Abstract
The introduction of a double-stranded RNA (dsRNA) into an organism to induce sequence-specific RNA interference (RNAi) of a target transcript has become a powerful technique to investigate gene function in nematodes and many organisms. Data provided here indicate that the inclusion of 1–2 mM spermidine and 50 mM octopamine and a 24 hr incubation period of nematodes in double-stranded RNA (dsRNA) soaking solutions resulted in a considerable increase in the percentage of nematodes that ingested dsRNA as compared to previous reports. This modified dsRNA soaking method was coupled with quantitative real-time RT-PCR (qRT-PCR) analyses to assess the potential silencing of the Heterodera glycines parasitism gene transcripts Hg-pel-1 and Hg-4E02 that are expressed within the esophageal gland cells of preparasitic H. glycines J2. The Hg-pel-1 transcript was most efficiently silenced with one dsRNA construct (ds267) at the highest dsRNA soaking concentration of 5.0 mg/ml, while the Hg-4E02 transcript was more efficiently silenced at the 2.5 mg/ml dsRNA concentration as compared to 5.0 mg/ml. A dsRNA construct (ds285) complementary to a different sequence within the Hg-pel-1 transcript than construct ds267 induced only minimal silencing of the Hg-pel-1 transcript at 2.5 mg/ml. The results suggest that both dsRNA concentration and sequence relative to the transcript targeted are critical for maximizing potential RNAi effects in parasitic nematodes.
Keywords: RNAi, SCN, gene silencing, Heterodera glycines, parasitism gene, plant-parasitic nematode, pectate lyase
The soybean cyst nematode (SCN) Heterodera glycines is a sedentary endoparasite of soybean roots that has become one of the major limiting factors of soybean production in the world (Lu et al., 2006; Wrather and Koenning, 2006). Plant-parasitic nematodes have evolved diverse parasitic relationships with their host plants to obtain nutrients that are necessary to support their development and reproduction (Davis et al., 2004). In H. glycines, proteins encoded by parasitism genes expressed in the esophageal gland cells are secreted through the nematode's stylet into plants to facilitate nematode intracellular migration within host roots and to mediate the modification of specific root cells into complex feeding cells required to support nematode growth and development (Davis et al., 2000). The secretions from sedentary endoparasites are particularly intriguing because of the complex changes in phenotype, function and gene expression that they modulate in the parasitized plant cell (Hussey et al., 2002). Over 50 parasitism genes have been cloned from H. glycines (Wang et al., 2001; Gao et al., 2003), but the true functions of many of these parasitism genes are unknown at present. Functional assays developed for C. elegans genes are being used to determine the function of genes in plant-pathogenic nematodes (Bird and Opperman, 1998; Geary and Thompson, 2001; Hashmi et al., 2003). These studies in C. elegans have immediate applicability to parasitic nematodes in cases where genes from the parasite have high similarity to C. elegans genes, as it may be inferred that the genes perform similar functions in the two organisms. However, very few of the putative parasitism genes identified to date have homologs in C. elegans and may even be species-specific (Davis et al., 2000; Geary and Thompson, 2001; Davis et al., 2004). Thus, the development of efficient assays for functional analysis of isolated parasitism genes encoding stylet secretions is essential for understanding the molecular genetics of the plant-nematode interactions (Davis et al., 2000; Hussey et al., 2002).
The obligate parasitic lifestyle of plant-parasitic nematodes makes the development of classical and molecular genetic techniques difficult with these organisms, and transitory DNA transformations have been obtained in only a very few reports (Hashmi et al., 1995; Jackstadt et al., 1999). The ability to introduce a genespecific double-stranded RNA (dsRNA) into an organism and achieve RNA interference (RNAi) of the complementary target transcript as first described in C. elegans (Fire et al., 1998) has provided a powerful gene-silencing technology for assays of gene function in multiple biological systems including plant-parasitic nematodes (Urwin et al., 2002; Novina and Sharp, 2004). The ability of nematodes to ingest dsRNA from their surrounding environment and induce gene-specific RNAi within the nematode (Timmons and Fire, 1998; Timmons et al., 2001) has lead to the development of a particularly useful technique, referred to as RNAi-by-soaking or RNAi soaking, that has promoted an initiative for genome-wide gene silencing in C. elegans (Tabara et al., 1998; Maeda et al., 2001). RNA interference was subsequently first reported in the plant-parasitic cyst nematodes Heterodera glycines and Globodera pallida (Urwin et al., 2002) by soaking preparasitic J2 in solutions containing the neurochemical octopamine to stimulate ingestion of dsRNA in vitro, a procedure modified from a protocol developed for C. elegans (Horvitz et al., 1982). The ability of ingested dsRNA to induce gene silencing in plant-parasitic nematodes has lead to recent exciting reports of plant host-derived RNAi that has dramatic effects on nematode parasitism of plants (Huang et al., 2006; Steeves et al., 2006; Yadav et al., 2006). While verification of dsRNA ingestion and target gene silencing continue to be technical challenges for parasitic stages of nematodes embedded within host tissues, direct monitoring of target gene silencing can be relatively efficiently achieved in plant-parasitic nematodes using the RNAi-soaking method for target genes that are expressed in the nematode life stages external to the plant host (Urwin et al., 2002; Bakhetia et al., 2005a; Chen et al., 2005; Fanelli et al., 2005; Rosso et al., 2005; Huang et al., 2006).
In this study, we used a modification of the RNAi soaking procedure of Urwin et al. (2002) to monitor the effects of dsRNA ingestion on gene expression in preparasitic J2 of H. glycines using quantitative real-time RT-PCR (qRT-PCR). We used the H. glycines pectate lyase (Hg-pel-1) (de Boer et al., 2002a) and Hg-4EO2 (Gao et al., 2003) genes as models since in situ hybridization (and preliminary qRT-PCR) studies revealed that these two genes are highly expressed in the subventral esophageal glands cells of preparasitic J2 of H. glycines, making them good candidates to quantitatively assess cyst nematode esophageal gland cell gene silencing by RNAi. We refer to both genes as “parasitism genes” although we recognize that Hg-4EO2 has not yet been conclusively proven to have a role in parasitism. Hg-4E02 encodes a predicted secreted peptide with conserved cysteine motifs found in pathogen elicitor molecules (Bos et al., 2003) and a functional nuclear localization signal in plant cells (Elling et al., 2007). Our experiments revealed that the treatment of H. glycines J2 by dsRNA homologous to Hg-pel-1 and Hg-4EO2 resulted in a significant, gene-specific reduction in the levels of cellular mRNA. We also demonstrate striking differences in silencing efficiency between different constructs synthesized along the length of our target gene.
Materials and Methods
Nematode culture and collection: Heterodera glycines in-bred line OP50 was propagated on roots of greenhouse soybean (Glycine max) plants (Goverse et al., 1994). J2 were hatched from eggs and extracted as previously described (Goellner et al., 2000). Briefly, to isolate H. glycines eggs, the cysts were gently crushed in a glass homogenizer, and the eggs were rinsed with water onto a 25-μm-pore sieve. Nematode eggs were stirred in a solution of 0.02% sodium azide for 30 min, rinsed with water on a 25-μm-pore sieve and then hatched over water and soybean root exudate at 28°C on a Baermann pan. After 2 d, hatched J2 were collected, rinsed with water on a 25-μm-pore sieve and resuspended in 100 μl water.
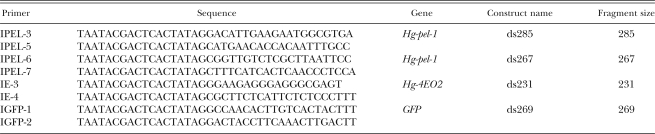
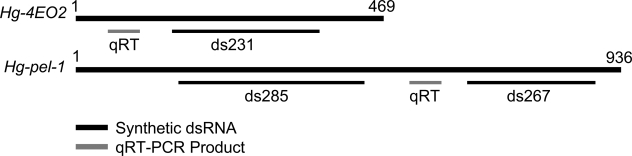
Gene silencing strategy: Hg-pel1-1 is an H. glycines gene that encodes a hydrolytic enzyme and is expressed in the subventral esophageal gland of preparasitic J2 (de Boer et al., 2002a). We have designed two different non-overlapping dsRNA (ds285 and ds267) within the coding region of this gene (Table 1, Fig. 1). The gene Hg-4EO2 encodes a protein of unknown function that contains a secretion signal peptide and a nuclear localization signal (Gao et al., 2003; Elling et al., 2007) and is expressed specifically within the subventral esophageal gland cells of H. glycines preparasitic J2 and parasitic life stages within host roots. We designed a single dsRNA (ds231) within the coding region of this gene. A dsRNA to a gene encoding a synthetic green fluorescent protein (GFP), not present in the nematode, was also designed as a negative control to assess the specificity of the dsRNA treatments. The size of all dsRNA was kept below 300 bp to facilitate ingestion induced in vitro by octopamine (Table 1). All experiments were conducted with freshly hatched J2.
Table 1.
Primers used to prepare dsRNA molecules for nematode soaking experiments.
Fig. 1.
Schematic of the Hg-4EO2 and Hg-pel-1 genes depicting the two dsRNA constructs and the target region of the quantitative real-time RT-PCR.
dsRNA synthesis: Full-length cDNA clones coding for Hg-pel-1 (AY026357) and Hg-4EO2 (AF473826) genes were used as template for RNA synthesis (Table 1). The plasmid DNA was isolated using the Wizard Plus miniprep DNA purification system (Promega, Madison, WI) and used as templates for PCR reactions using the primers shown in Table 1. A dsRNA negative control was included in the experiments by amplifying a fragment from a gene encoding GFP. The plasmid pPD114.108 (L3522), a GFP expression vector designed for C. el egans and obtained from Dr. Andrew Fire, Carnegie Institute of Washington, was used as a template for preparation of the negative control. The PCR amplification was performed as described previously (Wang et al., 2001) using the following cycle profile: 94°C for 2 min, followed by 35 cycles of 94°C 1 min, 69°C 40 sec, 72°C 1 min, and a final step of 72°C for 10 min. The PCR products were purified using a Qiagen PCR purification kit, and the quality and yield of the reactions were checked by agarose gel electrophoresis. Sense and antisense RNA were synthesized in a single reaction in vitro using the MEGAscript RNAi kit (Ambion, Austin, TX) according to manufacturer's instructions, except that the reactions were incubated for 16 hr to increase RNA yield. The amount and quality of dsRNA generated were estimated by ethidium bromide-staining and agarose gel following standard electrophoresis and quantified by spectrophotometry. The dsRNA products were ethanol precipitated, resuspended in nuclease-free water to a concentration of 10–15 μg/μl and stored at −80°C.
dsRNA treatments: The RNAi soaking method was developed by Tabara et al. (1998) for C. elegans and was modified by Urwin et al. (2002) for plant-parasitic nematodes with the addition of the neurotransmitter octopamine to induce feeding of the nematode outside the root. Maeda et al. (2001) have shown that the addition of gelatin and spermidine improved the efficiency of RNAi uptake in C. elegans. We applied the modifications of Urwin et al. (2002) and Maeda et al. (2001) for use in SCN. The soaking protocol involved dissolving of RNA in M9 soaking buffer (43 mM Na2HPO4, 22 mM KH2P04, 2 mM NaCl, 4.6 mM NH4Cl). Ten microliter aliquots of the nematode suspension containing 1,000 J2 were mixed with 5 to 10 μl of dsRNA solution (final concentrations of 5 mg/ml or 2.5 mg/ml), 50 mM octopamine (10 - 80 mM) (Q-0250, Sigma, St. Louis, MO), 0.2 mg/ml fluorescein isothiocyanate (FITC) Isomer I (0.1–1 mg/ml) (F-7250, Sigma), 0.05% gelatin, 1mM spermidine (1–4 mM) (S-2626, Sigma) and sufficient soaking buffer to achieve 30 μl total volume. Nematodes suspended in 30 μl of dsRNA solution were incubated in open tubes in a moist chamber for 24 hr at 28°C. Control treatments included nematodes suspended in the standard feeding components (FITC, octopamine, spermidine, gelatin and M9) without dsRNA and nematodes suspended in feeding components and dsRNA to GFP. For qRT-PCR assays, identical treatments were performed in parallel, except that FITC was not included in the soaking buffer. Each sample of FITC-treated nematodes was rinsed in deionized water three times by centrifugation (total washing time 1 hr) and monitored under the microscope for FITC uptake; motility was observed using a Nikon Eclipse E600 epifluorescence compound microscope (Nikon Instruments, Melville, NY). FITC ingestion under dsRNA feeding conditions was monitored, and photomicrographs of specimens were taken using a Nikon Eclipse E600 microscope equipped with RT-color SPOT digital camera (Diagnostic Instruments, Sterling Heights, MI).
RNA extraction, reverse transcription and PCR amplification: Only nematodes in dsRNA samples (minus FITC) that demonstrated at least 70% uptake and nematodes with high mobility (in their corresponding samples with FITC) were used for subsequent RNA extractions. After treatment, nematodes were thoroughly washed five times with nuclease-free water by centrifugation using standard procedures. One thousand preparasitic J2 were used for RNA extractions. J2 were pulverized using a microfuge and a micropestle as previously described (de Boer et al., 2002b), and RNA was extracted using the RNeasy mini Kit (Qiagen, Valencia, CA) following the manufacture's protocol for isolation of total RNA from animal tissue with the following modifications. After homogenizing the nematodes, 600 μL of RLT buffer (Qiagen) was added, and elution was performed in 40 μl of RNase-free water.
Trace amounts of genomic DNA were removed using RNase-Free DNase (Qiagen) and a Turbo DNA-free kit (Ambion). Reverse transcription reactions were performed using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). To ensure that the RT products originated from mRNA and not from DNA contamination, control reactions were included in which the reverse transcriptase was omitted (NRT). The resulting 60 μl cDNA reactions were stored at −80°C.
Quantitative real-time PCR experiments and product analysis: Endogenous mRNA levels were measured by two-step qRT-PCR based on the SYBR Green detection system using the ABI Prism 7000 Sequence Detection system and the SYBR Green PCR Kit (Applied Biosystems), according to the manufacturer's instructions. qRT-PCR primers were designed using Primer Express software (Applied Biosystems, Table 2). The qRT-PCR assay was designed to detect segments of the target mRNA external to the segment targeted by the dsRNA (Fig. 1). The subsequent real-time PCR reaction contained 3 μl of the cDNA in a total volume of 25 μl, consisting of 1x SYBR Green mix and 0.5 U of AmpErase uracil N-glycosylate (Applied Biosystems), 100 nM forward primer, 100 nM reverse primer and water to 25 μl. The reactions were performed in the MicroAmp 96-well plate capped with MicroAmp optical caps Perkin-Elmer (Applied Biosystems). PCR was performed using the following program: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The accuracy of the SYBR Green detection system was verified by using a heat-dissociation procedure at the end of each run using the Dissociation Curves application (Applied Biosystems) to ensure that only a single PCR product was amplified. For each treatment, identical samples were run in triplicate. Quantification of the transcript level was also normalized to the expression of the H. glycines beta actin gene (AF318603, Table 2). The beta actin gene is constitutively expressed in many tissues and has been reported to be a useful internal real-time PCR endogenous base line control in plant-parasitic nematodes (Painter and Lambert, 2003). The following control reactions were included: PCR negative control without cDNA template (NTC) to confirm that there were no nonspecific PCR products and a NRT control as previously described. Values were obtained from three independent experiments. To minimize interassay variation, samples with the same primer set were always amplified within one run. The entire experiment, beginning with dsRNA treatments, was performed three times. For each experiment, real-time PCR assays were performed two times, each containing triplicate assays of each treatment.
Table 2.
Primers used in real-time quantitative PCR experiments to quantify transcript levels.
Data analyses, such as the determination of the Threshold cycle (Ct), which represents the starting point of the exponential phase of PCR, and graphic presentation were carried out using the Sequence Detection Software v.1.07 (Applied Biosystems). The amount of target gene relative to the beta actin endogenous control was determined using the ΔΔCt method (Levak and Schmittgen, 2001) and expressed as 2−ΔΔ CT for graphic representation. Using five-fold serial dilutions of the purified RNA samples, tests of the dynamic range and amplification efficiency of each gene used in the qRT-PCR experiments were performed to confirm that the amplification efficiency of the target genes were approximately equal to the endogenous control gene.
Results
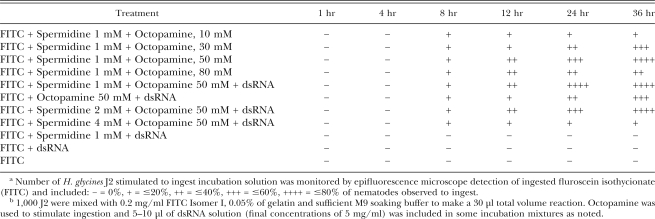
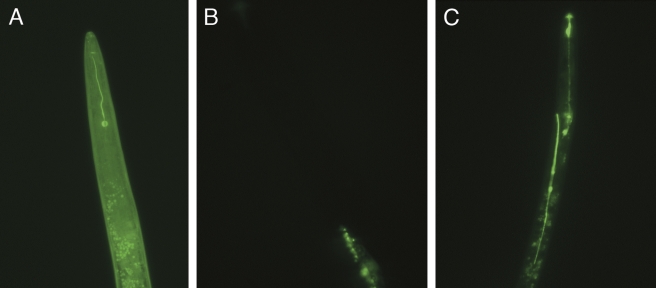
Demonstration of dsRNA uptake by H. glycines J2: To optimize the protocol to stimulate ingestion of RNAi soaking solutions by H. glycines J2 (Urwin et al., 2002), it was first necessary to demonstrate that dsRNA was taken up by nematodes in the soaking solution. FITC was used as a visual marker to detect ingestion of the soaking solution into the alimentary canal (stylet and pharyngeal lumen) of preparasitic H. glycines J2. Experiments were performed with various combinations of FITC (0.1–1 mg/ml), octopamine (10–80 mM), spermidine (1, 2 and 4 mM) and incubation times (1, 4, 8, 12, 24, 36 hr) (Table 3). Concentrations of FITC at 0.2 mg/ml provided sufficient FITC staining without excessive background staining (Fig. 2). Spermidine and gelatin have been shown to enhance RNAi activity in soaking experiments in other organisms (Maeda et al., 2001), and it proved helpful here for H. glycines, although it was not included in RNAi experiments described by Urwin et al. (2002). No FITC uptake was observed before 8 hr, and maximum uptake occurred between 24 and 36 hr of incubation. Octopamine concentrations of 50 mM produced the highest proportion of nematodes with FITC uptake. The lack of detection of ingested FITC in the absence of octopamine in these experiments was consistent with a previous report (Urwin et al., 2002) but differs from a recent report (Schroeder and MacGuidwin, 2007). Spermidine in the presence of octopamine increased the percentage of nematodes exhibiting FITC uptake, although spermidine concentrations of 4 mM or higher were toxic to the nematodes. During the feeding experiments, fluorescence was localized to the alimentary canal (Fig. 2A,C). On average, 70% of the nematodes ingested FITC at the 50 mM concentration of octopamine (Table 3). Viability of the nematodes was assessed after the soaking treatment. Nematodes were washed by centrifugation as previously described and monitored under the microscope. Under the optimized conditions described for RNAi soaking assays, J2 were highly mobile, indicating that the treatment was not toxic to the nematode. These experiments indicated that low concentrations of spermidine improved H. glycines J2 ingestion and viability in soaking solutions containing octopamine, gelatine and dsRNA, and that longer incubation time improved up-take of dsRNA.
Table 3.
Relative efficiencya of different RNAi soaking solutions to stimulate ingestion by preparasitic J2 of Heterodera glycines by soakingb in vitro as modified from Urwin et al. (2002).
Fig. 2.
Uptake of fluorescein isothiocyanate (FITC, 0.2 mg/ml) from incubation solutions by H. glycines J2 in the presence of 50 mM octopamine 24 hr after induction. A) Ingestion of FITC observed in the stylet and pharyngeal lumen; B) J2 soaked in FITC without octopamine showed no or very weak FITC uptake in the amphids; C) Detection of FITC in the J2 stylet, pharyngeal lumen and excretory/secretory system after octopamine treatment. a = amphids, e = excretory/secretory system, i = intestine, m = metacarpus, pl = pharyngeal lumen, s = stylet. A, B, Scale bar = 10 μm.
Quantification of gene transcripts in H. glycines: Before qRT-PCR experiments were performed, all RNA samples extracted from treated nematodes were treated with RNase-free DNase prior to reverse transcription and tested for the presence of DNA by PCR amplification assays using the designed primer pairs. No PCR products were detected when RNA was used as the template. Specificity of the qRT-PCR primers was analyzed by gel electrophoresis, which indicated that the primers amplified a single product with the expected length. A melting curve analysis was also performed, which resulted in a graph containing a single sharp peak indicating that all primers sets generated single PCR products without contaminating primer-dimers. Control experiments in which reverse transcriptase was excluded did not yield PCR products, indicating that the RNA samples contained no contaminating genomic DNA.
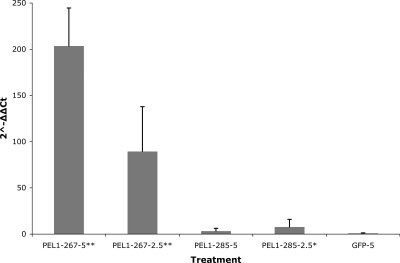
Effect of RNAi on target gene transcript levels: Preparasitic H. glycines J2 were treated with dsRNA molecules (Table 1, Fig. 1) corresponding to the target genes Hg-pel-1 and Hg-4EO2. The effectiveness of RNAi at inhibiting transcript accumulation was calculated using the ΔΔCt method (Levak and Schmittgen, 2001) using beta actin as an endogenous reference for normalization. The ds267 construct had the largest effect on Hg-pel-1 expression, showing a 203-fold and 89-fold reduction in transcript levels at concentrations of 5 mg/ml and 2.5 mg/ml, respectively, as compared to untreated nematodes (Fig. 3). Unlike construct ds267, the ds285 construct resulted in a three-fold decrease in transcript level at 5 mg/ml and a slightly higher seven-fold reduction at 2.5 mg/ml (Fig. 3). A small but statistically insignificant decrease in Hg-4EO2 transcript levels was observed using construct ds231 and a concentration of 5 mg/ml, but at 2.5 mg/ml a 51-fold decrease in transcript level was observed. In contrast, nematodes treated with a dsRNA construct corresponding to the GFP gene showed no significant change in expression of Hg-pel-1 mRNA levels, indicating that the Hg-pel-1 response observed was sequence-specific (Fig. 3).
Fig. 3.
Relative expression of Hg-pel-1 (AY026357) parasitism gene transcripts in preparasitic H. glycines J2 as measured by qRT-PCR. Freshly hatched J2 were stimulated to ingest either 2.5 or 5 mg/ml of dsRNA from soaking solutions for 24 hr prior to RNA extraction from nematodes and subsequent qRT-PCR. The ΔΔCt values for treatments were computed relative to the untreated control samples. The H. glycines beta actin (AF318603) gene was used as an endogenous reference for normalization between samples. Data are represented as fold reduction in expression levels (2−ΔΔCt) ± standard deviation of the mean based on duplicate qRT-PCR assays of three independent experiments. Asterisks indicate significantly reduced ΔCt values as compared to the untreated control using a Student's t-test (*P < 0.001; **P < 0.0001).
Discussion
This study provides direct evidence of RNAi-mediated gene silencing of parasitism genes expressed within the esophageal gland cells of H. glycines. Significant silencing of the Hg-pel-1 and Hg-4EO2 parasitism gene transcripts normally expressed in preparasitic H. glycines J2 was observed following incubation in solutions of dsRNA complementary to the target transcripts. A recent report of host-derived RNAi (Steeves et al., 2006) implies a similar mechanism to silence a H. glycines transcript encoding major sperm protein (MSP), but technical challenges associated with nematodes embedded in plant tissues precluded direct analyses of MSP gene silencing. Genes expressed in the pre-parasitic J2 stages of plant-parasitic nematodes have offered the advantage to directly assess effects of RNAi soaking on transcriptional activity of target genes in nematodes (Urwin et al., 2002; Bakhetia et al., 2005a; Chen et al., 2005; Fanelli et al., 2005; Rosso, et al., 2005), as was the case here. Indirect quantification of RNAi effects on gene expression in plant-parasitic nematodes has been reported through virtual Northerns (Urwin et al., 2002), mRNA in situ hybridizations (Rosso et al., 2005) and electrophoretic gels of RT-PCR products (Chen et al., 2005). The application of qRT-PCR allowed the direct quantification of RNAi effects observed in the H. glycines RNAi soaking experiments here, including dosage effects of the dsRNA solutions on the observed gene silencing. The use of qRT-PCR to monitor RNAi effects in plant-parasitic nematodes has also been recently reported for dual oxidase genes in H. glycines (Bakhetia et al., 2005a) and parasitism genes (Huang et al., 2006; Bakhetia et al., 2007).
Fluorescence of FITC was localized to the stylet and alimentary canal of preparasitic J2 that were incubated in RNAi soaking solution, indicating that soaking solution uptake occurred primarily through ingestion. We were able to optimize our RNAi soaking experiments to achieve a relatively high percentage of nematodes (70%) that ingested the soaking solution, in contrast to the 20% reported for H. glycenes by Urwin et al. (2002). An important observation is that silencing of genes is highly variable, and multiple replications of the silencing experiments can yield high variance among the replications (Bakhetia et al., 2005a). Natural variation among the biological samples may be exacerbated by the relatively small sample sizes (1,000 nematodes/experiment) that were used in this study. The improved rate of uptake observed in the experiments here was likely the result of increasing the incubation time to 24 hr and the concentration of octopamine to 50 mM, and the addition of (1–2 mM) spermidine and gelatin to the soaking solution. This improvement is of practical importance to reduce treatment variability and maintain viability of treated nematodes.
Gene silencing induced via RNAi soaking was effective at reducing expression levels of both genes that we tested, but the effect was dependent on a number of factors, including the dsRNA concentration and the position of the dsRNA construct within the target tran script. It has been suggested previously that the effectiveness of RNAi will depend on several variables: the target tissue, target gene, delivery point and the amount of RNA delivered (Fire, 1999; Orii et al., 2003). Overall, the ds267 construct targeting the Hg-pel-1 gene yielded the strongest inhibition of gene expression and was most effective at the highest concentration that was used (5 mg/ml). An opposite pattern was observed for the construct targeting the Hg-4EO2 gene, where the 2.5 mg/ml dose provided a relatively strong gene silencing effect and the 5.0 mg/ml dose resulted in minimal detectable silencing of the Hg-4E02 transcript. Thus, increased concentrations of dsRNA may not always improve the level of gene silencing, an effect that may be gene- and construct-dependent. Furthermore, the different dsRNA gene constructs varied in their ability to silence Hg-pel-1 as indicated by qRT-PCR. The ds285 construct minimally reduced expression of the Hg-pel-1 at either dosage as compared to the RNAi effect of construct ds267. Reports have indicated that the efficacy of silencing varies considerably among genes, some being more amenable to the technique than others (Fraser et al., 2000; Cutter et al., 2003; Zawadzki et al., 2006). It has also been shown that not all of the short interfering RNA (siRNA) synthesized along the length of a target gene in mammalian cell lines are equally effective for RNAi (Holen, 2002). The Hg-pel-1 results suggest that dsRNA efficacy is dependent on the complementary sequence position in the target gene since the size of both Hg-pel-1 dsRNA constructs are very similar, have similar GC content and lack repetitive sequences that would lead to the formation of hairpins or other secondary structures (data not shown).
Overall, this study provides significant evidence that dsRNA-mediated interference can be used to address questions related to parasitism gene function in H. glycines. Proteins secreted by esophageal glands play important roles in the establishment and development of plant-parasitic nematodes within their hosts (Davis et al., 2000). One such family of secreted proteins, pectate lyases, cleaves the alpha-1,4-galacturonosyl linkages of plant cell wall pectin polymers (Alghisi and Favaron, 1995). Pectin degrading enzymes have a crucial role in pathogenesis in a number of plant pathogen interactions (Alghisi and Favaron, 1995). The first example of non-symbiotic degradation of pectin in plant cell walls by an animal was provided by the identification of a pectate lyase in the cyst nematode G. rostochiensis (Popeijus et al., 2000). A pectate lyase has also been cloned from H. glycines; it possesses a predicted secretion signal and is expressed in the subventral gland of preparasitic J2 (de Boer et al., 2002a). Because pectic enzymes are necessary to break down the cell wall, it is assumed that Hg-pel-1 may have an essential role in pathogenesis. RNAi soaking of J2 has recently been used for functional analyses of the beta-1,4-endoglucanase (cellulase) proteins secreted from cyst nematodes (Chen et al., 2005), and the improvements in the RNAi soaking technique presented here hold promise to assess the roles of Hg-pel-1, Hg-4EO2 and other parasitism genes of plant-parasitic nematodes during infection of plant roots.
The RNAi soaking method may also provide an initial assay to screen nematode parasitism gene candidates for the ultimate goal of host-derived RNAi as has been recently reported (Huang et al., 2006; Steeves et al., 2006; Yadav et al., 2006). Optimal candidates for RNAi soaking will target parasitism genes that are expressed within the nematode during the soaking treatments, as opposed to parasitism genes that are induced to express only when the nematode enters the plants (Wang et al., 2001; Gao et al., 2003). Soaking plant-parasitic nematodes in dsRNA solutions allows ingestion of relatively large dsRNA (e.g., 400 bp) molecules (Urwin et al., 2002) through the nematode stylet, in contrast to dsRNA molecules that must be smaller than the approximately 40 kD size exclusion limit imposed for ingestion through the feeding tube of a sedentary endo-parasitic nematode within host roots (Davis et al., 2004; Bakhetia et al., 2005b). It is likely that the smaller siRNA produced by plant DICER enzyme cleavage of introduced dsRNA molecules (Novina and Sharp, 2004) are the molecules ingested by plant-parasitic nematodes to induce host-derived RNAi (Huang et al., 2006; Steeves et al., 2006). The variability in RNAi activity that is dependent upon target transcript sequence of the introduced dsRNA, as observed with Hg-pel-1 in the assays here, should be considered when designing constructs for host-derived RNAi. The assays optimized here for dsRNA-mediated gene silencing of target parasitic nematode transcripts in RNAi soaking experiments may provide an important initial screen for optimal dsRNA sequence to achieve maximal host-derived RNAi activity.
Footnotes
This research was supported by grants 2005–35604–15434 and 2006–35607–16601 of the National Research initiative of the United States Department of Agriculture and grant 5214 of the United Soybean Board. We thank Mr. Chris Miller (Applied Biosystems) for excellent technical support on qRT-PCR.
This paper was edited by Isgouhi Kaloshian
Literatured Cited
- Alghisi P, Favaron F. Pectin-degrading enzyme and plant parasitic interactions. European Journal of Plant Pathology. 1995;101:365–375. [Google Scholar]
- Bakhetia M, Charlton W, Atkinson HJ, McPherson M. RNA interference of dual oxidase in the plant nematode Meloidogyne incognita . Molecular Plant-Microbe Interaction. 2005a;18:1099–1106. doi: 10.1094/MPMI-18-1099. [DOI] [PubMed] [Google Scholar]
- Bakhetia M, Charlton WL, Urwin PE, McPherson MJ, Atkinson HJ. RNA interference and plant parasitic nematodes. Trends in Plant Science. 2005b;10:362–367. doi: 10.1016/j.tplants.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Bakhetia M, Urwin PE, Atkinson HJ. qPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host. Molecular Plant-Microbe Interactions. 2007;20:306–312. doi: 10.1094/MPMI-20-3-0306. [DOI] [PubMed] [Google Scholar]
- Bird DMcK, Opperman CH. Caenorhabditis elegans: A guide to parasitic nematode biology. Journal of Nematology. 1998;30:1–10. [PMC free article] [PubMed] [Google Scholar]
- Bos JIB, Armstrong M, Whisson SC, Torto T, Ochwo M, Birch PRJ, Kamoun S. Intraspecific comparative genomics to identify avirulence genes from Phytophthora . New Phytologist. 2003;159:63–72. doi: 10.1046/j.1469-8137.2003.00801.x. [DOI] [PubMed] [Google Scholar]
- Chen Q, Rehman S, Smant G, Jones JT. Functional analysis of pathogenicity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Molecular Plant-Microbe Interactions. 2005;18:621–625. doi: 10.1094/MPMI-18-0621. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA, Salcedo T, Estes AM, Good JM, Wood E, Hartl T, Maughan H, Strempel J, Wang B, Bryan AC, Dellos M. Molecular correlates of genes exhibiting RNAi phenotypes in Caenorhabditis elegans . Genome Research. 2003;13:2651–2657. doi: 10.1101/gr.1659203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EL, Hussey RS, Baum TJ. Getting to the roots of parasitism by nematodes. Trends in Parasitology. 2004;20:134–141. doi: 10.1016/j.pt.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Davis EL, Hussey RS, Baum TJ, Bakker J, Schots A, Rosso MN, Abad P. Nematode parasitism genes. Annual Review of Phytopathology. 2000;38:365–396. doi: 10.1146/annurev.phyto.38.1.365. [DOI] [PubMed] [Google Scholar]
- de Boer TJ, McDermott JP, Davis EL, Hussey RS, Popeijus H, Smant G, Baum JM. Cloning of a putative pectate lyase gene expressed in the subventral esophageal glands of Heterodera glycines . Journal of Nematology. 2002a;34:9–11. [PMC free article] [PubMed] [Google Scholar]
- de Boer JM, McDermott JP, Wang X, Maier T, Qiu F, Hussey RS, Davis EL, Baum TJ. The use of DNA microarrays for the developmental expression analysis of cDNAs from the oesophageal gland cell region of Heterodera glycines . Molecular Plant Pathology. 2002b;3:262–270. doi: 10.1046/j.1364-3703.2002.00122.x. [DOI] [PubMed] [Google Scholar]
- Elling AA, Davis EL, Hussey RS, Baum TJ. Active uptake of cyst nematode parasitism proteins into the plant cell nucleus. International Journal for Parasitology. 2007;37:1269–1279. doi: 10.1016/j.ijpara.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Fanelli E, Di Vito M, Jones JT, De Giorgi C. Analysis of chitin synthase function in a plant parasitic nematode, Meloidogyne artiellia, using RNAi. Gene. 2005;349:87–95. doi: 10.1016/j.gene.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Fire A. RNA-triggered gene silencing. Trends in Genetics. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gao BL, Allen R, Maier T, Davis EL, Baum TJ, Hussey RS. The Parasitome of the phytonematode Heterodera glycines . Molecular Plant-Microbe Interactions. 2003;16:720–726. doi: 10.1094/MPMI.2003.16.8.720. [DOI] [PubMed] [Google Scholar]
- Geary TG, Thompson DP. Caenorhabditis elegans: How good a model for veterinary parasites? Veterinary Parasitology. 2001;101:371–386. doi: 10.1016/s0304-4017(01)00562-3. [DOI] [PubMed] [Google Scholar]
- Goellner M, Smant G, de Boer JM, Baum TJ, Davis EL. Isolation of beta-1,4-endoglucanase genes of Globodera tabacum and their expression during parasitism. Journal of Nematology. 2000;32:154–165. [PMC free article] [PubMed] [Google Scholar]
- Goverse A, Davis EL, Hussey RS. Monoclonal antibodies that bind to the esophageal glands and stylet secretions of Heterodera glycines . Journal of Nematology. 1994;26:251–259. [PMC free article] [PubMed] [Google Scholar]
- Hashmi S, Ling P, Hashmi G, Reed M, Gaugler R, Trimmer S. Genetic transformation of nematodes using arrays of micromechanical piercing structures. BioTechniques. 1995;19:766–770. [PubMed] [Google Scholar]
- Hashmi S, Tawe W, Lustigman S. Caenorhabditis elegans and the study of gene function in parasites. Trends in Parasitology. 2003;17:387–393. doi: 10.1016/s1471-4922(01)01986-9. [DOI] [PubMed] [Google Scholar]
- Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Research. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz HR, Chalfie M, Trent C, Sulston JE, Evans PD. Seratonin and octopamine in the nematode Caenorhabditis elegans . Science. 1982;216:1012–1014. doi: 10.1126/science.6805073. [DOI] [PubMed] [Google Scholar]
- Huang G, Allen R, Davis EL, Baum TJ, Hussey RS. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proceedings of the National Academy of Sciences USA. 2006;103:14302–14306. doi: 10.1073/pnas.0604698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey RS, Davis EL, Baum TJ. Secrets in secretions: Genes that control nematode parasitism of plants. Brazilian Journal of Plant Physiology. 2002;14:183–194. [Google Scholar]
- Jackstadt P, Wilm TP, Zahner H, Hobom G. Trans formation of nematodes via ballistic DNA transfer. Molecular and Biochemical Parasitology. 1999;103:261–266. doi: 10.1016/s0166-6851(99)00089-4. [DOI] [PubMed] [Google Scholar]
- Levak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu P, Shannon JG, Sleper DA, Nguyen HT, Cianzo SR, Arelli R. Genetics of cyst nematode resistance in soybean PIs 467312 and 507354. Euphytica. 2006;149:259–265. [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Current Biology. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- Orii H, Mochii M, Watanabe K. A simple “soaking method” for RNA interference in the planarian Dugesia japonica . Development Genes and Evolution. 2003;213:138–141. doi: 10.1007/s00427-003-0310-3. [DOI] [PubMed] [Google Scholar]
- Painter JE, Lambert KN. Meloidogyne javanica chorismate mutase transcript expression profile using real-time quantitative RT-PCR. Journal of Nematology. 2003;35:82–87. [PMC free article] [PubMed] [Google Scholar]
- Popeijus H, Overmars H, Jones J, Blok V, Goverse A, Helder J, Schots A, Bakker J, Smant G. Degradation of plant cell walls by a nematode. Nature. 2000;406:36–37. doi: 10.1038/35017641. [DOI] [PubMed] [Google Scholar]
- Rosso MN, Dubrana MP, Cimboli N, Jaubert S, Abad P. Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Molecular Plant-Microbe Interaction. 2005;18:615–620. doi: 10.1094/MPMI-18-0615. [DOI] [PubMed] [Google Scholar]
- Schroeder NE, MacGuidwin AE. Incorporation of a fluorescent compound by live Heterodera glycines . Journal of Nematology. 2007;39:43–49. [PMC free article] [PubMed] [Google Scholar]
- Steeves RM, Todd TC, Essig JS, Trick HN. Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Functional Plant Biology. 2006;33:991–999. doi: 10.1071/FP06130. [DOI] [PubMed] [Google Scholar]
- Tabara H, Grishok A, Mello CC. RNAi in C. elegans: Soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court D, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans . Gene. 2001;26:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Urwin PE, Lilley CJ, Atkinson HJ. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Molecular Plant-Microbe Interactions. 2002;15:747–52. doi: 10.1094/MPMI.2002.15.8.747. [DOI] [PubMed] [Google Scholar]
- Wang X, Allen R, Ding X, Goellner M, Maier T, de Boer JM, Baum TJ, Hussey RS, Davis EL. Signal peptide-selection of cDNA cloned directly from the esophageal gland cells of the soybean cyst nematode Heterodera glycines . Molecular Plant-Microbe Interactions. 2001;14:536–544. doi: 10.1094/MPMI.2001.14.4.536. [DOI] [PubMed] [Google Scholar]
- Wrather JA, Koenning SR. Estimates of disease effects on soybean yields in the United States 2003 to 2005. Journal of Nematology. 2006;38:173–180. [PMC free article] [PubMed] [Google Scholar]
- Yadav BC, Veluthambi K, Subramaniam K. Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Molecular and Biochemical Parasitology. 2006;148:219–222. doi: 10.1016/j.molbiopara.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Zawadzki JL, Presidente PJA, Meeusen ES, De Veer MJ. RNAi in Haemonchus contortus: a potential method for target validation. Trends in Parasitology. 2006;22:485–489. doi: 10.1016/j.pt.2006.08.015. [DOI] [PubMed] [Google Scholar]