for histopathological observations. Embedded tissues were sectioned with a rotary microtome. Sections of 10 to 12 μm thickness were placed on glass slides, stained with safranin and fast-green, mounted permanently in 40% xylene solution of a polymethacrylic ester (Synocril 9122X, Cray Valley Products, NJ), examined microscopically and photographed (Johansen, 1940).
Results
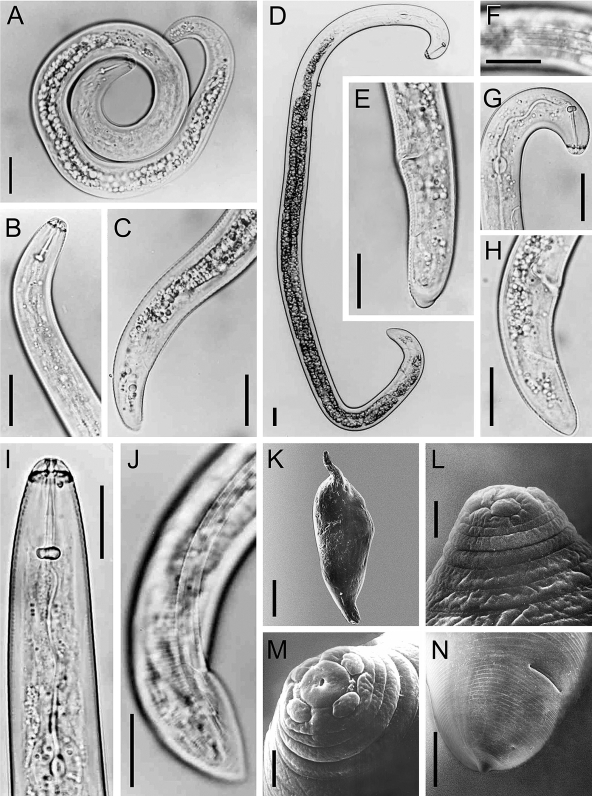
Nematode morphology: The morphometric values obtained in this study by LM and SEM observations on all life stages (juveniles, immature females, males and mature females) of the false-root-knot nematode population from Colonia Urquiza, La Plata, in the Buenos Aires province, were similar and correspond well to those characterizing N. aberrans s. l. (Fig. 1) and with the combined ranges reported by Inserra (1983), Manzanilla-López et al. (1997, 1999, 2002) and Sher (1970).
Fig. 1.
LM and SEM micrographs of different Nacobbus aberrans life stages. A–C) juvenile stage: entire body (A); anterior body portion and tail (B and C). D–G) immature female: entire body (D); vulva-tail region (E); lateral field (F); anterior and posterior body end (G,H). I,J) Male anterior and posterior body portion. K–N) SEM micrographs of adult females: entire body (K); anterior end in profile (L) and face view in (M); vulva terminus area (N) (Scale bars in K = 400 μm; all others = 25 μm).
Host status and nematode infection: Nacobbus aberrans in the Americas has a wide host range, including crops such as potato, tomato, sugar beet, chili pepper, beans, carrot, eggplant, squash, tobacco and other tuber crops. In our host-status tests, the nematode population from Colonia Urquiza infected and reproduced (producing egg masses) on lamuyo pepper, tomato and sugar beet, but not on potato; consequently, this population is assigned to the "sugar beet group."
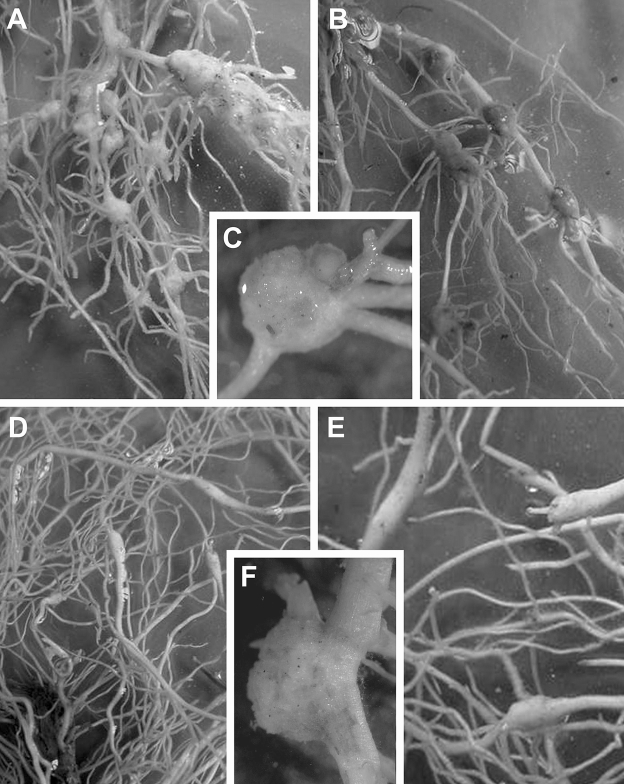
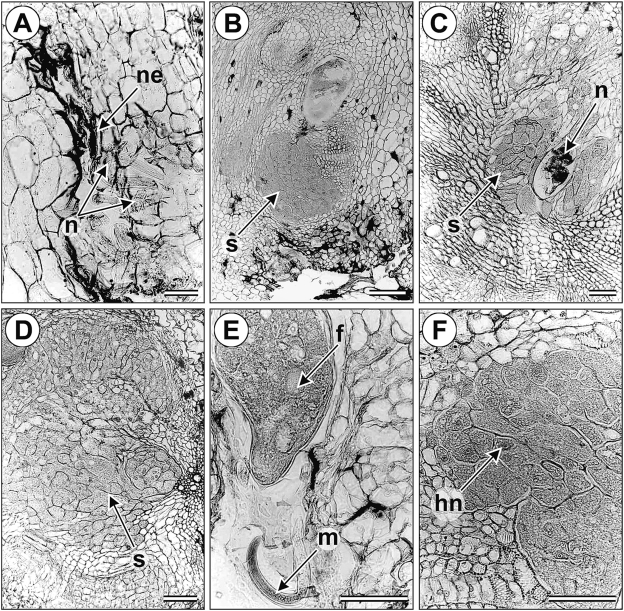
Histopathology of infected tomato roots: Nacobbus aberrans-infected tomato roots showed the typical rosary bead-like galls variable in size (Fig. 2), but larger (more than four-fold the root diameter) than those induced in lamuyo pepper (Fig. 2) and located commonly along the root axis but rarely on the root tip (Fig. 2). Numerous lateral roots arose from galled regions (Fig. 2). The infective J2 and males of N. aberrans invaded the cortical parenchyma of tomato roots and moved by intracellular migration, creating necrotic lesions and cavities in large areas of cortical tissues (Fig. 3A). Dense cytoplasm and hypertophic nuclei were observed in cortical cells near the nematode feeding. No feeding sites were observed associated with third- and fourth-stage juveniles (J3 and J4, respectively), which probably do not feed. A change to sedentary endoparasitim occurs when young and adult mature females of N. aberrans induce the formation of large, specialized feeding cells. These cells, grouped in a large syncytium (Fig. 3B–D), are derived by successive fusions of adjacent cells that are induced by the nematode feeding activity. These fusions are not complete; the cells maintain their individuality in the syncytium, which may involve more than 185 cells. Slight cell expansion and nucleus and nucleolus enlargement occur in the syncytium cells (Fig. 3D, F); these syncytia develop adjacent to the female's body as a spindle of interconnected cells. Mitotic stimulation occurs in the surrounding cells outside the syncytium, giving rise to the gall (Fig. 2). Accentuated asymmetry of root structure, fragmentation of the stele, hyperplasia of vascular parenchyma and abnormal proliferation of lateral roots are the most common histological alterations induced by the nematode.
Fig. 2.
Tomato root systems (A–C of tomato cv. Platense), (D–E of lamuyo pepper cv. Zafiro), with several so-called "rosary-like beads" induced by Nacobbus aberrans after 45 d nematode infection. Note at the insert (C) the large egg-mass, on the surface of large gall.
Fig. 3.
Anatomical changes induced by Nacobbus aberrans in tomato roots. Necrosis and cavities induced by juvenile stages in (A); large multicellular syncytia in B, C and D. Posterior female body and male into the galled root tissues in (E); Syncytial mononucleate cells showing hypertrophied nuclei and nucleoli. Abbreviations used: ne = necrosis; S = syncytium; N = nematode; f = Female; Hn = hypertrophied nucleus; m = male. (Scale bar = 50 μm)
Molecular characterization, cloning and sequencing: The length of the ITS regions (including 206 bp of the 3′ end of the 18S and 102 bp of the 5′ end of the 26S) was 922 bp for the juvenile stages and males for A3 population under study. The ITS1 and ITS2 lengths were 248 to 250 bp and 207 bp, respectively.
The ITS amplified fragments for N. aberrans isolates present in the database varied between 922 and 951 bp. There were length differences in the ITS1 that varied from 248 to 298 bp, whereas the ITS2 varied from 183 to 207 bp.
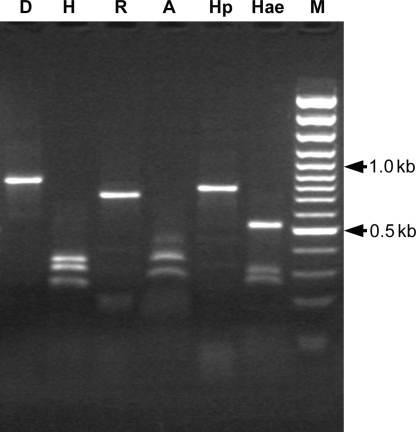
The amplified ITS products were directly digested with six restriction enzymes (see Materials and Methods) which revealed identical cleavage patterns for most of the individual nematodes (Fig. 4). Only two individual nematodes showed different patterns: one specimen with the enzymes Hae III and Rsa I. Hae III showed extra restriction sites compared to the expected profile, thus no distinguishable bands can be appreciated. Indeed, Rsa I has just one more site compared to the representative restriction profile. The ITS amplicon of the second specimen remained undigested by Hae III (data not shown).
Fig. 4.
Restriction patterns of the amplified ITS region of individual male specimen of Nacobbus aberrans from Argentina. A: Alu I; D: Dde I; Hae: Hae III; H: Hinf I; Hp: Hpa II; R: Rsa I; and M: 100 bp DNA ladder.
In addition to the main RFLP pattern for each nematode, there were also faint secondary bands visible in some digests for the enzyme Alu I (Fig. 4). In order to determine the intra-population variability of this Argentinean population of N. aberrans, cloning and sequencing of the ITS fragments from three individual nematode males were performed. The sequence analysis of several clones for each specimen revealed very little or no intraspecific variation either in size or in sequence. Small dinucleotide repeats, or degenerate versions, such as (CA)n, (GA)n and (GT)n, were detected in both ITS. Intra-individual heterogeneity was always observed at the dinucleotide (CA)n, the repeat number of which was ≤8 and resulted in the main polymorphic microsatellite. Stretches of (A)n and (T)n were also present and are considered to represent microsatellites. Intra-population nucleotide dissimilarity ranged from 0.22% to 1.42%.
Pairwise comparisons of the ITS sequences of the N. aberrans A3 population along with published ITS sequences from different populations of N. aberrans displayed a nucleotide dissimilarity ranging from 0.9% to 14.5%. When the ITS1 and ITS2 sequences were compared with the corresponding sequences from the database, the level of nucleotide differences among populations ranged from 0.4% to 14.85% for ITS1 and from 0.5% to 26.7% for ITS2.
The multi-alignment of the most representative N. aberrans sequences from the database (15 sequences) along with those used in this study showed the presence of many insertion-deletion regions (indels) of varying sizes (1–20 bp) present in both ITS1 and ITS2. Those indels clearly contributed to the length differences observed in both ITS 1 and ITS2 among isolates. The multi-alignment, not presented here, is available on request.
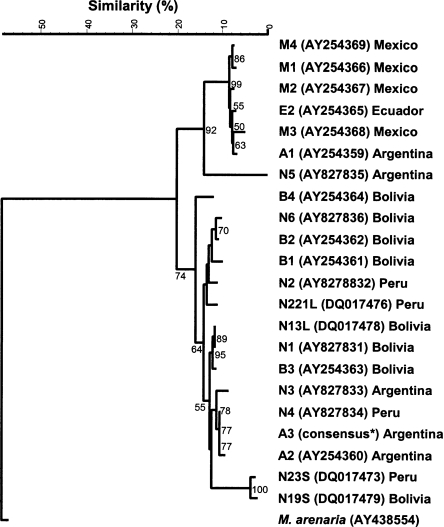
Phylogenetic analysis by Maximum Parsimony, by using M. arenaria sequence as outgroup, generated a tree comprised of three main groups (Fig. 5). One group contained the A3 population under study together with A2 and N3 populations from Argentina and N4 from Peru. A separate group, close to the previous one, included the N19S and N23S sequences, which were clones obtained from Nl and N2 populations, respectively. Another group included Bolivian and Peruvian isolates which were closely related to the isolates of the previous group containing N4 from Peru and A2, A3 and N3 from Argentina. A third group contained Mexican, Ecuadorian and two Argentinean (Al and N5) isolates. Only the optimum MP tree obtained is shown in Figure 5.
Fig. 5.
Maximum parsimony tree resulting from analysis of alignments of internal transcribed spacer ITS sequences of Nacobbus aberrans populations from different geographic areas and Meloidogyne arenaria as outgroup. * = the consensus sequence of A3 population was identified from several ITS sequences of A3 population. Bootstrap supports more than 50% are given for appropriate clade.
The 5.8S rDNA gene was 157 bp in length, and the sequences obtained are identical to those of Fl and F2 populations from Peru reported by Ibrahim et al. (1997). Twenty-three variable nucleotide sites were detected among all populations, and the nucleotide differences ranged from 0 to 6%.
The other PCR fragment of the rDNA obtained corresponded to the D3 expansion domain, 340 bp long, of the 26S rDNA, and it is less constrained than the core structure sequences. A BLAST search at NCBI was performed using our D3 sequence as query showing a 96% of similarity with the D3 region of N. aberrans present in the database (Al-Banna et al., 1997).
Discussion
Accurate identification and pathogenic characterization of plant-parasitic nematodes is an essential first step in designing effective control measures. This is especially important for the false root-knot nematode, N. aberrans, because of its severe pathogenicity in several vegetable crops and to efficiently implement phytosanitary quarantine measures. In fact, specific primers for detection of Nacobbus spp. in soil and in potato tubers are already available (Atkins et al., 2005).
Based on morphometrical and morphological studies, the false root-knot nematode population from Colonia Urquiza, La Plata, in the Buenos Aires province (Argentina) matches with populations reported by Sher (1970), Inserra (1983), Doucet (1989), Doucet and Di Rienzo (1991), and Manzanilla-López et al. (1997, 1999) for N. aberrans. Usually, the migratory stages of N. aberrans are confused with other endoparasitic nematode species, particularly Pratylenchus spp. and Meloidogyne spp. Nevertheless, N. aberrans can be easily distinguished from Pratylenchus nematodes by the combination of the following characters: long dorsal overlap of the pharyngeal glands, single ovary, terminal bursa and the short tail length. Although N. aberrans induce root swellings similar to those of Meloidogyne spp., it is also easy to distinguish from Meloidogyne by morphological differences (pharyngeal-intestinal glands, number of female ovaries, immature female shape) and feeding behavior (type of root galls and host-parasite responses).
The ITS-RFLP analyses revealed that the restriction patterns obtained were similar to those reported for A2 and N3 populations of N. aberrans, both from Argentina (Anthoine and Mugniéry, 2005). In particular, the restriction enzymes Dde I, Hinf I and Hpa II produced identical patterns to N4 population of N. aberrans from Peru, whereas Alu I enzyme showed in A3 population one restriction site less than the N4 population (Anthoine and Mugniéry, 2005). However, A2 and N3 populations from Argentina produced similar restriction patterns to N4 and our population (A3) by using the previous enzymes. Reid et al. (2003) used a primer set internal to those used in this study, thus amplifying a shorter ITS containing region. Therefore, only the internal fragments showed the same lengths in both N4 and A3 populations. When the enzyme Alu I was used, faint secondary bands were visible in some digests despite the extended digestion period used (Fig. 4). Intra-and inter-population differences in RFLP can be explained by the existence of differences in restriction sites in ITS sequences and/or the appearance of additional ITS haplotypes with different sequences. Heterogeneity in ITS regions is widely reported among plant-parasitic nematodes and also among N. aberrans isolates (Zijlstra et al., 1995; Blok et al., 1998; Hugall et al., 1999; Subbotin et al., 2000; Waeyenberge et al., 2000; Reid et al., 2003; Anthoine and Mugniéry, 2005).
The sequences obtained from different cloned ITS amplicons showed a very low level of nucleotide variation inside the A3 population under study. The pairwise comparison of these sequences with those of N. aberrans isolates from the database showed high level of nucleotide differences (0.9%–14.5%) with respect to that recorded for a wide range of species of nematodes that usually is ≤1 (Gasser and Newton, 2000). These findings support two hypotheses: on one hand, that N. aberrans s. l. species is characterized by substantial intra-specific variation in the ITS region as also reported by Reid et al. (2003) or, that on the other hand, the observed dissimilarities (0.4%–14.5% for ITS1, 0.5%–26.7% for ITS2) could reflect the level of nucleotide differences in closely related species belonging to this genus with insufficient time to differentiate (Manzanilla-López et al., 2002; Nadler, 2002).
In addition, the alignment of the most representative N. aberrans sequences from the database along with our sequences revealed a lot of indels in both ITS1 and ITS2. When considering the indels, three hypothetical sequence groups are clearly detected (Fig. 6, bordered by dotted lines) as previously reported (Reid et al., 2003; Anthoine and Mugniéry, 2005). One hypothetical sequence group is consistent with our sequences together with N3 and A2 populations from Argentina and N4 population from Peru; the second hypothetical sequence group includes Bl, B2, B3, B4, Nl and N6 populations from Bolivia and N2 population from Peru; and the third hypothetical sequence group includes N5 and Al populations from Argentina and Mexican and Ecuadorian populations. These results confirm the existence of two distinct genotypes for N. aberrans isolates from Argentina as already described by Reid et al. (2003) and our A3 population groups as well, with A2 and N3 populations suggesting a shared geographical origin.
Fig. 6.
Portion of the multi-alignment of different isolates of Nacobbus aberrans. The alignment corresponds to a region located at the 5’ end of the ITS1. The outlined horizontal lines mark the three hypothetical groups; the upper sequence group is the first one, the intermediate the second one and last sequence group the third one. The accession numbers of each isolate are: Al: AY254359; A2: AY254360; Bl: AY254361; B2: AY254362; B3: AY254363; B4: AY254364; E2: AY254365; Ml: AY254366; M2: AY254367; M3: AY254368; M4: AY254369; Nl: AY827831; N2: AY8278832; N3: AY827833; N4: AY827834; N5: AY827835; N6: AY827836; N13L: DQ017478; N19S: DQ017479; N211L: DQ017476; and N23S: DQ017473.
The third hypothetical sequence group displayed remarkable homogeneity among each other but high sequence variability compared to the other two groups. In contrast, the first and second hypothetical sequence groups showed lower variability among each other, mainly at the microsatellite level, suggesting that the isolates belonging to the first and second hypothetical groups are characterized by intra-population sequence variability (Fig. 6). The phylogenetic tree obtained with the MP analysis (Fig. 5) generated a tree with a topology similar to those obtained by Reid et al. (2003) and Anthoine and Mugniéry (2005) which confirmed the presence of three main groups among N. aberrans isolates. One group contained the Mexican, Ecuadorian and two Argentinean (A1 and N5) populations. In fact, recently these authors demonstrated that the N5 population was able to cross with other populations, but the progenies obtained were always infertile and nonviable. On the basis of the crossing results, Anthoine and Mugniéry (2006) proposed that the N5 population should be considered as a separate species. Thus, based on this and the previous findings, the N. aberrans isolates showing almost identical ITS sequences with regard to N5 population (Fig. 5 and 6), we suggest that all these isolates should be also considered as a separate species from those N. aberrans isolates included in the first and second groups. N19S and N23S sequences which did not cluster according to geographical origin suggest a high level of intra-population sequence variability. cob the relatively long branch observed in the tree also reflects this high level of intra-population variability.
In order to assess the variability of the N. aberrans A3 population, the D3 region of the 26S rDNA was also sequenced. The D3 region is characterized by lower evolution rate compared to the ITS-containing region, thus allowing the study of the divergence between closely related species (Kaplan et al., 2000; Olson and Littlewood, 2002). There was no variation between D3 sequences of the A3 population under study. The Blast search revealed the presence of just one D3 sequence of N. aberrans (Al-Banna et al., 1997) in the database, and the pairwise comparison revealed that there was 4% variability. Because the number of D3 sequences is yet low, it could be of great interest to sequence this region in those isolates for which the ITS sequences are already available.
Results on the host-status test of the population of N. aberrans from Colonia Urquiza, La Plata, support its identity within the "sugar beet group," and this confirms what could be inferred from the geographical origin of the population, since the La Plata region corresponds to what Manzanilla-López et al. (2002) refer to as lowlands of Argentina. The histopathological responses induced by the infection of N. aberrans in tomato were characterized by syncytia with dense cytoplasm, hypertrophied nuclei and nucleoli and partial cell wall dissolution. Comparison of these results with those reported in chili pepper (Castillo and Marban-Mendoza, 1984; Manzanilla-López et al., 2002) indicates that nematode parasitism and histopathology are similar in both hosts. Our results also confirm that the infective J2 and males of N. aberrans behave like Pratylenchus nematodes, invading the cortical parenchyma of tomato roots and moving by intracellular migration creating necrotic lesions and cavities in large area of cortical tissues.
In summary, both ITS-RFLP and sequence analyses clearly discriminate the different isolates. In addition, sequence data are strongly supportive of the existence among N. aberrans isolates of three distinct groups, each one containing different isolates coming from different geographical areas and hosts, thus suggesting the existence of species that have shared a common ancestor quite recently.
Footnotes
This paper was edited by Zafar Handoo.
Literature Cited
- Al-Banna L, Williamson V, Gardner SL. Phylogenetic analysis of nematodes of the genus Pratylenchus using nuclear 26S rDNA. Molecular Phylogenetic and Evolution. 1997;7:94–102. doi: 10.1006/mpev.1996.0381. [DOI] [PubMed] [Google Scholar]
- Anthoine G, Mugniéry D. Variability of the ITS rDNA and identification of Nacobbus aberrans (Thorne, 1935) Thorne and Allen, 1944 (Nematoda: Pratylenchidae) by rDNA amplification. Nematology. 2005;7:503–516. [Google Scholar]
- Anthoine G, Mugniéry D. Crossing experiments with South American populations of Nacobbus aberrans (Thorne, 1935) Thorne and Allen, 1944 (Nematoda: Pratylenchidae) Nematropica. 2006;36:67–77. [Google Scholar]
- Atkins SD, Manzanilla-López RH, Franco J, Peteira B, Kerry BR. A molecular diagnostic method for detecting Nacobbus in soil and in potato tubers. Nematology. 2005;7:193–202. [Google Scholar]
- Baldwin JG, Cap GB. Systematics of Nacobbus, the false root-knot nematode. In: Gommers FJ, Maas PWT, editors. Nematology from molecule to ecosystem. Invergowrie, Scotland: European Society of Nematologists; 1992. pp. 101–112. [Google Scholar]
- Blok VC, Malloch G, Harrower BE, Phillips MS. Intraspecific variation in ribosomal DNA in populations of the potato cyst nematode Giobodera pallida . Journal of Nematology. 1998;30:262–274. [PMC free article] [PubMed] [Google Scholar]
- CABI/EPPO. Quarantine pests for Europe. Second edition. Wallingford, UK: CABI Publishing; 1997. [Google Scholar]
- Castillo PG, Marban-Mendoza N. Histopatologia y desarrollo de Nacobbus aberrans Thorne y Allen, 1944 en raices de Capsicum annuum y C. baccatum . Agrociencia. 1984;56:85–93. [Google Scholar]
- Coolen WA. Methods for extraction of Meloidogyne spp. and other nematodes from roots and soil. Root-knot nematodes (Meloidogyne species). Systematics, biology, and control. In: Lamberti F, Taylor CE, editors. New York: Academic Press; 1979. pp. 317–329. [Google Scholar]
- De Bruijn N, Stemerding S. Nacobbus serendipiticus, a plant parasitic nematode new to The Netherlands. Netherlands Journal of Plant Pathology. 1968;74:227–228. [Google Scholar]
- De Luca F, Reyes A, Grunder J, Kunz P, Agostinelli A, De Giorgi C, Lamberti F. Characterization and sequence variation in the rDNA region of six nematode species of the genus Longidorus (Nematoda) Journal of Nematology. 2004;36:147–152. [PMC free article] [PubMed] [Google Scholar]
- Doucet ME. The genus Nacobbus Thorne and Allen, 1944 in Argentina. 1. Study of a population of N. aberrans (Thorne, 1935) Thorne and Allen, 1944 on Chenopodium album L. from Rio Cuarto, Province of Córdoba. Revue de Nematologie. 1989;12:17–26. [Google Scholar]
- Doucet ME, Di Rienzo JA. El genero Nacobbus Thorne and Allen, 1944 en Argentina. 3. Caracterización morfológica y morfométrica de poblaciones de N. aberrans (Thorne, 1935) Thorne and Allen , 1944. Nematropica. 1991;21:19–35. [Google Scholar]
- Esser RP. A water agar en face technique. Proceedings of the Helminthological Society of Washington. 1986;53:254–255. [Google Scholar]
- Franklin MT. Nacobbus serendipiticus n. sp., a root galling nematode from tomatoes in England. Nematologica. 1959;4:286–293. [Google Scholar]
- Gasser RB, Newton SE. Genomic and genetic research on bursate nematodes: Significance, implications and prospects. International Journal for Parasitology. 2000;30:509–534. doi: 10.1016/s0020-7519(00)00021-7. [DOI] [PubMed] [Google Scholar]
- Hugall A, Stanton J, Moritz C. Reticulate evolution and the origins of ribosomal internal transcribed spacer diversity in apomictic Meloidogyne . Molecular Biology and Evolution. 1999;16:157–164. doi: 10.1093/oxfordjournals.molbev.a026098. [DOI] [PubMed] [Google Scholar]
- Ibrahim SK, Baldwin JG, Roberts PA, Hyman BC. Genetic variation in Nacobbus aberrans: An approach toward taxonomic resolution. Journal of Nematology. 1997;29:241–249. [PMC free article] [PubMed] [Google Scholar]
- Inserra, R.N. 1983. Development and pathogenicity of the false root-knot nematode Nacobbus aberrans on sugar beet and its relationships with Heterodera schachtii and Meloidogyne hapla on sugar beet. Ph.D. Dissertation, Utah State University, USA 78 pp.
- Johansen DA. Plant microtechnique. New York: McGraw-Hill Book Co; 1940. [Google Scholar]
- Kaplan DT, Thomas WK, Frisse LM, Sarah JL, Stanton JM, Speijer PR, Marin DH, Opperman CH. Phylogenetic analysis of geographically diverse Radopholus similis via rDNA sequence reveals a monomorphic motif. Journal of Nematology. 2000;32:134–142. [PMC free article] [PubMed] [Google Scholar]
- Kirjanova ES, Lovanova NA. A potato parasite. Zashchita Rastenii. 1975;9:49. [Google Scholar]
- Manzanilla-López RH, Costilla MA, Doucet M, Franco J, Inserra RN, Lehman PS, Cid del Prado-Vera I, Souza RM, Evans K. The genus Nacobbus Thorne and Allen, 1944 (Nematoda: Pratylenchidae): Systematics, Distribution, Biology and Management. Nematropica. 2002;32:149–227. [Google Scholar]
- Manzanilla-López R, Evans K, Bridge J. Plant diseases caused by nematodes. In: Cheng ZX, Chen SY, Dickson DW, editors. Nematology Advances and Perspectives. Vol. 2. Nematode Management and Utilization. Wallingford, UK: CABI Publishing; 2004. pp. 637–716. [Google Scholar]
- Manzanilla-López RH, Halford P, Russell M, Evans K, Cid Del Prado-Vera I, Rowe J. Characterization of Nacobbus aberrans from Mexico and South America. Nematropica. 1997;27:114. [Google Scholar]
- Manzanilla-López RH, Harding S, Evans K. Morphometric study on twelve populations of Nacobbus aberrans (Thorne, 1935) Thorne & Allen, 1944 (Nematoda: Pratylenchidae) from Mexico and South America. Nematology. 1999;1:477–498. [Google Scholar]
- Nadler SA. Species delimitation and nematode biodiversity: Phylogenies rule. Nematology. 2002;4:615–625. [Google Scholar]
- Nicholas KB, Nicholas HB, Jr, Deerfield DW., II GeneDoc: Analysis and visualization of genetic variation. EMBNEW NEWS. 1997;4:1–4. http://www.psc.edu/biomed/genedoc.
- Olson PD, Littlewood DTJ. Phylogenetics of the Monogenea—evidence from a medley of molecules. International Journal for Parasitology. 2002;32:233–244. doi: 10.1016/s0020-7519(01)00328-9. [DOI] [PubMed] [Google Scholar]
- Prasad SK, Khan E, Chawla L. New records of nine nematode genera from the Indian Union. Indian Journal of Entomology. 1965;27:360–361. [Google Scholar]
- Reid A, Manzanilla-López RH, Hunt DJ. Nacobbus aberrans (Thorne, 1935) Thorne and Allen, 1944 (Nematoda: Pratylenchidae): A nascent species complex revealed by RFLP analysis and sequencing of the ITS-rDNA region. Nematology. 2003;5:441–451. [Google Scholar]
- Sher SA. Revision of the genus Nacobbus Thorne and Allen 1944 (Nematoda: Tylenchoidea) Journal of Nematology. 1970;2:228–235. [PMC free article] [PubMed] [Google Scholar]
- Subbotin S, Halford PD, Warry A, Perry RN. Variations in ribosomal DNA sequences and phylogeny of Globodera parasitising solanaceous plants. Nematology. 2000;2:591–604. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne CD. The sugar beet nematode and other indigenous nemic parasites of shad-scale. Journal of Agricultural Research. 1935;51:509–514. [Google Scholar]
- Thorne CD, Allen MW. Nacobus dorsalis, nov. gen. nov. spec. (Nematoda: Tylenchidae) producing galls on the roots of alfilaria, Erodium cicularium (L.) L'Her. Proceedings of the Helminthological Society of Washington. 1944;11:27–31. [Google Scholar]
- Vrain CT, Wakarchuck DA, Levesque AG, Hamilton RI. Intraspecific rDNA restriction fragment length polymorphism in the Xiphinema americanumgrowp . Fundamental and Applied Nematology. 1992;15:563–573. [Google Scholar]
- Waeyenberge L, Ryss A, Moens M, Pinochet J, Vrain TC. Molecular characterisation of 18 Pratylenchus species using rDNA Restriction Fragment Length Polymorphism. Nematology. 2000;2:135–142. [Google Scholar]
- Yin KC, Feng ZX. The investigation of plant nematodes. Acta Phytophylactica Sinica. 1981;8:122–123. [Google Scholar]
- Zamudio GV. Nematode control on tomato (Lycopersicon esculentum Mill) in the Valsequillo Valley, Puebla, Mexico. Nematropica. 1983;13:123. [Google Scholar]
- Zijlstra C, Lever AEM, Uenk BJ, Van Silfhout CH. Differences between ITS regions of isolates of root-knot nematodes Meloidogyne hapla and M. chitwoodi . Phytopathology. 1995;85:1231–1237. [Google Scholar]