Abstract
A PCR-based diagnostic assay was developed for early detection and identification of Aphelenchoides fragariae directly in host plant tissues using the species-specific primers AFragFl and AFragRl that amplify a 169-bp fragment in the internal transcribed spacer (ITS1) region of ribosomal DNA. These species-specific primers did not amplify DNA from Aphelenchoides besseyi or Aphelenchoides ritzemabosi. The PCR assay was sensitive, detecting a single nematode in a background of plant tissue extract. The assay accurately detected A. fragariae in more than 100 naturally infected, ornamental plant samples collected in North Carolina nurseries, garden centers and landscapes, including 50 plant species not previously reported as hosts of Aphelenchoides spp. The detection sensitivity of the PCR-based assay was higher for infected yet asymptomatic plants when compared to the traditional, water extraction method for Aphelenchoides spp. detection. The utility of using NaOH extraction for rapid preparation of total DNA from plant samples infected with A. fragariae was demonstrated.
Keywords: Aphelenchoides fragariae, detection, diagnosis, foliar nematode, ITS1, method, NaOH, ornamental host, PCR, rDNA
Foliar nematodes, Aphelenchoides spp., are an economically damaging and frequently encountered pest in the foliage plant and nursery industries (Richardson and Grewal, 1993; LaMondia, 2001). Foliar nematodes are distributed across the globe on a wide range of nursery-grown crops including numerous ferns, foliage and flowering plants, and herbaceous and woody perennials (Goodey, 1933; Crossman and Christie, 1936; Stokes, 1966; Juhl, 1978; Decker, 1989; Mor and Speigel, 1993; Daughtrey et al., 1995; Knight et al., 1997, 2002) Aphelenchoides fragariae and A. ritzemabosi are the most common parasitic nematode species detected on the aerial parts or shoots of ornamental plants, but A. besseyi has also been reported (Marlatt, 1966; Anonymous, 2004; Kahn, 2004; Perez and Fernandez, 2004; Oliveira and Kubo, 2006). An increase in the number of ornamental plant samples diagnosed with foliar nematodes was recorded between 1990 and 2000 at the NC State University Plant Disease and Insect Clinic (http://www.ces.ncsu.edu/depts/ent/clinic/). Similarly, the number of perennial plants diagnosed with foliar nematodes submitted to the University of Tennessee Plant and Pest Diagnostic Center increased between 2000 and 2005 (Windham et al., 2005). The cultivation and sale of herbaceous perennial plants is an increasing multimillion-dollar industry in the US (USDA National Agricultural Statistics Service, 2006). As the market demand for perennial plants continues to grow, foliar nematodes are likely to become an even larger threat to the nursery industry due to an increase in the interstate and international movement of ornamental crops, as well as extremely limited management options. There is currently no practical or highly effective method for managing foliar nematodes in ornamental crops (LaMondia, 1999; Jagdale and Grewal, 2002; Warfield and Parra, 2003; Jagdale and Grewal, 2004; Warfield et al., 2004a, 2004b, 2004c).
Aphelenchoides spp. feed ectoparasitically or as endoparasites on the epidermis, mesophyll and parenchyma tissues of leaves or fronds (Maggenti, 1981; Decker, 1989; Volvas et al., 2005). Symptoms typically appear as vein-delimited lesions or blotches due to the inability of the nematode to cross the major leaf veins once inside the plant (Goodey, 1933; Southey, 1993; Chizhov et al., 2006). Chlorosis, necrosis, leaf tattering and defoliation are common symptoms caused by foliar nematode feeding and can render a plant unmarketable.
The expression of symptoms may take from weeks to months depending on the host plant and environmental conditions, making early detection of foliar nematodes problematic. Growers are faced with the constant risk of introducing foliar nematode-infected yet asymptomatic plant materials into their production facilities whereby rapid spread of the nematodes to adjoining, healthy nursery stock can occur (Lehman, 1996; Warfield et al., 2004a).
Currently, the most practical and effective control strategy is early detection and exclusion of this pest from growing facilities, including the use of nematodefree propagative stock. The lack of effective controls has generated the need to develop an accurate, highly sensitive and practical method to screen stock plants and incoming plant materials for the presence of foliar nematodes prior to symptom development. Water extraction is the traditional method used for the detection of foliar nematodes; however, this method relies upon the ability of the nematode to emerge from the host tissue (Esser and Riherd, 1981; Jagdale and Grewal, 2002). Low populations of foliar nematodes or certain life stages of the nematode, such as the eggs, may go undetected. In addition, microscopic examination and accurate morphologic identification of any extracted nematodes requires a trained specialist.
The variability of the ribosomal DNA (rDNA) sequence within the noncoding internal transcribed spacer (ITS) regions has previously been used for differentiation of plant-parasitic nematode species and discrimination of isolates (Ibrahim et al., 1994; Ferris et al., 1995; Cherry et al., 1997; Powers et al., 1997; Iwahori et al., 1998; Beckenbach et al., 1999). More recently, discrimination among plant-parasitic nematodes using species-specific PCR primer pairs targeting specific ITS regions has been used for identification purposes or as a rapid diagnostic tool for Bursaphelenchus spp. (Matsunaga and Togashi, 2004; Cao et al., 2005), Heterodera spp. (Szalanski et al., 1997; Subbotin et al., 2001), Ditylenchus spp. (Kerkoud et al., 2007), Meloidogyne spp. (Zijlstra et al., 1995; Qui et al., 2006) and Xiphinema spp. (Wang et al., 2003; Oliveira et al., 2005). There is currently no molecular-based assay available for species-specific identification and detection of A. fragariae.
A NaOH extraction method for the rapid preparation of plant samples for PCR (Wang et al., 1993) has been successfully applied for quick extraction of DNA from fungal-infected plant tissues (Ristaino et al., 1998) and bryophytes (Werner et al., 2002). In this paper, we describe the development of a practical and sensitive PCR diagnostic protocol using species-specific primers for both identification and early detection of A. fragariae directly in host plant tissue and the utility of using NaOH extraction for rapid preparation of DNA from plant samples infected with this nematode. We demonstrate the sensitivity and accuracy of this detection assay using field samples comprised of a diversity of naturally infected ornamental host plants collected at several locations in North Carolina.
Materials and Methods
Morphological identification of Aphelenchoides species: Conventional “water extraction” of foliar nematodes from infected leaf tissues included incubation of coarsely chopped plant samples placed in a petri dish containing a shallow volume of water to promote nematode emigration from host tissues (Jagdale and Grewal, 2002). The extracted nematodes were identified using a compound light microscope equipped with differential interference contrast imaging (DIC) to observe characteristics unique to each species. When available, at least three adult females from each sample were examined to corroborate accurate species identification. Nematodes were considered representative of the genus Aphelenchoides based on the morphological characteristics outlined by Hunt (1993). In addition to a large metacorpus, which is a distinctive feature of all nematodes belonging to the Suborder Aphelenchina, each nematode was examined for three species-specific characteristics as previously described by Sanwal (1961) and Hunt (1993). For A. fragariae, the cephalic region appears high and almost continuous with the body, the tail ends in a blunt point and the oocytes are in a single-file line. For A. besseyi, the cephalic region appears rounded and slightly wider than the body; the tail ends in a three to four pointed terminus (mucro) and the oocytes are in two to four rows. For A. ritzemabosi, the cephalic region is hemispherical and slightly wider than the body, the tail ends with two to four mucro and oocytes are in multiple rows.
DNA extraction from A. fragariae: Nematodes propagated on alfalfa plants grown in agar-based monoxenic culture (originally isolated from Reiger begonia ‘Schawbenland Red’ collected in Ashtabula County, OH, by R. Riedel, The Ohio State University; unpublished) on Gamborg's B-5 media (Sigma-Aldrich, St. Louis, MO) in 100- xl5-mm plastic petri dishes were used as a positive control. The cultures were maintained in a plant growth chamber at 26°C with a 12-hr d and 12-hr night. A minimum of 23 d after subculture, the entire plant-nematode-agar culture was broken apart using a metal spatula, inverted onto a Kimwipelined mesh screen suspended above a shallow bowl of water (Baermann pan) and covered with foil for 48 hr to allow the nematodes to emerge from the tissue and migrate into the water. The extraction suspension was centrifuged in a 1.5-ml microcentrifuge tube at 8,000 rpm for 1 min to pellet the nematodes. The supernatant was removed, and the process was repeated until all of the extraction water was collected, spun and removed. The nematode pellet, composed of mixed stages of juveniles and adults, was transferred to a 2.0 ml conical tube filled halfway with 1-mm glass beads and 180 μl of lysis buffer ATL (Qiagen, Inc., Valencia, CA). Nematodes were subsequently disrupted in a Mini-BeadBeater-1 (Biospec Products, Inc., Bartlesville, OK) for 10 sec at 2,500 rpm, and total genomic DNA was extracted using a DNeasy Blood and Tissue Kit (Qiagen, Inc.) following the Animal Tissues Spin Protocol supplied by the manufacturer. The DNA was eluted in 50 μl of kit elution buffer and stored at −20°C. Nucleic acids were not quantified prior to use in PCR amplifications.
PCR primer design for A. fragariae: Universal primers rDNA2 (5′-TTGATTACGTCCCTGCCCTTT-3′) described in Vrain et al. (1992) and rDNA1.58S (5′-ACGAGCCGAGTGATCCACCG-3′) described in Cherry et al. (1997) were used to amplify the partial sequence of the 18S rRNA gene, the full sequence of the ITS1 region and a partial sequence of the 5.8S rRNA gene from template DNA extracted from cultured A. fragariae. PCR amplifications were performed in 50-μl reactions containing 3.0 μl of total DNA, 1.5 units of Platinum Taq DNA Polymerase (Invitrogen, Inc., Carlsbad, CA), lx PCR buffer (20 mM Tris-HCL [pH 8.4], 50 mM KC1), 1.5 mM MgCl2, 200 μM of each dNTP, 0.2 μM of each oligonucleotide primer and sterile molecular-grade water to volume. PCR products were separated on a 1.5% agarose gel in lx TBE. The resulting 450-bp fragment was cut from the gel and purified using a MinElute Gel Extraction Kit (Qiagen, Inc.). Purified DNA was sent to the DNA Analysis Facility of the Iowa State University Office of Biotechnology for sequencing.
DNA sequence of the ITSl region from cultured A. fragariae obtained with the universal primers was aligned using Clustal X program (Chenna et al., 2003) with other isolates of A. fragariae ITSl and Aphelenchoides spp. sequences available from GenBank. It was also compared to the closest nucleotide sequences in GenBank to determine potential similarity among other nematode species, insects, plants, or plant pathogens using BLASTn (Altschul et al., 1997). From the combined analyses, forward primer AFragFl (5′-GCAAGTGCTATGCGATCTTCT-3′) and reverse primer AFragRl (5′-GCCACATCGGGTCATTATTT-3′) were designed from unique sequences near the ends of the aligned A. fragariae ITSl region using Primer3 (Rozen and Skaletsky, 2000). Primer3 code is available online at http://primer3.sourceforge.net. Primers AFragFl and AFragRl were rated for amplification success based on potential secondary structural problems including hairpins, dimers and palidromes using Net Primer (PREMIER Biosoft International, Palo Alto, CA) and predicted to amplify a PCR product 169 bp in length.
PCR amplification using species-specific primers: Optimized and reproducible PCR amplifications using primers AFragFl and AFragRl were performed in 25-μl reactions containing 2.0 μl of total DNA, 1.25 units of Platinum Taq DNA Polymerase (Invitrogen Corp.), lx PCR buffer (20 mM Tris-HCL [pH 8.4], 50 mM KC1), 1 mM MgCl2, 200 μM of each dNTP, 0.4 μM of each oligonucleotide primer and sterile molecular-grade water to volume. A reaction without DNA template was always included as a negative control. Reactions were carried out in a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems, Foster City, CA) programmed according to the following: an initial denaturation step at 94°C for 2 min followed by 40 cycles, each consisting of denaturation at 94°C for 1 min, annealing at 53°C for 40 sec and extension at 72°C for 1 min, with a single final extension at 72°C for 10 min. The thermal cycler was set to hold at 4°C after the program was complete. Ten microliter aliquots of the PCR products were added to 2 μl of 6x dye marker solution (0.25% bromophenol blue, 30% glycerol) and separated on a 1.5% agarose gel containing 0.15 μg/ml ethidium bromide buffered in lx TBE, run at 100 Vfor 1 hr and visualized under UV light.
DNA sequencing of the ITSl region for comparison of A. fragariae isolates: To detect potential variability in the ITSl region among A. fragariae isolates collected in North Carolina, DNA extracted from seven independent isolates morphologically identified as A. fragariae was used as PCR template with the universal primers rDNA2 and rDNA1.58S, and the products were subsequently sequenced as described above. Sequences of three of the seven isolates have been deposited in GenBank under the accession numbers EU219917 to EU219919. Sequence of the full ITSl region, including the predicted 169-bp ITSl fragment obtained with primers AFragFl and AFragRl, was aligned and compared with the ITSl consensus sequence of A. fragariae (GenBank Accession AF119049).
Specificity of the A. fragariae-specific PCR assay: The primer specificity of AFragFl and AFragRl was tested in PCR reactions using template DNA extracted from several cultured nematode species, including Meloidogyne incognita, Heterodera schachtii, Pratylenchus penetrans, Caenorhabditis elegans, Ditylenchus dipsaci, A. besseyi and A. ritzemabosi. Primer specificity of AFragFl and AFragRl was also evaluated against DNA extracted from common nursery and greenhouse pests and pathogens including obscure mealybug (Pseudococcus affinis), whiteflies (Trialeurodes spp.) and spider mites (Tetranychus sp.) collected from NC State University's greenhouse facility, and aphids (Microsiphum spp. and Myzus persicae) collected in a landscape perennial bed located in Raleigh, NC. DNA from these pests was extracted using the Qiagen DNeasy Blood and Tissue Kit according to the manufacturer's Animal Tissues Protocol. The DNA was eluted in two steps using 50 μl of the elution buffer supplied by the manufacturer and stored at −20°C. Cheryle O'Donnell at the University of California, Davis, kindly provided DNA of Western flower thrips (Frankliniella occidentalis). DNA was also extracted from mycelia of the fungal pathogen Botrytis cinerea, grown overnight in spore-inoculated V8 broth and extracted using a PUREGENE DNA Purification Kit for Yeast and Gram-positive Bacteria (Gentra Systems, Minneapolis, MN). DNA from pure, bacterial cultures including Xanthomonas campestris pv. zinniae and a Pseudomonas sp. (isolated from Lantana camara) was extracted and purified using the DNeasy Blood and Tissue Kit (Qiagen, Inc.) and evaluated for cross-reactivity with primers AFragFl and AFragRl. Extracted DNA from several non-infected host plants including Lantana camara ‘New Gold’, Salvia microphylla, Verbena ‘Snowflurry’, Heuchera ‘Frosted Violet’, Dahlia x ‘Bishop of Canterbury’ and Asplenium nidus was used as template in the PCR-reaction mixture to determine if the A. fragariae-specific primers had any cross-reactivity to plant rDNA.
Sensitivity of the A. fragariae-specific PCR assay: DNA was extracted from decreasing numbers of nematodes collected from a monoxenic culture of A. fragariae. In the first assay, nematodes were hand picked from the extraction suspension and pooled in groups of 50, 20, 15, 10, 5, 4, 3, 2 and 1 individual nematode for DNA extraction using the Qiagen DNeasy Blood and Tissue Kit. For the second assay, DNA from a single A. fragariae nematode was extracted together with increasing numbers of 6-mm leaf disks (from 1 to 20 disks) collected from uninfected A. nidus leaf tissue using a paper punch. To avoid a potential reduction in DNA yield and purity, the leaf disks were weighed to determine when the maximum amount of plant tissue (100 mg) recommended by the manufacturer was reached. This second assay was repeated two more times using leaf disks removed from non-infected Heuchera ‘Frosted Violet’ and Dahlia x ‘Bishop of Canterbury’ leaves. DNA extracted from leaf disks without the addition of a nematode was used as a negative control. The third assay varied the number of individual nematodes but kept the number of leaf disks constant. Increasing increments of nematodes from 1 to 1,000 were hand picked and combined with three non-infected A. nidus leaf disks, and total DNA was extracted using the Qiagen DNeasy Plant Mini Kit. Extracted DNA from each sensitivity assay was used as template in PCR-reaction mixtures with the A. fragariae-specific primer set, AFragFl and AFragRl.
DNA extraction, PCR and sequencing of other Aphelenchoides spp.: Three additional Aphelenchoides species (A. ritzemabosi, A. besseyi and Aphelenchoides sp.) were used in the present study. To obtain DNA, several crushed nematode specimens were transferred to a microcentrifuge tube containing 16 μl double-distilled water, 2 μl 10X PCR buffer and 2 μl Proteinase K (600 μg/ml) (Promega, Madison, WI). The tubes were incubated at 65°C for 1 hr followed by 95°C for 15 min. Detailed protocols for PCR, cloning and automated sequencing were performed as previously described (Tanha Maafi et al., 2003). Primers TW81 (5′-GTTTCCGTAGGTGAACCTGC-3′) and AB28 (5′-ATATGCTTAAGTTCAGCGGGT-3′) were used for amplification and sequencing of the ITS rDNA fragment. Sequences of these three species have been deposited in GenBank under the accession numbers EU186067 through EU18071.
Detection of foliar nematodes in field samples of naturally infected ornamental plant hosts: Ferns, herbaceous perennials and woody ornamental plants exhibiting foliar nematode symptoms were collected at wholesale nurseries, retail garden centers and outdoor landscapes in North Carolina from 2004 to 2007. Symptomatic leaf tissue from each plant sample was assayed for the presence of foliar nematodes using both water extraction and PCR-amplification of plant-nematode DNA extract using primers AFragFl and AFragRl. One symptomatic leaf per plant was cut in half. One half was thinly sliced with scissors and placed in a 60- x 15-mm petri dish containing just enough water to cover the tissue. Extraction dishes were incubated at room temperature for 24 to 48 hr and examined under a dissecting microscope to confirm the presence of nematodes. Nematodes in the petri dishes were pelleted by centrifugation as described above and fixed with 10% formaldehyde for later identification to species using light microscopy to examine morphological features. A maximum 1.0 mg of leaf tissue was removed from the second half of the leaf sample and placed in a 2-ml conical tube containing two 5-mm glass beads. The tissue was frozen in liquid nitrogen by immersing the tube for 30 sec and subsequently disrupted in a Mini-BeadBeater-1 (Biospec Products, Inc.) for 10 sec at 2,500 rpm. This process was repeated three times. Ground tissue was processed for DNA extraction using a DNeasy Plant Mini Kit (Qiagen, Inc.) following the manufacturer's protocol. Total genomic DNA was eluted in a final volume of 100 μl elution buffer and stored at −20°C. Extracted DNA from each plant sample was used as template in PCR-reaction mixtures with the A. fragariae-specific primer set.
NaOH extraction of A. fragariae template DNA from infected host plants: NaOH lysis was tested as an alternative method to the Qiagen DNeasy Plant Mini Kit for rapid extraction of template DNA from foliar nematode-infected plant samples. Six-millimeter diam. leaf disks were removed from healthy and nematode-infected (symptomatic) host plants using a paper punch. The sampled host plants included Abelia x grandiflora ‘Kaleidoscope’, Anemia tomentosa (Hairy Flowering Fern), A. nidus (Bird's Nest Fern), Buddleja davidii ‘Nanho Purple’ (Butterfly Bush), Disporum smilacinum ‘Aureovariegata’ (Dwarf Variegated Fairy Bells), Hosta ‘Golden Tiara’, Lantana camara ‘Miss Huff’, Salvia greggii ‘Maraschino’ and Tolmiea menziesii (Piggyback Plant). Three leaf disks from each set, healthy vs. infected, were extracted in water to confirm the presence or absence of nematodes. Three leaf disks were used for DNA extraction using the DNeasy Plant Mini Kit (Qiagen, Inc.), and the final three leaf disks were subjected to NaOH lysis. For the NaOH extraction method, the three leaf disks were briefly ground for 5 to 10 sec in 150 μl of 0.5N NaOH using a polypropylene Pellet Pestle (Kimble/Kontes, Vineland, NJ) to disrupt the cells, after which 5 μl was transferred immediately into a sterile 1.5 ml microcentrifuge tube containing 495 μl of 100 mM Tris-HCl, pH 8.3 (Wang et al., 1993). Serial dilutions of the NaOH-extracted DNA were prepared using 100 mM Tris-HCL, pH 8.3. Two or 3 μl of the dilute DNA suspension was added to 25 μl of PCR master mix. Total DNA extracted using the NaOH method and the Qiagen DNAeasy Plant Mini Kit was used as template in PCR-reaction mixtures with the A. fragariae-specific primer set.
Comparison of the traditional water extraction method and PCR-based assay for early detection of A. fragariae in nursery samples: Four nursery blocks of naturally infected herbaceous perennials or ferns were sampled biweekly for the presence of foliar nematodes over a 2 mon period beginning 31 May 2006. The nursery blocks were located in a commercial nursery in North Carolina. Eighteen plants with symptoms suspect for foliar nematode infection were removed by the grower from a block of 54 Disporum smilacinum ‘Aureovariegata’ (Dwarf Variegated Fairy Bells) plants and were subsequently isolated in a separate greenhouse. The remaining 36 asymptomatic Disporum plants were consolidated into a 0.6 m2 area and sampled as one nursery block. The 18 isolated, symptomatic Disporum plants were sampled as a second 0.3 m2 nursery block. A third nursery block containing 36 symptomatic and asymptomatic Oxalis regnellii (False Shamrock) plants in a 0.6 m2 area was sampled (data not presented). The fourth nursery block sampled consisted of 24 symptomatic and asymptomatic Mildella nitidula ferns in a 0.35 m2 area. Plants in each nursery block were hand-watered.
Each plant in the selected nursery blocks was sampled at four dates. Symptomatic leaves or fronds were sampled if present, otherwise, asymptomatic leaves were arbitrarily selected. One leaf was removed from the bottom, middle and top third of each D. smilacinum and O. regnellii plant. Three fronds from each M. nitidula fern were sampled by removing one frond from the exterior, middle and interior of the plant. Leaves or fronds collected from each plant were bulked and processed as one sample. Plant samples were placed in an insulated ice chest for transport back to the laboratory. Samples were either immediately processed or stored at 4°C for processing within 24 hr.
Plant samples were assayed for the presence of foliar nematodes using both water extraction and the A. fragariae-specinc PCR assay. Two 6-mm diam. leaf disks were removed from each leaf or frond using a paper punch for a total of 6 leaf disks/sample (Esser and Riherd, 1981). Three leaf disks, one from each leaf or frond in the bulked sample, were used for the water extraction assay, and the other three leaf disks were used in the PCR detection assay. The recovery of a single foliar nematode from the water extraction assay or the presence of a 169-bp PCR amplification product was considered a positive detection.
The proportion of plant samples infected with foliar nematodes was determined for each detection method (water extraction vs. PCR assay) for each nursery block sampled at each sampling date. A paired t-test was performed on the raw data to compare the sensitivity of the two detection methods for the early detection of foliar nematodes in host plant tissue.
Results
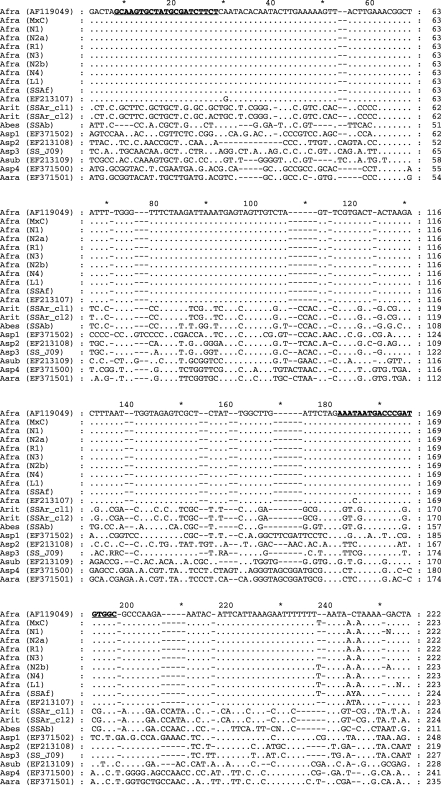
Aphelenchoides fragariae primer design and ITS1 sequence alignment: Divergence in 3' DNA sequence between aligned ITS1 sequences of A. fragariae, A. besseyi and A. ritzemabosi was detected within the ITS1 region (Fig. 1). DNA sequence of the internal 169-bp ITS1 fragment generated with primers AfragFl and AfragRl was identical among seven independent isolates of A. fragariae (confirmed morphologically) collected in North Carolina and the consensus ITS1 sequence of A. fragariae (GenBank Accession AF119049).
Fig. 1.
Nucleotide sequence alignment of partial fragment of the ITSl-rRNA gene of Aphelenchoides fragariae, GenBank Accession AFl 19049 with A. fragariae isolates (Afra) from monoxenic culture (MxC); from naturally infected host plants from six different nursery (N), retail (R) or landscape (L) locations in North Carolina including × Heucherella ‘Sunspot’ (Nl) (EU219917), Dryopteris remota (N2a), Abelia × grandiflora ‘Edward Goucher’ (Rl), Lantana camara ‘Miss Huff’ (N3) (EU219918), Arachnoides simplicior ‘Variegata’ (N2b), Buddleja davidii ‘Petite Plum’ (N4) (EU219919), Hosta sp. (LI); A. fragariae isolate, Pteris longifolia, Moscow, Russia (SSaf) (EU186070); A. fragariae isolate, Aconitum napellus, Netherlands (EF213107); A. ritzemabosi isolate with two ITS clones, Sambucus racemosa, Moscow, Russia (SSAr_cll and SSAr_cl2)(EU186067, EU186068); A. besseyi isolate, rice, Krasnodar region, Russia (SSAb)(EU186069); Aphelenchoides sp. 1, tulip, Netherlands (Aspl) (EF371502); Aphelenchoides sp. 2, tulip, originally identified as A. ritzemabosi, Netherlands (Asp2) (EF213108); Aphelenchoides sp. 3, University of California-Riverside collection (SS-J09) (EU186071); A. subtenuis, Crocus vernus, Netherlands (Asub) (EF213109); Aphelenchoides sp. 4, Narcissus, originally identified as A. besseyi, Netherlands (Asp4) (EF371500); A. arachidis, Sternbergia, Netherlands (Aara) (EF371501). Aphelenchoides fragariae-speciiic primers (AfragFl and AfragRl) are in bold type and underlined. Identities with the A. fragariae consensus sequence are indicated by a period; gaps introduced to maximize alignment are marked by hyphens.
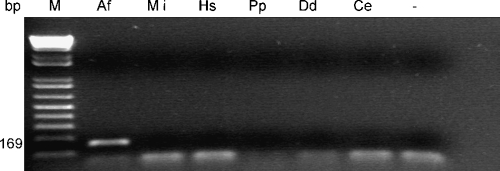
Specificity of the A. fragariae-specific PCR assay: The AfragFl and AfragRl primer pair reproducibly directed amplification within the rDNA-ITSl region of A. fragariae, generating a 169-bp product using total genomic DNA extracted from individual nematodes, populations of nematodes and leaf tissue samples naturally infected with A. fragariae (Figs. 2–5). PCR-reaction mixtures containing primers AFragFl and AFragRl did not produce an amplification product using extracted DNA from the nematodes species M. incognita, H. schachtii, P. penetrans, C. elegans, D. dipsaci (Fig. 2), A. besseyi or A. ritzemabosi (data not shown). The negative controls, without template DNA, produced no amplification product.
Fig. 2.
Agarose gel of PCR products obtained using, Aphelenchoides fragariae species-specific primers AfragFl and AfragRl. Templates of total DNA were extracted from Af: A. fragariae, Mi: Meloidogyne incognita; Hs: Heterodera schachtii, Pp: Pratylenchus penetrans, Dd: Ditylenchus dipsaci; and Ce: Caenorhabditis elegans. Lane M contains DNA molecular marker, and lane “-” contains the PCR reaction without DNA template.
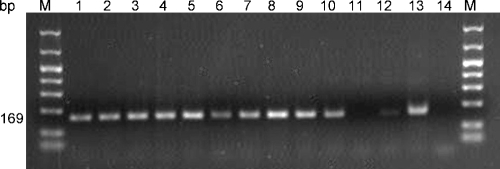
Fig. 5.
Comparison of tissue grinding duration on the amplification of DNA by PCR. Duplicate samples of three naturally infected leaf disks of Hosta ‘August Moon’ and Hosta ‘Midas Touch’ were ground in 0.5N NaOH for 10, 20 or 60 sec. Lanes 1 and 2: ‘August Moon’ 10 sec; Lanes 3 and 4: ‘August Moon’ 20 sec; Lanes 5 and 6: ‘August Moon’ 60 sec; Lanes 7 and 8: ‘Midas Touch’ 10 sec; Lanes 9 and 10: ‘Midas Touch’ 20 sec; Lanes 11 and 12: ‘Midas Touch’ 60 sec; Lane 13: known A. fragariae DNA; Lane 14: NaOH instead of DNA template. Lane M contains DNA molecular size marker.
Primers AfragFl and AfragRl also failed to produce a PCR product with template DNA extracted from obscure mealybug, whiteflies, spider mites, aphids, thrips, B. cinerea, X. campestris pv. zinniae and the Pseudomonas sp. Extracted DNA from healthy host plants showed no cross-reactivity with primers AFragFl and AFragRl based on the failure to produce a PCR product with this primer set.
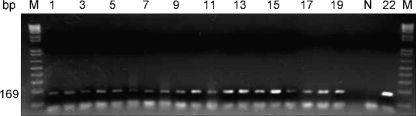
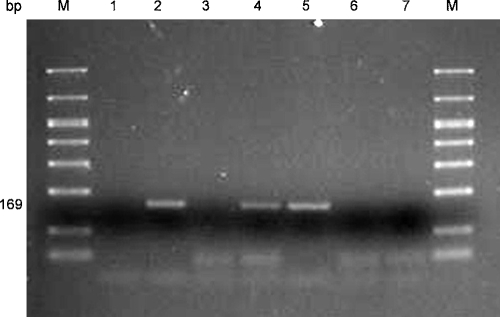
Sensitivity of the A. fragariae-specific PCR assay: DNA was successfully PCR-amplified from one individual A. fragariae nematode, extracted alone, or extracted together with increasing amounts of leaf tissue ranging from 1 to 20 leaf disks (20 leaf disks =146 mg) removed from A. nidus, Heuchera or Dahlia leaves. The PCR assay using primers AfragFl and AfragRl also successfully detected a single nematode, up to the experimental maximum of 1,000 pooled nematodes, within a background consisting of three A. nidus leaf disks (Fig. 3).
Fig. 3.
Sensitivity of Aphelenchoides fragariae species-specific PCR primers. Three 6-mm diam. Asplenium nidus (Bird's Nest Fern) leaf disks were spiked with increasing increments of individual A. fragariae ranging from 1 to 1,000 nematodes. Lane numbers 1 to 20 correspond to the number of nematodes in each DNA extraction (1 to 19 nematodes are shown). No amplification product was obtained with the 20 nematode extraction due to a processing error (Lane 20). Lane M contains DNA molecular size marker, lane N contains the PCR reaction without DNA template and lane 22 contains the PCR reaction with known A. fragariae DNA.
Morphological identification of nematode isolates from naturally infected plant samples collected in North Carolina: More than 120 foliar nematode-infected ferns and herbaceous and woody perennials representing 26 different plant families were collected across eight counties from eight wholesale nurseries, three retail garden centers and two landscape plantings in North Carolina (Table 1). All plant samples displayed symptoms of foliar nematode damage at the time of collection. Based on morphology, 112 of the 115 nematode isolates examined were identified as A. fragariae (Sanwal, 1961; Hunt, 1993). Three nematode isolates deviated in one or more characteristics from the original descriptions of A. fragariae. The shape of the cephalic region of the nematodes extracted from Cheilanthes wrightii appeared more similar to published descriptions of A. ritzemabosi. The shape of the tail region and the number of mucro on the tail of nematodes extracted from Chrysanthemum x ‘Color Echo’ and Dahlia x ‘Bishop of Canterbury’ did not match the original descriptions of A. fragariae. Despite these morphological differences, the DNA amplified from these three isolates using the AFragFl and AFragRl primer set was 169 bp in length and had 100% nucleotide sequence identity with the corresponding 169-bp ITS1 sequence generated by DNA extracted from the A. fragariae monoxenic culture and the Gen-Bank consensus sequence (data not shown).
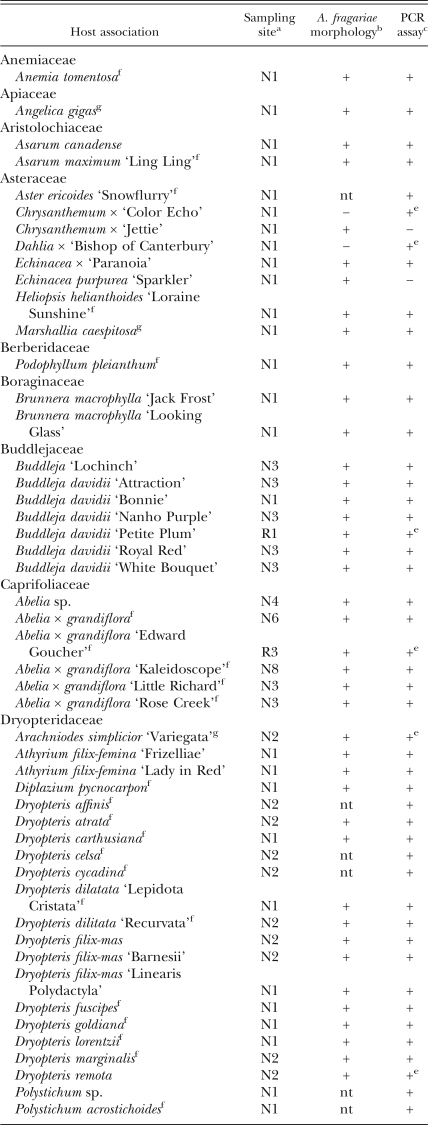
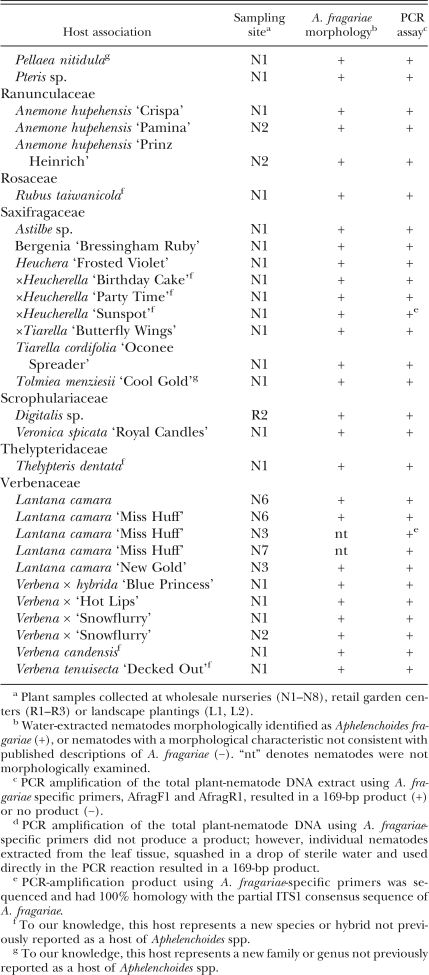
Table 1.
Foliar nematode-infected ferns, ornamental herbaceous and woody perennial plants collected from 2004 to 2007 in nurseries, retail garden centers and landscape plantings in North Carolina, listed alphabetically by plant family.
Detection of A. fragariae among naturally infected plant samples collected in North Carolina using species-specific primers: Total genomic DNA containing both plant and nematode DNA produced a 169-bp fragment for 119 of the 127 plant samples when PCR-amplified with primers AFragFl and AFragRl (Table 1). Eight of the plant samples identified as infected by A. fragariae, based on the morphology of the extracted nematodes, failed to produce a PCR-amplification product using the species-specific primers and plant-nematode DNA extract as template. These eight infected plant samples included six different Salvia species, a Chrysanthemum hybrid and Echinacea purpurea (Purple Coneflower).
NaOH lysis as an alternative method for DNA extraction: NaOH lysis was compared to the Qiagen Dneasy Plant Mini Kit for efficiency in the extraction of total DNA directly from foliar nematode-infected plant tissue. DNA was extracted from both healthy and foliar nematode-infected host plants using duplicate plant samples for each extraction method. DNA extracted from the healthy plants using either DNA extraction method failed to produce a product when PCR-amplified with AFragFl and AFragRl. DNA extracted from the naturally infected host plants of the same nine species produced the expected 169-bp fragment when PCR-amplified with the A. fragariae primer set using either extraction method (Fig. 4). A dilution series of NaOH-extracted DNA from infected A. nidus plants showed sensitivity within the range tested (1:100 to 1: 100,000-fold dilution) when PCR-amplified with primers AfragFl and AfragRl.
Fig. 4.
Comparison of DNA extraction methods for detection of Aphelenchoides fragariae in naturally infected Asplenium nidus (Bird's nest fern) plant tissue using species-specific PCR primers. Lane 1: healthy A. nidus extracted with Qiagen Dneasy Plant Mini Kit; Lane 2: infected A. nidus extracted with Qiagen Dneasy Plant Mini Kit; Lane 3: healthy A. nidus extracted by NaOH lysis; Lane 4: infected A. nidus extracted by NaOH lysis; Lane 5: known A. fragariae DNA template; Lane 6: PCR reaction using NaOH instead of DNA template; Lane 7: PCR reaction using water instead of DNA template. Lane M contains DNA molecular size marker.
Comparison of traditional water extraction and the PCR-based assay for early detection of A. fragariae in nursery samples: Disporum smilacenum asymptomatic block: All 36 plants in the nursery block were initially asymptomatic, with the number of symptomatic plants increasing at each sampling date for a total of 16 plants exhibiting foliar nematodes symptoms during the 2-mon sampling period. Foliar nematodes were detected at the first sampling date in 6% and 53% of the D. smilacenum samples using water extraction and PCR, respectively. At the second sampling date, A. fragariae were detected in 22% of the samples using water extraction, compared to 78% of the samples using the PCR-based detection assay. Foliar nematodes were detected in 22% of the samples by water extraction at the third sampling date compared to 53% detected by the PCR-assay. At the fourth and final sampling date, 33% and 31% of the plant samples were positive for foliar nematodes by water extraction and PCR, respectively. Across all four sampling dates, foliar nematodes were detected in a significantly (P = 0.04) higher percentage of samples using the PCR-assay compared to the water extraction method of detection.
Isolated Disporum smilacenum nursery block: Seven of 18 plants in the isolated nursery block of D. smilacinum had foliar nematode symptoms at the first sampling date. By the fourth and final sampling date, all 18 plants had evidence of foliar nematode damage based on visual symptoms during the 2-mon sampling period. Foliar nematodes were detected in 56% of the plant samples using water extraction compared to 94% of the samples using the PCR assay at the first sampling date. At both the second and third sampling dates, 56% of the samples were positive for foliar nematodes using the water extraction method compared to 72% of the samples using the PCR detection assay. At the fourth sampling date, 78% and 72% of the plant samples were positive for foliar nematodes using the water extraction method or the PCR detection method, respectively. Collectively across all four sampling dates, there was no significant difference (P = 0.06) between detection methods in the percentage of plants in which foliar nematodes were detected.
Mildella nitudula nursery block: Twenty-three of 24 plants in the nursery block had symptoms suspected as foliar nematode damage at the first sampling date. Foliar nematodes were detected in 42% of the samples using the water extraction method compared to 50% using the PCR-based assay at the first sampling date. At the second sampling date, 50% of the samples were positive for foliar nematodes by the water extraction compared to 79% by PCR. The number of samples that tested positive for foliar nematodes increased to 79% for the water extraction method and 100% by the PCR method at the third sampling date. The number of A. fragariae positive samples at the final sampling date was 75% for samples assayed by water extraction compared to 54% of the samples assayed by PCR. All 24 plants had what appeared to be foliar nematode damage at the final sampling date. Collectively across all four sampling dates, there was no significant difference (P = 0.07) between detection methods in the percentage of plants in which foliar nematodes were detected.
Discussion
We have developed a sensitive and rapid PCR-based diagnostic assay that enables accurate identification and early detection of A. fragariae directly in infected host plant tissue by amplification of a species-specific rDNA sequence within the ITS1 region using a single primer pair, AFragFl and AFragRl. The accuracy and reliability of the PCR protocol was demonstrated by the analysis of field samples consisting of more than 120 ornamental host plants naturally infected with A. fragariae. In the rare cases (8 samples, 6 from infected Salvia plants) where infected plant tissue extracts produced no PCR products, amplification products were obtained when the foliar nematodes were first isolated from those tissues prior to DNA extraction. The specificity of the AFragFl and AFragRl primer pair was demonstrated by the inability of the primer pair to amplify any product from DNA extracted from other plant-parasitic nematode species, healthy host plant species, species of common fungal and bacterial pathogens and common insect and mite pests. Since a single A. fragariae individual within a plant tissue extract could be detected by the PCR assay, detection of these nematodes within asymptomatic host plant tissue is possible. The ability to detect A. fragariae DNA within a mixture of host plant and nematode DNA eliminates the need to extract nematodes from plant tissues prior to DNA extraction and does not rely on a trained specialist for nematode identification.
Aphelenchoides fragariae was the only Aphelenchoides species recovered from the ornamental plant samples collected in North Carolina; however, DNA of A. besseyi and A. ritzemabosi extracted from nematodes from rice and elderberry, respectively, collected in Russia failed to produce a product when PCR-amplified using our A. fragariae-specific primers. Based on sequence comparisons of the ITS1 region of A. fragariae with other Aphelenchoides species, our primer set would not be expected to amplify a product from A. arachidis or other plant-parasitic and fungal-feeding Aphelenchoides spp. Therefore, the potential exists to develop additional primer sets that would be specific to other Aphelenchoides species for use in PCR, multiplex PCR or real-time PCR diagnostic assays.
Although our primers would fail to detect plant samples infected with A. ritzemabosi or A. besseyi, A. fragariae was the most prevalent and only foliar nematode species found on cultivated, ornamental crops sampled over a 3-yr period in North Carolina. Only three Aphelenchoides isolates from the samples assayed in the current study could not be positively identified as A. fragariae using morphology, yet these three isolates produced a 169-bp PCR product with primers AFragFl and AFragRl. Our findings are consistent with data from Esser et al. (1988), in which A. fragariae was found with the highest frequency of occurrence on foliar nematode-infected ornamental hosts sampled in Florida. Based on 470 foliar nematode-infected ornamental hosts, Esser et al. (1988) reported 24 occurrences of A. besseyi, 379 occurrences of A. fragariae and 20 occurrences of A. ritzemabosi. The additional 47 reported samples in Florida were from A. nidus (Bird's Nest Fern) and Narcissus sp. collected in the early 1930s, which were infected with A. subtenuis (Esser et al., 1988). Aphelenchoides subtenuis is perhaps more common in Denmark, England and Holland, where it has been found in narcissus bulbs, as well as in crocus, tulip and gigantic onion (Decker, 1989). Aphelenchoides subtenuis has also been found in corms and pseudostems of gladioulus in Iran (Deimi et al., 2006). Aphelenchoides blastophthorus has been found on Scabiosa caucasica in England, in addition to a number of other herbaceous plant species including Anemone hepatica, Begonia, Dipsacus fullonum (fuller's teasel), narcissus, Trollius europaeus (globe flower) and Viola in Denmark, England, and Holland (Decker, 1989). The occurrence of A. blastophthorus in buds of Convallaria (lily of the valley) imported as rhizomes from the Netherlands has been reported (Karnkowski, 2004).
Among the ornamental crop samples collected in North Carolina, we have identified 11 genera and one plant family (Oxalidaceae) that, to our knowledge, have not been previously reported as hosts of Aphelenchoides spp. These plant genera, each associated with A. fragariae, included Agastache, Angelica, Arachniodes, Cheilanthes, Marshallia, Mildella, Oxalis, Pellaea, Pyrrosia, Tolmiea and Tovara. In addition, we have identified 50 species of commercially grown ornamental crops that, to our knowledge, have not been previously reported as hosts of Aphelenchoides spp. Each of these species, plus an additional four hybrids, was found in association with A. fragariae.
Nucleotide sequence comparison of seven A. fragariae isolates collected from different geographic locations in North Carolina showed that the rDNA partial ITS1 sequence amplified by our species-specific primers had 100% homology to the reported ITS1 sequence of an A. fragariae isolate collected in Shizuoka, Japan, by Iwahori et al. (1998), to the ITS1 sequence amplified from DNA extracted from the monoxenic culture of A. fragariae (isolate collected in Ohio) and to the ITS1 sequence of an A. fragariae isolate collected from ferns in Russia (Chizhov et al., 2006). An additional ITS1 sequence has recently been reported from an A. fragariae isolate collected in the Netherlands (GenBank Accession EF213107). The ITS1 sequence of EF213107 has two base pair substitutions compared to the A. fragaraie consensus sequence of Iwahori et al. (1998); one substitution is within the 169-bp region amplified by our species-specific primer pair, and the other is located within the reverse primer (AFragRl), four bp from the end. Neither substitution would be expected to impede amplification by the AFragFl and AFragRl primer pair. These sequence comparisons further demonstrate the utility of our primer set in discriminating A. fragariae isolates among more geographically separated populations of this nematode species.
DNA extraction from foliar nematode-infected host plants using NaOH lysis was fast, simple and reproducible across the host plant species assayed. However, we found that the duration of grinding during tissue disruption was a critical step in the successful amplification of extracted DNA. DNA extraction using excessive grinding of the tissue (typically more than 20 seconds) resulted in the failure to consistently amplify a product with our primer pair (Fig. 5). Using a higher dilution of the DNA extract reduced this problem, presumably by lowering the concentration of PCR inhibitors in the extract. NaOH-based DNA extraction from A. fragariae-infected Salvia plants produced successful PCR products with the AFragFl and AFragRl primers, while DNA extracted from infected Salvia tissue using the Qiagen Dneasy Plant Mini Kit did not. While the NaOH lysis protocol for DNA extraction has not been tested as extensively as the Qiagen Dneasy Plant Mini Kit for sensitivity and reproducibility, implementation would greatly reduce both the overall cost of our PCR-based assay and the time required to perform the assay.
The relative efficiency of foliar nematode detection using our PCR-based assay was significantly higher compared to conventional water extraction when assaying asymptomatic plant tissue. This is attributed to the low population of nematodes in the asymptomatic plant tissue and to the inability of certain nematode life stages to migrate from the plant tissue during water extraction. Once the plant tissue showed evidence of foliar nematode damage, detection using water extraction or PCR was equally efficient in the detection of this pest. A decrease in the number of positive detections by either method was observed in some of the nursery blocks at the later two sampling dates. Because the sampling method required removal of infected (symptomatic) plant tissue at each sampling date, the amount of secondary inoculum was subsequently reduced, limiting the number of new leaf infections. In addition, symptoms attributed to foliar nematode damage could have been caused by other pathogens or factors. Botrytis cinerea commonly infects plants previously infected with foliar nematodes (LaMondia, 2001), and bacteria are often found in association with foliar nematode-damaged leaves. In clinical diagnostic situations, plants are not routinely assayed for the presence of foliar nematodes unless a plant is exhibiting symptoms. Our results provide evidence that infected yet asymptomatic plant material may inadvertently spread foliar nematodes throughout the nursery trade through both local and long-distance shipments. Our sensitive PCR-based detection assay detected a higher proportion of foliar nematode-infected plants in asymptomatic vs. symptomatic plant tissue, allowing for earlier detection of nematodes prior to symptom development. Early detection of foliar nematodes would be advantageous in screening propagative stock plants and incoming plant materials to help prevent the spread or introduction of foliar nematodes within or into a growing facility. We are currently working on the validation of a sampling protocol to determine the number of plants in a nursery block that would need to be sampled in order to detect A. fragariae if present. Once introduced into a growing facility, foliar nematodes have the potential to spread with no effective means of control. Therefore, early detection and eradication are critical for the management of foliar nematodes in plant nurseries.
The genus Aphelenchoides contains more than 140 nominal species, many of which are inadequately morphologically characterized and are difficult to distinguish (Hunt, 1993). Although such species as A. fragariae, A. besseyi, A. ritzemabosi, A. subtenuis and A. blastophorus can be differentiated using a range of morphological features including head shape, number of incisures, posterior uterine branch length, mucro structure on the tail tip, and others (Allen, 1952), molecular approaches using PCR-RFLP (Ibrahim et al., 1994) and PCR with species-specific primers as developed in this paper for A. fragariae could offer a more accurate and practical solution for Aphelenchoides species differentiation. Our A. fragariae-specific PCR assay has application in plant disease diagnostic clinics in assisting growers with screening plant material for early detection of foliar nematodes, in regulatory laboratories where accurate identification is required for phytosanitary permits and as a tool to help study A. fragariae biology to improve and develop more effective foliar nematode management strategies for control of this pest in ornamental plant nurseries.
Footnotes
This research was supported in part by grants from the American Floral Endowment, Fred C. Gloeckner Foundation and the Horticultural Research Institute. The authors acknowledge James Sugar, Kelly Konczal and Kim Sugar for technical support, Christine A. Casey for statistical assistance and helpful discussion, Byron Adams of Brigham Young University for providing the universal primers and David Harshman of Clemson University for providing the monoxenic nematode culture. A portion of an M.Sc. thesis by the first author.
This paper was edited by Andrea Skantar.
Literatured Cited
- Allen JW. Taxonomic status of the bud and leaf nematodes related to Aphelenchoides fragariae (Ritzema Bos 1891) Proceedings of the Helminthological Society of Washington. 1952;19:108–120. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous. Aphelenchoides besseyi . EPPO Bulletin. 2004;34:303–308. [Google Scholar]
- Beckenbach K, Blaxter M, Webster JM. Phylogeny of Bursaphelenchus species derived from analysis of ribosomal internal transcribed spacer DNA sequences. Nematology. 1999;1:539–548. [Google Scholar]
- Cao AX, Liu XZ, Zhu SF, Lu BS. Detection of the pinewood nematode, Bursaphelenchus xylophilus, using a real-time polymerase chain reaction assay. Phytopathology. 2005;95:566–571. doi: 10.1094/PHYTO-95-0566. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry T, Szalanski AL, Todd TC, Powers TO. The internal transcribed spacer region of Belonolaimus (Nemata: Belonolaimidae) Journal of Nematology. 1997;29:23–29. [PMC free article] [PubMed] [Google Scholar]
- Chizhov VN, Chumakova OA, Subbotin SA, Baldwin JG. Morphological and molecular characterization of foliar nematodes of the genus Aphelenchoides: A. fragariae and A. ritzemabosi (Nematoda: Aphelenchoididae) from the Main Botanical Garden of the Russian Academy of Sciences, Moscow. Russian Journal of Nematology. 2006;14:179–184. [Google Scholar]
- Crossman L, Christie JR. A list of plants attacked by the leaf nematode (Aphelenchoides fragariae) Plant Disease Reporter. 1936;20:155–165. [Google Scholar]
- Daughtrey ML, Wick RL, Peterson JL. St. Paul, MN: American Phytopathological Society; 1995. Compendium of flowering potted plant diseases. [Google Scholar]
- Decker H. Leaf-parasitic nematodes. In: Sveshnikova NM, editor. Plant nematodes and their control (phytonematology) New York: E.J. Brill; 1989. pp. 354–368. [Google Scholar]
- Deimi AM, Tanha Maafi Z, Palomares Rius JE, Castillo P. Aphelenchoides subtenuis (Cobb, 1926) Steiner & Buhrer, 1932 (Nematode: Aphelenchoididae) from Iran with morphological and morphometric characterization. Nematology. 2006;8:903–908. [Google Scholar]
- Esser RP, O'Bannon JH, Clark RA. Florida Department of Agricultural and Consumer Services, Division of Plant Industry. Nematology Circular 160; 1988. Procedures to detect foliar nematodes for annual nursery or out of state inspections. [Google Scholar]
- Esser RP, Riherd CC. Distribution of Aphelenchoides fragariae in leaves of Ficus elastica and Asplenium nidus . Plant Disease. 1981;65:425–426. [Google Scholar]
- Ferris VR, Miller LI, Faghihi J, Ferris JM. Ribosomal DNA comparisons of Globodera from two continents. Journal of Nematology. 1995;27:273–283. [PMC free article] [PubMed] [Google Scholar]
- Goodey T. New York: E.P. Dutton and Company; 1933. Plant-parasitic nematodes and the diseases they cause. [Google Scholar]
- Hunt DJ. Wallingford: CAB International; 1993. Aphelenchida, Longidoridae and Trichodoridae: Their systematics and bionomics. [Google Scholar]
- Ibrahim SK, Perry RN, Burrows PR, Hooper DJ. Differentiation of species and populations of Aphelenchoides and of Ditylenchus angustus using a fragment of ribosomal DNA. Journal of Nematology. 1994;26:412–421. doi: 10.1163/003525994x00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahori H, Tsuda K, Kanzaki N, Izui K, Futai K. PCR- RFLP and sequencing analysis of ribosomal DNA of Bursaphelenchus nematodes related to pine wilt disease. Fundamental and Applied Nematology. 1998;21:655–666. [Google Scholar]
- Jagdale GB, Grewal PS. Identification of alternatives for the management of foliar nematodes in floriculture. Pest Management Science. 2002;58:451–458. doi: 10.1002/ps.472. [DOI] [PubMed] [Google Scholar]
- Jagdale GB, Grewal PS. Effectiveness of a hot water drench for the control of foliar nematodes Aphelenchoides fragariae in floriculture. Journal of Nematology. 2004;36:49–53. [PMC free article] [PubMed] [Google Scholar]
- Juhl MV. Liste over vaertplanter for bladnematoden Aphelenchoides ritzemabosi . Ugeskrift for Agronomer, Hortonomer, Forst-kandidater og Licentiater. 1978;123:183–186. [Google Scholar]
- Kahn MR. Observations on foliar nematode, Aphelenchoides besseyi, in tuberose (Polianthes tuberosa L.) Annals of Plant Protection Sciences. 2004;12:106–109. [Google Scholar]
- Karnkowski W. Frequent occurrence of Aphelenchoides blastophthorus Franklin, 1952 (Nematoda-Aphelenchoididae) in imported Convallaria rhizomes. Progress in Plant Protection. 2004;44:791–794. [Google Scholar]
- Kerkoud M, Esquibet M, Plantard O, Avrillon M, Guimier C, Franck M, Léchappé J, Mathis R. Identification of Ditylenchus species associated with Fabaceae seeds based on a specific polymerase chain reaction of ribosomal DNA-ITS regions. European Journal of Plant Pathology. 2007;118:323–332. [Google Scholar]
- Knight KWL, Barber CJ, Page GD. Plant-parasitic nematodes of New Zealand recorded by host association. Supplement to the Journal of Nematology. 1997;29:640–656. [PMC free article] [PubMed] [Google Scholar]
- Knight KWL, Hill CF, Sturhan D. Further records of Aphelenchoides fragariae and A. ritzemabosi (Nematoda: Aphelenchida) from New Zealand. Australasian Plant Pathology. 2002;31:93–94. [Google Scholar]
- LaMondia JA. Efficacy of insecticides for control of Aphelenchoides fragariae and Ditylenchus dispaci in flowering perennial ornamentals. Supplement to the Journal of Nematology. 1999;31:644–469. [PMC free article] [PubMed] [Google Scholar]
- LaMondia JA. Nematodes: Common and important problems in the ornamental and landscape industry. Nematology Newsletter. 2001;47:1–4. [Google Scholar]
- Lehman PS. Florida Department of Agriculture and Consumer Services, Division of Plant Industry. Nematology Circular 216; 1996. Dispersal modes for foliar nematodes. [Google Scholar]
- Maggenti A. New York: Springer-Verlag; 1981. General Nematology. [Google Scholar]
- Marlatt RB. Ficus elastica: A host of Aphelenchoides besseyi in a subtropical climate. Plant Disease Reporter. 1966;50:689–691. [Google Scholar]
- Matsunaga K, Togashi K. A simple method for discriminating Bursaphelenchus xylophilus and B. mucronatus by species-specific polymerase chain reaction primer pairs. Nematology. 2004;6:273–277. [Google Scholar]
- Mor M, Spiegel Y. Ruscus hypophyllum: A new host for Aphelenchoides fragariae . Journal of Nematology. 1993;25:312–313. [PMC free article] [PubMed] [Google Scholar]
- Oliveira CMG, Fenton B, Malloch G, Brown DJF, Neilson R. Development of species-specific primers for the ectoparasitic nematode species Xiphinema brevicolle, X. diffusum, X. elongatum, X. ifacolum and X. longicaudatum (Nematoda: Longidoridae) based on ribosomal DNA sequences. Annals of Applied Biology. 2005;146:281–288. [Google Scholar]
- Oliveira CMG, Kubo RK. Foliar nematodes (Aphelenchoides spp.) on begonia in Brazil. Revista Brasileira de Horticultura Ornamental. 2006;12:134–137. [Google Scholar]
- Perez A, Fernandez E. New hosts of Aphelenchoides besseyi (Christie, 1942) in Cuba. Fitosanidad. 2004;8:45–46. [Google Scholar]
- Powers TO, Todd TC, Burnell AM, Murray PCB, Fleming CC, Szalanski AL, Adams BA, Harris TS. The rDNA internal transcribed spacer region as a taxonomic marker for nematodes. Journal of Nematology. 1997;29:441–450. [PMC free article] [PubMed] [Google Scholar]
- Qui JJ, Westerdahl BB, Anderson C, Williamson VM. Sensitive PCR detection of Meloidogyne arenaria, M. incognita, and M. javanica extracted from soil. Journal of Nematology. 2006;38:434–441. [PMC free article] [PubMed] [Google Scholar]
- Richardson PN, Grewal PS. Nematode pests of glasshouse crops and mushrooms. In: Evans K, Trudgill DL, Webster JM, editors. Plant parasitic nematodes in temperate agriculture. Wallingford: CAB International; 1993. pp. 501–544. [Google Scholar]
- Ristaino JB, Madritch M, Trout CL, Parra G. PCR amplification of ribosomal DNA for species identification in the plant pathogen genus Phytophthora . Applied and Environmental Microbiology. 1998;64:948–954. doi: 10.1128/aem.64.3.948-954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. New Jersey: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sanwal KC. A key to the species of the nematode genus Aphelenchoides Fischer, 1894. Canadian Journal of Zoology. 1961;39:143–148. [Google Scholar]
- Southey JF. Nematode pests of ornamental and bulb crops. In: Evans K, Trudgill DL, Webster JM, editors. Plant Parasitic Nematodes in Temperate Agriculture. Wallingford: CAB International; 1993. pp. 463–500. [Google Scholar]
- Stokes DE. Florida Department of Agriculture and Consumer Services, Division of Plant Industry. Nematology Circular 6; 1966. A foliar nematode problem of birds-nest fern. [Google Scholar]
- Subbotin SA, Peng D, Moens M. A rapid method for the identification of the soybean cyst nematode Heterodera glycines using duplex PCR. Nematology. 2001;3:365–371. [Google Scholar]
- Szalanski AL, Sui DD, Harris TS, Powers TO. Identification of cyst nematodes of agronomic and regulatory concern by PCR-RFLP of ITS1. Journal of Nematology. 1997;29:255–267. [PMC free article] [PubMed] [Google Scholar]
- Tanha Maafi Z, Subbotin SA, Moens M. Molecular identification of cyst-forming nematodes (Heteroderidae) from Iran and a phylogeny based on the ITS sequences of rDNA. Nematology. 2003;5:99–111. [Google Scholar]
- USDA National Agricultural Statistics Service. Floriculture Crops 2005 Survey. [16 September 2007];2006 http:www.nass.usda.gov.
- Volvas N, Minuto A, Garibaldi A, Troccoli A, Lamberti F. Identification and histopathology of the foliar nematode Aphelenchoides ritzemabosi (Nematoda: Aphelenchoididae) on basil in Italy. Nematology. 2005;7:301–308. [Google Scholar]
- Vrain TC, Wakarchuk DA, Levesque AC, Hamilton RI. Intraspecific rDNA restriction fragment length polymorphism the Xiphinema americanum group. Fundamental and Applied Nematology. 1992;15:563–573. [Google Scholar]
- Wang H, Qi M, Cutler AJ. A simple method of preparing plant samples for PCR. Nucleic Acids Research. 1993;21:4153–4154. doi: 10.1093/nar/21.17.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bosselut N, Castagnone C, Voisin R, Abad P, Esmenjaud D. Multiplex polymerase chain reaction identification of single individuals of the Longidorid nematodes Xiphinema index, X. diversicaudatum, X. vuittenezi, and X. italiae using specific primers from ribosomal genes. Phytopathology. 2003;93:160–166. doi: 10.1094/PHYTO.2003.93.2.160. [DOI] [PubMed] [Google Scholar]
- Warfield CY, Dudley JB, Hight PA. Evaluation of chemical and cultural methods for management of foliar nematodes on woody ornamental crops in nurseries. Southern Nursery Association Research Conference. 2004a;49:208–211. [Google Scholar]
- Warfield CY, Parra GR. Fungicide and Nematicide Tests 58:OT021; 2003. Evaluation of pesticides for control of foliar nematodes on lantana, 2002. [Google Scholar]
- Warfield CY, Parra GR, Dudley JB. Fungicide and Nematicide Tests 59:OT003; 2004b. Efficacy of miticides and a disinfestant for foliar nematode control on lantana and buddleia, 2003. [Google Scholar]
- Warfield CY, Parra GR, Hight PA. Fungicide and Nematicide Tests 59:OT001; 2004c. Evaluation of miticides and a surface disinfestant for control of foliar nematodes on abelia, 2003. [Google Scholar]
- Werner O, Ros RM, Guerra J. Direct amplification of NaOH extraction: Two rapid and simple methods for preparing bryophyte DNAfor polymerase chain reaction (PCR) Journal of Bryology. 2002;24:127–131. [Google Scholar]
- Windham AS, Conlon HP, Windham M. Foliar nematodes and perennial plants: The silent epidemic. Southern Nursery Research Association Conference. 2005;50:282–284. [Google Scholar]
- Zijlstra C, Lever AEM, Uenk BC, Van Silfhout CH. Differences between ITS regions of isolates of root-knot nematodes Meloidogyne hapla and M. chitwoodi . Phytopathology. 1995;85:1231–1237. [Google Scholar]