Abstract
Root-knot nematodes are a major group of plant-parasitic nematodes, but their sister group within the Tylenchida remains to be identified. To find the sister group and for any investigation of the evolutionary biology of the genus Meloidogyne, it would be useful to identify the most basal species within Meloidogyninae. Meloidogyne spartinae, a root-knot nematode parasitic on cordgrass (Spartina spp.), constitutes a potentially interesting early diverging (or at least highly divergent) root-knot nematode because it was originally described in a different genus, Hypsoperine (and later Spartonema), due to its unique anatomy and biology (although it was later put in synonymy by some, but not all, taxonomists). We have sequenced the whole 18S rDNA of this species and compared it to other sequences of this region that are available in GenBank for numerous Meloidogyne species. Phylogenetic analysis unambiguously locates the branch corresponding to M. spartinae as a lately diverging species, more closely related to M. maritima, M. duytsi or the M. ardenensis-hapla group. Thus, the distinction of a separate genus (Hypsoperine or Spartonema) for this species is not justified.
Keywords: Molecular biology, taxonomy, phylogeny, Tylenchida, Meloidogyninae, M. spartinae, 18S ribosomal DNA
Because they constitute major pests among phytoparasitic nematodes, the root-knot nematodes (Meloidogyne spp.) have received growing interest among plant nematologists. Two species of this genus, M. incognita (Kofoid and White, 1919) and M. hapla Chitwood, 1949, will soon be the first plant-parasitic nematode species to have their genomes sequenced (Bird, 2005; Castagnone-Sereno, 2006). Their homogenous morphology, wide host range (with numerous species being polyphagous) and reproductive biology, often characterized by parthenogenesis (eliminating the possibility of conducting experimental crosses to investigate species limits and relationships), have motivated the use of molecular tools to identify species within this speciosegenus (at least 79 species already described [Siddiqi, 2000]), and to establish their phylogenetic relationships (Castagnone-Sereno et al., 1993; Hugall et al., 1994, 1999; Chen et al., 2003, Skantar and Carta, 2004; Scholl and Bird, 2005). Recent investigations of the molecular phylogeny of Tylenchida based on 18S sequences have deeply modified the conception of the phylogeny and taxonomy of Meloidogyninae. In particular, several studies suggest that the family Heteroderidae, which was considered until now as being formed by two sister sub-families (Heteroderinae and Meloidogyninae), is a polyphyletic group (Baldwin et al., 2004; Holterman et al., 2006, Subbotin et al., 2006). While the Heteroderinae are nested within the Hoplolaimidae, Meloidogyninae correspond to a distantly related group nested within the Pratylenchidae. However, the sister group of Meloidogyninae remains to be identified (Subbotin et al., 2006). To locate the sister group, and for any investigation of the evolutionary biology of the genus Meloidogyne, it would be useful to identify the most basal species within Meloidogyninae. However, to date, Hypsoperine is the only genus other than Meloidogyne that has been recognized within the Meloidogyninae. This genus was first distinguished from Meloidogyne spp. by Sledge and Golden (1964) for Hypsoperine graminis Sledge and Golden, 1964, a North American species parasitic on St. Augustine grass, Stenotaphrum secundatum (Walt.) Kuntze, and for H. acronea Coetzee, 1956 (Coetzee, 1956), an African species found on Sorghum bicolor (L.) Moench spp. bicolor. The authors “believe that this form represents a new genus which occupies a position between Heterodera and Meloidogyne, being closer to the latter;” they highlight that “the posterior portion of the female body is drawn out into a distinct rounded protuberance on which are situated vulva and anus” and the presence of a “thick cuticle.” In 1965, Rau and Fassuliotis described Hypsoperine spartinae Rau and Fassuliotis, 1965, which is associated with Spartina alterniflora Loisel., although they consider that “some of the characters of their new species do not correspond with the Hypsoperine as outlined by Sledge and Golden.” This species presents the largest dimensions of the second-stage juvenile within the Meloidogyninae, a very thin female cuticle, no gelatinous matrix (the eggs are deposited free in the gall) (Eisenback and Hirschmann, 2001), the smallest haploid chromosome number of the subfamily (Triantaphyllou, 1987), and anobligatory amphimictic mode of reproduction (Triantaphyllou, 1990). In 1969, a last species, H. ottersoni Thorne, 1969, which infests reed canarygrass, Phalaris arundinacea L., was added to the genus Hypsoperine (Thorne, 1969).
In 1968, Whitehead synonymized Hypsoperine with Meloidogyne, and this opinion was also shared by Jepson (1987), Luc et al. (1988) and Eisenback and Triantaphyllou (1991). However, there was no consensus on the taxonomic status of the Hypsoperine genus (see for example Allen and Sher, 1967, or Triantaphyllou, 1990, who reported that “[its] generic status has been a controversial subject among nematode taxonomists”). This genus was considered valid by Golden in 1971 and by Handoo et al. in 1993, while Siddiqi (1986) split the genus in two, with H. acronea, H.graminis Sledge and Golden, 1964, H. megriensis Poghosyan, 1971, H. mersa Siddiqi and Booth, 1991, H. ottersoni and H. propora Spaull, 1977, staying in the genus Hypsoperine, while H. spartinae was considered a separate genus named Spartonema. In 2000, Siddiqi synonymized Hypsoperine with Meloidogyne but still considered that Spartonema spartinae (to which was added S. kikuyensis) should be kept in a distinct genus (Siddiqi, 2000).
Aside from the purely taxonomical issue of the genus name of the root-knot nematode producing galls on Spartina alterniflora and evaluating the validity of the genus Hypsoperine or Spartonema, we were mainly concerned with the phylogenetic position of this species relative to the other Meloidogyninae species. In particular, several characteristics concerning its morphology, its cytology or its reproductive mode suggest that it could be a rather divergent species of Meloidogyne and, hence, could be an interesting taxon to: (i) constitute an outgroup for a phylogeny of the Meloidogyne species, (ii) avoid long-branch attraction in broader phylogenetic investigations including Meloidogyninae and other Tylenchida families or sub-families and (iii) identify the sister group of the Meloidogyninae. For those reasons, we sequenced the 18S gene of the root-knot nematode associated with Spartina (to our knowledge, the first sequence for this species available in GenBank), added this sequence to a large data set of other Meloidogyninae species already available in GenBank and conducted phylogenetic analyses to identify the location of this species within the phylogenic tree of the Meloidogyninae.
Materials and Methods
Roots of S. alterniflora were collected 19 June 2005, from Cattus Island County Park near Toms River in Ocean County, NJ. Galls on the roots were dissected, and the nematodes (females, males and juveniles) extracted were identified morphologically using the keys and diagnostic characters provided by Rau and Fassuliotis (1965), Whitehead (1968) and Eisenback and Hirschmann (2001). In particular, the identification of the host plant (on which no other root-knot nematodes have been reported to date), the size of the juveniles (>750 μm, the longest among root-knot nematodes) and their characteristic long bulbous tail easily allowed the identification of the species (photographs available upon request).
Two primers (SSU-tyl-F GAAACTGCGTACGGCTCATT and SSU-tyl-R GGTTCAAGCCACTGCGATTA) allowing amplification of the entire 18S gene (1,664 bp) were designed from an alignment of 18S complete sequences of 62 Tylenchida species available in GenBank. DNA was extracted as described by Plantard and Porte (2003) except that one female was used instead of one juvenile. PCR reactions were done in 10 μl final volume containing 1 μl of DNA extraction, 1X reaction buffer (containing 10 mM tris-HCl, pH 8.3, 50 mM KCl), 2.5 mM MgCl2, 0.8 mM dNTP, 1μM of each primer, 0.5 unit Taq polymerase (AmpliTaq, Applied Biosystems). Amplifications were performed with a PTC-100 (MJ Research, Inc.) thermocycler using the following program: (i) one denaturation cycle at 94°C for 1 min, (ii) 30 cycles with 50 sec at 94°C, 50 sec at 56°C and 1 min 50 sec at 72°C, (iii) final step at 72°C for 5 min. PCR amplifications were checked with a 1% agarose gel electrophoresis. The amplified DNA was excised from the gel using MinElute Gel extraction Kit (Qiagen) and cloned into E. coli TOP10 strain with TA Cloning kit and pCR2.1 plasmid (Invitrogen). The transformants were screened by PCR with M13 universal primers (M13 forward-GTAAAACGACGGCCAGT, M13 reverse-AACAGCTATGACCATG) in 30 μl final volume containing the selected clone in 10 μl of sterilized distilled water, 1X reaction buffer (pH 8.5), 1.5 mM MgCl2, 0.8 mM dNTP, 0.46 μM of each primer, 0.025 unit Taq polymerase (GoTaq Flexi, Promega). Amplifications were performed with a PTC-100 (MJ Research, Inc.) thermocycler and the following program was used: (i) one denaturation cycle at 96°C for 5 min, (ii) 30 cycles with 1min at 96°C, 1min at 55°C and 3 min at 72°C, (iii) final step at 72°C for 5 min. PCR amplifications were checked with a 0.8% agarose gel electro phoresis. The amplicons were cleaned using a sephadex G50 matrix (Amersham Biosciences) before completing the sequencing reaction (Macrogen, Inc.). Both strands were sequenced by the use of seven sequencing primers, including several internal primers, and this sequence is available in GenBank under the accession number EF189177.
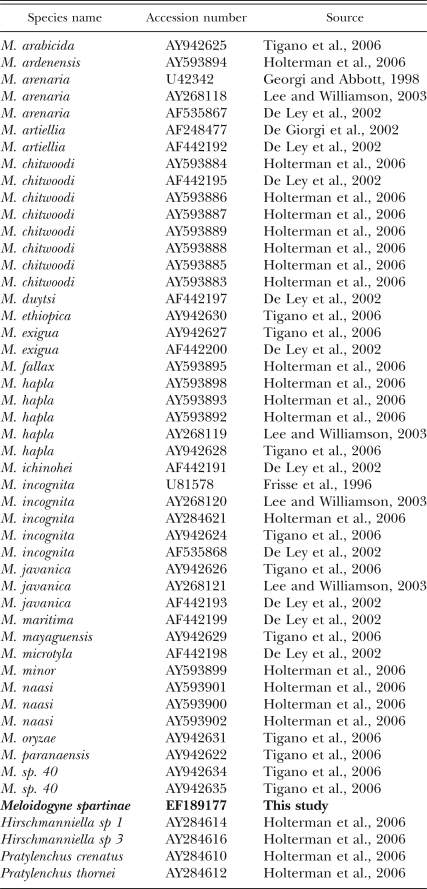
As many 18S sequences of Meloidogyne spp. as possible were retrieved from GenBank using BLASTN and our sequence as a request (Table 1). Only the longest sequences (i.e., more than 1,500 nucleotides) were kept. Based on the recent finding concerning the location of the Meloidogyninae within the Pratylenchidae (Holterman et al., 2006; Subbotin et al., 2006), four outgroups from the Pratylenchidae family were used (two sequences from the genera Hirschmanniella and Pratylenchus). The total data set contained 56 sequences, to which was added our new sequence. Sequences that did not exhibit any polymorphism with another sequence within the data set were discarded (seven sequences). Alignments were conducted with Clustalx with default options. Phylogenetic reconstructions using distance-based methods (Neighbor Joining using Kimura-2-parameters, Juke & Cantor, Tamura-Nei or logdet distances) were conducted with MEGA3 software (Kumar et al., 2001). For phylogenetic analyses based on Maximum Likelihood (ML), the appropriate model was selected using Modeltest (Posada and Crandall, 1998) and the likelihood settings of the selected model were used in PAUP*4b.10 (Swofford, 2002). To evaluate alternative topologies where M. spartinae was constrained to belong to different groups, Kishino-Hasegawa (KH) tests were conducted using PAUP*4b.10 (two-tailed KH test with 1,000 bootstrap replicates and an estimated RELL distribution).
Table 1.
List of the species from which the 18S nucleotide sequence has been used in this study.
Results
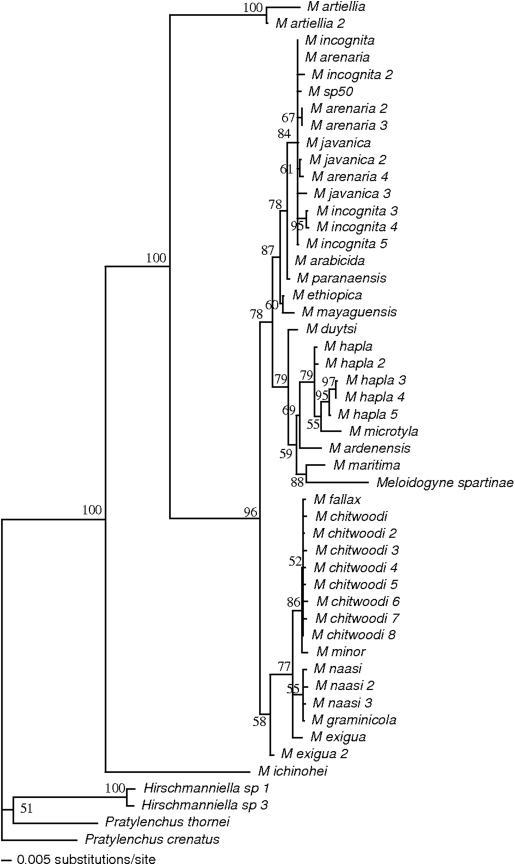
According to both hierarchical likelihood ratio tests and the Akaike information criterion, the selected ML model using Modeltest was the GTR+I+G model (General Time Reversible model, with a proportion of in variable sites and among-site rate variation defined by a gamma distribution). Among the outgroups, while a sister group relationship between the two Hirshmaniella species was always observed (BV 99 or 100%), the two Pratylenchus species were much more distantly related, with a sister group relationship between those two congeneric species in half of the analyses but never with a high BV (35–52%; Fig. 1). The Meloidogyne genus always formed a monophyletic group (BV 100% in the ML tree), except for the most basal and divergent taxa, M. ichinohei Araki, 1992, which clustered as the sister group of the Pratylenchid outgroups in some NJ trees. The remaining Meloidogyne species, including the root-knot species associated with Spartina, always formed a monophyletic group with BV at 96% (ML) or 99 or 100% (NJ).
Fig. 1.
Phylogeny of the Meloidogyne genus based on the 18S sequence. Maximum Likelihood tree obtained with PAUP*4b10, based on the GTR+G+ I model and likelihood settings determined by Modeltest. Only bootstrap values (based on 1,000 replications) superior to 50 are shown.
Within those Meloidogyne species, a first clade corresponded to M. artiellia Franklin, 1961, whereas all the other species formed a separate clade with BV at 99 or 100%. Within this second clade (which includes all the species except M. ichinohei and M. artiellia), three to six groups could be recognized, depending on which data set (pairwise or complete deletion, default or manually corrected alignment) or distance was used. A relatively well supported group (BV from 69% in the ML tree to 77–95% in the NJ trees) was formed by M. ardenensis Santos, 1968, M. microtyla Mulvey et al., 1975, and M. hapla (hereafter called the M. ardenensishapla group). Meloidogyne spartinae was carried by a relatively long branch; this is due to a high number of singletons (25) found in this sequence relative to all other Meloidogyne species. However, careful examination of the sequence at each singleton position confirmed that those differences were not base-calling errors. In most cases, M. spartinae and M. maritima Jepson, 1987, were sister groups (and in particular in all the analyses based on pairwise deletion; BV ranging from 62–73% in the NJ trees and 88% in the ML tree), and this clade is the sister group of the M. ardenensishapla group. Meloidogyne duytsi Karssen et al., 1998, is more closely related to M. maritima, M. spartinae or the M. hapla-ardenensis group and thus appears either as the sister group of those species (BV 79% in ML tree), or as a sister group of the remaining species. All the remaining Meloidogyne species formed two last groups of species. A first group contained two well supported clades; the first clade contained M. arenaria (Neal, 1889) Chitwood, 1949, M. incognita, M. javanica (Treub, 1885) Chitwood, 1949, M. mayaguensis Rammah & Hirschmann, 1988, M. paranaenis Caeirno et al., 1996, M. ethiopica Whitehead, 1968, and M. arabicida Lopez and Salizar, 1989 (BV ranging from 87% in the ML tree to 94–100% in the NJ trees), and the second clade corresponded to M. exigua Göldi, 1892, M. naasi Franklin, 1965, M. graminicola, M. chitwoodi Golden et al., 1980, M. fallax Karssen, 1996, and M. minor Karssen et al., 2004 (BV ranging from 58% in the ML tree to 83–95% in the NJ trees or 77% in the ML tree to 99–100% in the NJ trees when excluding one M. exigua sequence). When 10 alternative topologies (placing M. spartinae with M. artiellia, or with M. ichinohei, or with the outgroups, etc.) were tested using the KH test, the highest likelihood score was obtained for the topology corresponding to M. spartinae clustering with the other Meloidogyne species, M. artiellia being the sister-group of this first cluster, then M. ichinohei being the sister-group of this second cluster (−ln L = 10,786.62; all other topologies tested with a significantly lower likelihood score, KH–test: P < 0.05).
Discussion
While the phylogenetic relationships among all the Meloidogyne species are not fully resolved (in particular, the relative position of the three to six groups described above is not resolved; but see a more comprehensive study based on multiple gene analysis of EST data to confidently locate the position of the groups containing M. chitwoodi, M. hapla, M. incognita and M. javanica, [Scholl and Bird, 2005]), our analysis unambiguously places M. spartinae within the genus Meloidogyne. The exact location of M. spartinae is not fully resolved, but it is certainly not close to the early diverging species M. ichinohei or M. artiellia. The basal position of those two species (to which should probably be added the recently described M. baetica but for which an 18S sequence is not available to date [Castillo et al., 2003; Subbotin et al., 2006]) has already been stated by several authors (De Giorgi et al., 2002; De Ley et al., 2002; Tigano et al., 2005). Among the more lately diverging Meloidogyne species, M. spartinae is neither related to the exigua-chitwoodi-fallax group nor to the arenaria-javanica-incognita group. Meloidogyne spartinae is more closely related to the M. ardenensishapla group, M. maritima or M. duytsi. Concerning the ecology of M. spartinae, it is interesting to note that those last two species are also associated with Poaceae species restricted to the sea shore, namely Ammophila arenaria (L.) Link and Thinopyrum junceiforme (A. & D. Löve) A. Löve (previously known as Elymus farctus) in Europe (with a vicariant species, M. sasseri, associated with A. breviligulata Fern. on the east coast of the US, but for which no 18S sequence is available [Handoo et al., 1993]), like Spartina spp., although cordgrass has a different ecology because it is found in the intertidal zone and not on sand dunes.
Meloidogyne graminis is another Meloidogyne species once included in the Hypsoperine genus (cf. Sledge and Golden, 1964). Because a short 18S sequence (637 bp) of Meloidogyne graminis is available in GenBank (Powers et al., 2005), this species could also be compared to M. spartinae. Phylogenetic analysis (data not shown) provides a topology similar to that previously described, with M. graminis being the sister group of M. spartinae (although with a weak bootstrap value of 47%, probably due to the shorter sequence), close to M. maritima or the M. hapla-ardenensis group. Thus, the close relation ship between M. graminis and M. spartinae was correctly identified by the authors that described those species, but their supposedly distant relationship with other root-knot nematodes leading to the description of a separate genus (Hypsoperine by Sledge and Golden, 1964, or Spartonema by Siddiqi, 1986) was erroneous.
Thus, our analysis, which is based on molecular investigations, confirms the statements made by Triantaphyllou (1987) based on anatomical observations of the structure of the reproductive system and by Eisenback and Hirschmann (2001) based on morphological investigations that M. spartinae is a member of the genus Meloidogyne. Our results confirm the synonymyzation of Spartonema with Meloidogyne as previously proposed by Jepson (1987). Contrary to the hypotheses made by several authors (Triantaphyllou, 1987, 1990; Eisenback and Hirschmann, 2001) that the atypical chromosome number of this Meloidogyne species (a unique feature shared only with M. kikuyensis) represents the ancestral form, our results suggest instead that it is a derived character state arising from the 13 to 19 chromosomes of most other Meloidogyne species. Finally, because of the internal position of M. spartinae within the Meloidogyne phylogenetic tree, our results highlight; (i) the importance of the highly divergent species within the Meloidogyne genus (which are M. artiellia, M. ichinohei and potentially M. baetica) for investigations on the evolutionary biology of root-knot nematodes, and (ii) the necessity of getting additional sequences of other poorly known Meloidogyne species (including species like M. spartinae that are not associated with crops) such as M. kikuyensis.
Footnotes
The authors thank Didier Mugniéry (UMR “BiO3P”) for helpful discussion concerning the identification and morphology of M. spartinae, Alain Buisson (LNPV, Rennes) for photographs, Malika Ainouche (Université de Rennes 1) and Jean-Yves Rasplus (CBGP, Montpellier) for his help in phylogenetic analysis.
This paper was edited by Zafar Handoo.
Literature Cited
- Allen MW, Sher SA. Taxonomy problems concerning the phytoparasitic nematodes. Annual Review of Phytopathology. 1967;5:247–264. [Google Scholar]
- Baldwin JG, Nadler SA, Adams BJ. Evolution of plant parasitism among nematodes. Annual Review of Phytopathology. 2004;42:83–105. doi: 10.1146/annurev.phyto.42.012204.130804. [DOI] [PubMed] [Google Scholar]
- Bird DM. Model systems in agriculture: Lessons from worms. Annals of Applied Biology. 2005;146:147–154. [Google Scholar]
- Castagnone-Sereno P. Genetic variability and adaptive evolution in parthenogenetic root-knot nematodes. Heredity. 2006;96:282–289. doi: 10.1038/sj.hdy.6800794. [DOI] [PubMed] [Google Scholar]
- Castagnone-Sereno P, Piotte C, Uijthof J, Abad P, Wajnberg E, Vanlerberghe-Masutti F, Bongiovanni M, Dalmasso A. Phylogenetic relationships between amphimictic and parthenogenetic nematodes of the genus Meloidogyne as inferred from repetitive DNA analysis. Heredity. 1993;70:195–204. [Google Scholar]
- Castillo P, Vovlas N, Subbotin S, Troccoli A. A new root-knot nematode, Meloidogyne baetica n. sp (Nematoda: Heteroderidae), parasitizing wild olive in Southern Spain. Phytopathology. 2003;93:1093–1102. doi: 10.1094/PHYTO.2003.93.9.1093. [DOI] [PubMed] [Google Scholar]
- Chen P, Roberts PA, Metcalf AE, Hyman BC. Nucleotide substitution patterning within the Meloidogyne rDNA D3 region and its evolutionary implications. Journal of Nematology. 2003;35:404–410. [PMC free article] [PubMed] [Google Scholar]
- Coetzee V. Meloidogyne acronea, a new species of root-knot nematode. Nature. 1956;177:899–890. doi: 10.1038/177899a0. [DOI] [PubMed] [Google Scholar]
- De Giorgi CD, Veronico P, Luca FD, Natilla A, Lanave C, Pesole G. Structural and evolutionary analysis of the ribosomal genes of the parasitic nematode Meloidogyne artiellia suggests its ancient origin. Molecular and Biochemical Parasitology. 2002;124:91–94. doi: 10.1016/s0166-6851(02)00161-5. [DOI] [PubMed] [Google Scholar]
- De Ley IT, De Ley P, Vierstraete A, Karssen G, Moens M, Vanfleteren J. Phylogenetic analyses of Meloidogyne small subunit rDNA. Journal of Nematology. 2002;34:319–327. [PMC free article] [PubMed] [Google Scholar]
- Eisenback JD, Hirschmann H. Additional notes on the morphology of Meloidogyne spartinae (Nematoda: Meloidogynidae) Nematology. 2001;3:303–312. [Google Scholar]
- Eisenback JD, Triantaphyllou H. Root-knot nematode: Meloidogyne sp. and races. In: Nickle WR, editor. Manual of agricultural nematology. New York: Marcel Decker; 1991. pp. 191–274. [Google Scholar]
- Georgi LL, Abbott AG. Variation in ribosomal genes in Meloidogyne arenaria . Fundamental and Applied Nematology. 1998;21:685–694. [Google Scholar]
- Golden AM. Classification of the genera and higher categories of the order Tylenchida (Nematoda) In: Zuckerman BM, Mai WF, Rohde RA, editors. Plant Parasitic Nematodes, vol. 1. Morphology, Anatomy, Taxonomy and Ecology. New York: Academic Press,; 1971. pp. 191–232. [Google Scholar]
- Handoo ZA, Huettel RN, Golden AM. Description and SEM observations of Meloidogyne sasseri n. sp. (Nematoda, Meloidogynidae), parasitizing beachgrasses. Journal of Nematology. 1993;25:628–641. [PMC free article] [PubMed] [Google Scholar]
- Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Molecular Biology and Evolution. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- Hugall A, Moritz C, Stanton J, Wolstenholme DR. Low, but strongly structured mitochondrial DNA diversity in root-knot nematodes (Meloidogyne) Genetics. 1994;136:903–912. doi: 10.1093/genetics/136.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugall A, Stanton J, Moritz C. Reticulate evolution and the origins of ribosomal internal transcribed spacer diversity in apomictic Meloidogyne . Molecular Biology and Evolution. 1999;16:157–164. doi: 10.1093/oxfordjournals.molbev.a026098. [DOI] [PubMed] [Google Scholar]
- Jepson SB. Farnham Royal, UK: Commonwealth Agricultural Bureaux; 1987. Identification of root-knot nematodes (Meloidogyne species) p. 265. [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Luc M, Maggenti AR, Fortuner R. A reappraisal of Tylenchina (Nemata). 9. The family Heteroderidae Filip'ev & Schuurmans Stekhoven, 1941. Revue de Nematologie. 1988;11:159–176. [Google Scholar]
- Plantard O, Porte C. Isolation and characterization of microsatellite loci in the sugar beet cyst nematode Heterodera schachtii . Molecular Ecology Notes. 2003;3:139–141. [Google Scholar]
- Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Powers TO, Mullin PG, Harris TS, Sutton LA, Higgins RS. Incorporating molecular identification of Meloidogyne spp. into a large-scale regional nematode survey. Journal of Nematology. 2005;37:226–235. [PMC free article] [PubMed] [Google Scholar]
- Rau GJ, Fassuliotis G. Hypsoperine spartinae n. sp., a gall-forming nematode on the roots of smooth cordgrass. Helminthological Society. 1965;32:159–162. [Google Scholar]
- Scholl EH, Bird DM. Resolving tylenchid evolutionary relationships through multiple gene analysis derived from EST data. Molecular Phylogenetics and Evolution. 2005;36:536–545. doi: 10.1016/j.ympev.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Siddiqi MR. Farnham Royal, Slough, UK: Commonwealth Agricultural Bureaux; 1986. Tylenchida: Parasites of plants and insects. [Google Scholar]
- Siddiqi MR. 2nd edition. Farnham Royal, Slough, UK: Commonwealth Agricultural Bureaux; 2000. Tylenchida: Parasites of plants and insects. [Google Scholar]
- Skantar AM, Carta LK. Molecular characterization and phylogenetic evaluation of the Hsp90 gene from selected nematodes. Journal of Nematology. 2004;36:466–480. [PMC free article] [PubMed] [Google Scholar]
- Sledge EB, Golden AM. Hypsoperine graminis (Nematoda: Heteroderidae), a new genus and species of plant-parasitic nematode. Helminthological Society. 1964;31:83–88. [Google Scholar]
- Subbotin SA, Sturhan D, Adams BJ, Powers TO, Mullin PG, Chizhov VN, Vovlas N, Baldwin JG. Molecular phylogeny of the order tylenchida: Analysis of nuclear ribosomal RNA genes. Journal of Nematology. 2006;38:296–296. [Google Scholar]
- Swofford DL. Sunderland, MA: Sinauer Associates; 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. [Google Scholar]
- Thorne G. Hypsoperine ottersoni sp. n. (Nemata, Heteroderidae) infesting Canary grass, Phalaris arundinacea (L.) reed in Wisconsin. Proceedings of the Helminthological Society of Washington. 1969;36:98–102. [Google Scholar]
- Tigano MS, Carneiro R, Jeyaprakash A, Dickson DW, Adams BJ. Phylogeny of Meloidogyne spp. based on 18S rDNA and the intergenic region of mitochondrial DNA sequences. Nematology. 2005;7:851–862. [Google Scholar]
- Triantaphyllou AC. Cytogenetic status of Meloidogyne (Hypsoperine) spartinae in relation to other Meloidogyne species. Journal of Nematology. 1987;5:84–87. [PMC free article] [PubMed] [Google Scholar]
- Triantaphyllou AC. Cytogenetic status of Meloidogyne kikuyensis in relation to other root-knot nematodes. Revue de Nematologie. 1990;13:175–180. [Google Scholar]
- Whitehead AG. Taxonomy of Meloidogyne (Nematodea:Heteroderidae) with descriptions of four new species. Transactions of the Zoological Society of London. 1968;31:263–401. [Google Scholar]