Abstract
Fluorescence recovery after photobleaching has been an established technique of quantifying the mobility of molecular species in cells and cell membranes for more than 30 years. However, under nonideal experimental conditions, the current methods of analysis still suffer from occasional problems; for example, when the signal/noise ratio is low, when there are temporal fluctuations in the illumination, or when there is bleaching during the recovery process. We here present a method of analysis that overcomes these problems, yielding accurate results even under nonideal experimental conditions. The method is based on circular averaging of each image, followed by spatial frequency analysis of the averaged radial data, and requires no prior knowledge of the shape of the bleached area. The method was validated using both simulated and experimental fluorescence recovery after photobleaching data, illustrating that the diffusion coefficient of a single diffusing component can be determined to within ∼1%, even for small signal levels (100 photon counts), and that at typical signal levels (5000 photon counts) a system with two diffusion coefficients can be analyzed with <10% error.
INTRODUCTION
Due to the small length scales, diffusion is the dominating transport mechanism both within cells and in the surrounding cell membrane (1). Diffusion analysis has therefore become essential to characterize the mobility of molecules in these systems (2–5). Fluorescence recovery after photobleaching (FRAP) is probably the most well-established method of characterizing the diffusion of fluorescently labeled molecules in liquid crystalline membranes (4–7). In this method a small spatially confined area of the fluorescent molecules, fluorophores, is bleached by a geometrically confined, high-intensity light pulse. The recovery of the bleached area is subsequently studied as a function of time, yielding information about the mobility and rate of diffusion of the studied molecular components. Several different methods of analyzing FRAP data have been suggested since the introduction of the technique in the 1970s (8–10).
In a pioneering article, Axelrod et al. (8) monitored the total change in fluorescence over the initially bleached area due to the influx of fluorescently labeled molecules. By using an analytical expression for the recovery of the fluorescently-labeled molecules, assuming a Gaussian (8) or circular (11) profile of the illumination/bleaching profile, they obtained an effective diffusion coefficient describing the area studied. Later, Gordon et al. (12) refined the fitting algorithm to extract two diffusing components from the data. However, two-component recovery required more than a 10-fold increase in the signal/noise ratio (SNR) to give the same accuracy in the determined values as from single component recovery (12). Thus, the accurate determination of multiple diffusion coefficients requires a high SNR in the FRAP data. These methods also require that the initially bleached area has a certain shape, which must be carefully characterized to yield accurate values of the diffusion coefficients (6,8). Furthermore, when monitoring only the recovery of the total fluorescence intensity from the bleached area, no information is provided about the actual distribution of the fluorescently labeled molecules inside the studied area. This will, in turn, exclude information contained in the recovery profiles of the images (13,14). Temporal and spatial variations in the emitted intensity, due to drift, bleaching during the recovery process, or uneven illumination of the image, for example, can also lead to erroneous values of the estimated diffusion coefficient, which cannot easily be corrected-for when only the total intensity during recovery is monitored (13,15).
In the mid-1980s, different groups started to use video cameras to measure FRAP, thus obtaining spatial information on the bleached spot and its surroundings as a function of time (16,17). This opened up new possibilities with respect to data analysis. It also made it possible to discern anisotropic from isotropic diffusion (16). Different approaches of describing the two-dimensional fluorescence intensity profiles obtained in these experiments have been proposed. Kapitza et al. (16) used the intensity of the central part of the bleached spot to determine the effective lateral diffusion coefficient of the fluorescently labeled molecules. However, later methods have been proposed that use more of the spatial information contained in the images, by fitting a curve to the spatial data in each image (13,15,18). The main shortcoming of these curve-fitting approaches is that the bleaching profile must follow the function used for the curve fits throughout the recovery process (19).
One method that avoids the problem of assuming a certain bleaching profile is based on a Fourier transform of the acquired images (14,20). However, when evaluating experimental data, Tsay and Jacobson (14) noticed that their method was more sensitive to noise than traditional methods of FRAP analysis (see above) employing the total intensity of the bleached spot to characterize the diffusion. Furthermore, since the Fourier transforms are performed on the acquired images only, not on the entire system, the analyzed data must be restricted in time (18). The reason is that in the Fourier transform method, it is assumed that the intensity outside the image does not change during recovery. This is a correct assumption provided that the bleached spot is completely contained within the images, but is not valid when the bleached spot has diffused out to the edge of the image. As a consequence, the Fourier transform method is limited to short times or large fields of view. This, in turn, can lead to severe limitations, especially in the analysis of systems with multiple, i.e., fast- and slow-diffusing, components.
In this article, we present an improved method of FRAP analysis, which efficiently overcomes some of the critical limitations of the currently available methods of analysis. The method is an alternative version of the Fourier transform method, but is based on spatial frequency analysis of circularly averaged radial profiles instead of the entire image. The one-dimensional radial data are also compensated for a net influx of fluorescently labeled molecules from outside the field of view, as well as for temporal variations in illumination during the recovery process. Taken together, these steps were shown to result in a highly accurate and robust method which efficiently suppresses noise in the data.
To validate the accuracy and sensitivity of the method, data were analyzed for various simulated and experimentally obtained series of FRAP images. FRAP data were simulated with the program Comsol Multiphysics 3.4, which uses the finite element method to simulate the time evolution resulting from the diffusion of an initial concentration of molecules. Poisson-distributed noise was added to the simulated data to resemble typical experimental conditions. The effect of different SNRs was investigated, as well as deviations from circular symmetry of the bleached area. Experimental data for the diffusion of different components anchored or inserted into supported phospholipid bilayers (SPBs) on glass were obtained using a conventional FRAP setup. SPBs have gained an important role as model systems for cell membranes (21,22), and diffusion analysis of various SPB systems is currently under extensive study by us and others (23–25). As a special case, we investigated the diffusion of 35-nm lipid vesicles tethered to a SPB via cholesterol-tagged DNA. In addition to being a system with an exciting potential for bioanalysis of transmembrane proteins (23,26,27), it also constitutes a system with two simultaneously diffusing and identically fluorescently labeled components: the tethered vesicles and the lipids in the bilayer. Since a low level of noise has been shown to be vital for the successful analysis of two diffusing components (12), this was a suitable system for a comparison between the new and previously used methods.
THEORETICAL BASIS OF THE ANALYSIS
For a plane, isotropic system, the recovery of fluorescently-labeled molecules after photobleaching can be modeled with Fick's second law (28),
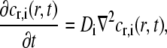 |
(1) |
where r is the radial distance from the center of the bleached spot, t the time, and Di the characteristic diffusion coefficient of the ith diffusing component. The variable cr,i(r,t) is the relative concentration, cr,i(r,t) = ci(r,t)/ci,eq, where ci is the concentration and ci,eq is the equilibrium concentration of unbleached, fluorescently labeled molecules of the ith type. In Eq. 1, it has been assumed that the initial concentration of fluorescently labeled molecules after photobleaching has circular symmetry. Furthermore, the influence of convection and bleaching while monitoring the recovery process is assumed to be negligible. The local diffusion coefficient Di, in Eq. 1, is also assumed to be isotropic. This is not always the case, since the local diffusion coefficient can also show anisotropic behavior (2). However, many of the lateral transport modes in cell membranes have been found to exhibit isotropic behavior (2,16), which would be the overall situation in a nonordered system. The extension to anisotropic diffusion is therefore not considered in this work.
An analytical solution to Eq. 1 can only be obtained for some special cases, such as when the bleached spot has a Gaussian intensity profile. However, in the general case, the Hankel transform can be used to determine the solution to Eq. 1, where a system much larger than the initial dimensions of the bleached spot is assumed. A Hankel transform of Eq. 1 shows that the transform fi(k,t) has the following general dependence on t and the spatial frequency k,
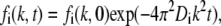 |
(2) |
where fi(k,t) at the spatial frequency k is defined as (29,30)
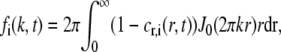 |
(3) |
where J0 is the 0th order Bessel function. The integral in Eq. 3 contains the expression 1-cr,i(r,t) instead of cr,i(r,t) to get a converging integral. The Hankel transform fi(k,t) will thus contain information about the spatial distribution of the fluorescently labeled molecules, where small and large values of k indicate how the concentration changes over large and small distances, respectively.
The relative concentration cr,i(r,t) in a FRAP experiment can be related to the profile of the fluorescent light intensity emitted by the ith component. This means that the Hankel transform fi(k,t) can be determined from a FRAP image when there is only one type of diffusing component present in the system. The diffusion coefficient Di is then given by Eq. 2. For a system with different diffusing components, the profile of the fluorescent light intensity will instead be the sum of the intensities of all the fluorescently labeled molecules in the system. In this case, Eq. 2 can be rewritten as
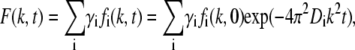 |
(4) |
where the parameter γi is the fraction of the total fluorescent light intensity emitted from the ith component before bleaching, Ipre,i:
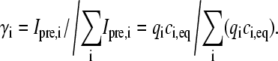 |
(5) |
The parameter qi is a proportionality constant between the concentration ci(r,t) and the intensity of the fluorescent light emitted by the ith component. Equation 4 is the basis of the FRAP analysis presented in this work.
MATERIALS AND METHODS
Simulated FRAP data
Equation 1 was used to simulate images typically obtained from a FRAP experiment. The program Comsol Multiphysics 3.4 (Comsol AB, Stockholm, Sweden), which is based on the finite element method, was used to solve Eq. 1. This allows the recovery of an artificially bleached spot to be simulated. Only one diffusing component was included in each simulation. In cases where multiple diffusion coefficients were determined, the solutions for each different component were superimposed to yield the total concentration profiles, assuming that the different types of diffusing components did not interact with each other. The value at t = 0 was set to: c(r,0) = ceqexp(−K × exp(−r2/w2)) for all simulations, except when analyzing deviations from circular symmetry. This expression describes the amount of fluorescently-labeled molecules after bleaching with a Gaussian-shaped light intensity distribution (8). It was further assumed in all simulations that ceq = 1, K = 2, and w = 5 μm.
The simulated area was chosen to be a square with the dimensions 300 × 300 μm, with the center of the square situated at (x,y) = (0,0). The net flux of fluorescently labeled molecules over the edges of the simulated area was assumed to be negligible, which means that the boundary condition chosen was c = ceq. The solution of Eq. 1 was computed at a time interval of 2 s from 0 to 78 s, where the maximum time was chosen to ensure that c = ceq at the boundaries of the simulated system. The simulated values of the recovery were then scaled up with a factor Ipre, after which Poisson-distributed noise was added. The value of Ipre was chosen to be 5000 unless otherwise stated. This value is comparable to the fluorescence intensity (in photon counts) measured with the charge-coupled device (CCD) camera during a typical experiment in this study.
The images were transformed into a 16-bit image sequence consisting of 512 × 512 pixels to mimic experimentally acquired data. The distance between two pixels was set to 0.267 μm, equal to the pixel size in the CCD camera used in the experimental part, and the images were centered on the bleached area. This was done with a script written in MATLAB 2007b (The MathWorks, Natick, MA) and the images were stored as 32-bit .tif files.
Experimental procedure
The experiments were divided into two parts. In the first part the diffusion of different types of fluorescently labeled molecules in a SPB was studied. All these experiments were carried out with an egg yolk phosphatidylcholine (egg PC) bilayer with a single type of diffusing species: either lissamine rhodamine B 1,2-dihexadecanoyl-sn-glycero-3-phosphatidylethanolamine (rhodamine-DHPE) or 2-(12-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)dodecanoyl-1-hexadecanoyl-sn-glycero-3-phosphocholine (NBD C12-HPC) lipids incorporated into the SPB. The diffusion coefficients of both these systems have been thoroughly studied previously (31–34). In the second part, a system consisting of two diffusing components was studied: 35 nm (diameter) lipid vesicles, tethered to the SPB as described previously (23), using cholesterol-DNA anchors, and individually labeled lipids in the SPB. Both the lipids in the vesicles and in the bilayer were labeled with rhodamine. The FRAP setup for the experiments was based on an inverted Eclipse TE2000-U microscope (Nikon, Melville, NY) with an iXon EMCCD camera (Andor Technology, Belfast, Northern Ireland) for the recording of the images. Each image consisted of 512 × 512 pixels with a pixel size of 0.267 × 0.267 μm in all experiments. A Kr-Ar mixed gas ion laser was used for bleaching, while the recovery was monitored with a super-high-pressure mercury lamp. The width of the bleached spot, which had a Gaussian profile, was in the range 10–20 μm for the different experiments.
Vesicle preparation
Egg PC from Avanti Polar Lipids (Alabaster, AL) was dissolved in methanol (VWR International, Stockholm, Sweden) to a lipid concentration of 25 mg/mL. A lipid mixture consisting of 99 wt % egg PC and 1 wt % rhodamine-DHPE from Invitrogen (Carlsbad, CA) was subsequently prepared with a total lipid mass of 5 mg/mL. The solvent was evaporated under a flow of nitrogen for 1 h. A mixture of 100 mM NaCl, 10 mM tris[hydroxymethyl]aminomethane (TRIS), and 1 mM ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA) from Sigma-Aldrich (St. Louis, MO) with a pH of 8.0 was used as buffer during the experiments, unless otherwise stated. One mL of buffer was added to the lipids and the mixture was passed 11 times through a membrane with a pore size of 100 nm (Whatman, Maidstone, UK) via a Mini-Extruder (Avanti Polar Lipids). The extrusion procedure was repeated with a membrane with a pore size of 30 nm (Whatman), yielding vesicles with an estimated mean diameter of 35 nm (23). Following the same procedure as described above, 35-nm vesicles consisting of 98 wt % egg PC and 2 wt % NBD C12-HPC (Invitrogen) were also made at a total lipid concentration of 5 mg/mL.
Incorporation of cholesterol-tagged DNA
For the experiments with tethered lipid vesicles, cholesterol-tagged DNA (cDNA) was incorporated into the 35-nm diameter lipid vesicles to anchor the vesicles to the SPB. Double-cholesterol DNA was used to firmly tether the cDNA to the lipid vesicles and the SPB (23). To accomplish this, two 30-mer cDNA strands (cDNAA: 5′-TGG-ACA-TCA-GAA-ATA-AGG-CAC-GAC-GGA-CCC-cholesterol-3′ and cDNAC: 5′-TAT-TTC-TGA-TGT-CCA-AGC-CAC-GAG-TTC-CCC-cholesterol-3′) were hybridized separately with shorter 15-mer cDNA strands (cDNAB: 5′-cholesterol-CCC-TCC-GTC-GTG-CCT-3′ and cDNAD: 5′-cholesterol-CCC-GAA-CTC-GTG-GCT-3′) to produce double-cholesterol anchors with an overhang of 15 basepairs. All cDNA was obtained from MedProbe (Oslo, Norway). The overhang from cDNAA+B is complementary to the overhang from cDNAC+D and can subsequently hybridize to produce a strong bond. The hybridization of both cDNAA+B and cDNAC+D was performed at a ratio of 5:6 (30-mer/15-mer), to ensure that all functional anchors are double-cholesterol anchors. The concentration of hybridized cDNA was adjusted to 5 μM with TE buffer (10 mM TRIS, 1 mM EDTA, pH = 8.0). cDNAA+B was incorporated into the vesicles at a molar ratio of 1:2 (23.8 nM cDNAA+B/47.6 nM vesicles) for 30 min. The following parameters were assumed in calculating the lipid vesicle concentration of 35 nm vesicles from the concentration of lipids (clipid = 0.4 mg/ml): mlipid = 760.09 g/mol, Alipid = 0.7 nm2, and hSPB = 5 nm, where mlipid is the molar mass of a lipid, Alipid the cross-sectional area of a lipid, and hSPB the thickness of a lipid bilayer (23). cDNAC+D at a concentration of 60 nM was in turn incorporated into the bilayer for a period of 30 min before rinsing with buffer and adding the vesicles anchored with cDNAA+B.
Bilayer formation on glass
The SPBs were formed on glass slides (25 mm diameter and 0.13–0.16 mm in thickness, Menzel-Gläser, Braunschweig, Germany) which had been cleaned with piranha (3:1 sulfuric acid/30% hydrogen peroxide) (VWR International) for 15 min and then thoroughly rinsed with deionized Milli-Q water (Millipore, Billerica, MA). The cleaned glass slide was placed in a custom-made flow cell, with a volume of 350 μL. A prepared solution of unilamellar vesicles was diluted with buffer to a lipid concentration of 100 μg/mL and added to the flow cell. The flow cell was encapsulated in aluminum foil to prevent bleaching, and the bilayer was allowed to form on the glass slides for a period of 30 min to 1 h. The solution in the flow cell was then replaced with buffer.
Microscopy setup
The flow cell was mounted on an Eclipse TE2000-U microscope (Nikon) and a 60× magnification (NA = 1.00) water immersion objective (Nikon) was used for observation. The measurements were made at ambient temperature, 20–25°C. The fluorescence images were collected with an iXon 512 × 512 pixel EMCCD camera (Andor Technology) cooled to −70°C during the recordings. With the 60× magnification objective, the camera pixel size was 0.267 × 0.267 μm. A TRITC/FITC filter cube (Nikon) was used to filter out the excitation light and the emission light from the experiments with rhodamine/NBD fluorophores. A Kr-Ar mixed gas ion laser (Stabilite 2018, Spectra-Physics Lasers, Mountain View, CA) was used at wavelengths of 531 nm/488 nm to bleach the rhodamine/NBD fluorophores, while the recovery of fluorescently labeled molecules was monitored with a super-high-pressure mercury lamp (Nikon). The illumination profile of the lamp was adjusted to be as homogeneous as possible over the entire field of view and symmetric around the center of the image. Images were acquired at 2-s intervals and the exposure time was set to 0.1 s. The opening and closing of the lamp was controlled by a shutter (Ludl, Hawthorne, NY) to minimize bleaching between the acquisitions of images. The shutter open time was set to 225 ms, with the camera exposure in the center of this interval to avoid missing frames. A series of prebleaching images was acquired to correct for nonuniform illumination and an image was also taken with the light source turned off to correct for dark counts in the CCD camera. The area was then bleached with the laser, adjusted to produce a centered circularly symmetric bleached spot with suitable dimensions and an approximate Gaussian shape. Recovery was then monitored by switching to the mercury lamp. The images were saved and converted into 32-bit .tif files. FRAP data from different regions of the SPB was acquired to improve statistics.
Description of the FRAP analysis
A summary of the different steps used in the Hankel transform method for the analysis of FRAP data is presented below. All steps were implemented in MATLAB 2007b.
To eliminate the effects of an uneven illumination profile, Iip(x,y), the relative fluorescent intensity, Ir(x,y,t), was defined as
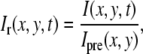 |
(6) |
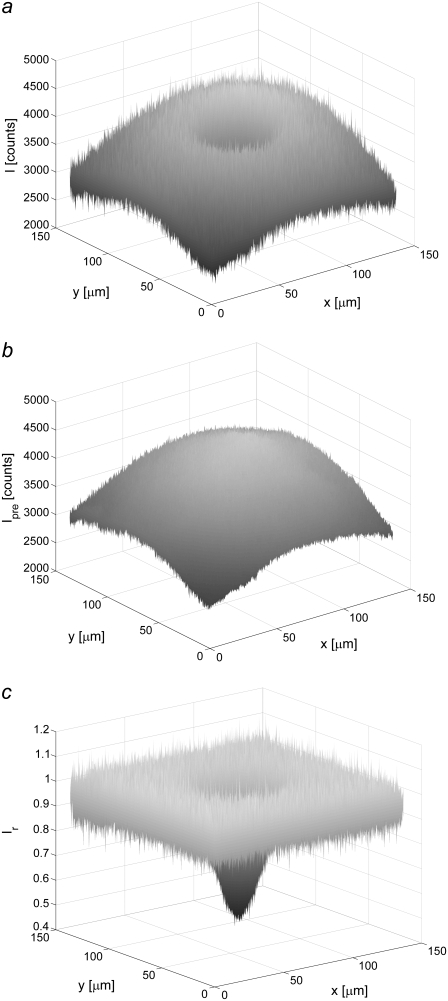
where I(x,y,t) is the fluorescent intensity detected by the CCD camera at the position (x,y) a time t after bleaching (see Fig. 1 a), and where Ipre(x,y) is the fluorescent intensity before bleaching (see Fig. 1 b). It is assumed that the dark counts arising from the detector and the intensity due to ambient light have been subtracted from both I(x,y,t) and Ipre(x,y). A typical profile of Ir(x,y,t) is shown in Fig. 1 c, which, in contrast to the raw data shown in Fig. 1, a and b, displays a flat background surrounding the bleached area. The relation between the relative intensity, Ir(r,t), and the relative concentration of labeled fluorescent molecules, cr,i(r,t), is assumed to be represented by
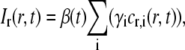 |
(7) |
where γi is the intensity fraction of the ith component (see Eq. 5). β(t) is a measure of the total intensity in the studied system after photobleaching and does not depend on the diffusive properties of the various components. Parameters influencing β(t) are instead temporal intensity variations such as drift, bleaching, and fluctuations in the light illumination (see below). In Eq. 7 it has further been assumed that all diffusing components are labeled with the same type of fluorophores, thus making β(t) identical for all diffusing components.
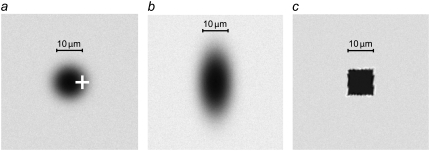
FIGURE 1.
Images illustrating the various steps used in the compensation of uneven illumination in the imported FRAP images. (a) Image of the intensity with a bleached spot, (b) image taken before bleaching, and (c) a compensated version of the bleached image, Ir(x,y,t), defined in Eq. 6.
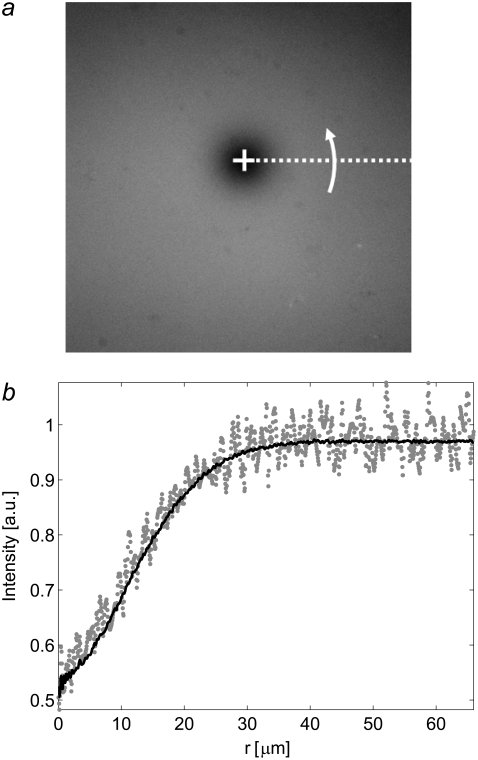
The noise in the intensity profiles was reduced by circular averaging of the data around the center of the bleached spot, as shown in Fig. 2 a. First the center of mass of Ir(x,y,t) within an encircled region containing the bleached spot was determined. The angular averaging of Ir(x,y,t), yielding Ir(r,t), was then performed for each image in the interval 0 < r < R (see Fig. 2 b), where R is the shortest distance between the center of mass and the edge of the image. The radial values were chosen such that they could be directly evaluated with the numerical Hankel transform (30).
FIGURE 2.
(a) The center of mass of the bleached spot (open cross). The intensity at each radial value was obtained by averaging the radial values obtained by revolving the dashed line in the figure. (b) Comparison between the intensity, Ir(x,y,t), along a line from the center and out to edge of the image (shaded dots), with an averaged radial profile, Ir(r,t), calculated by assuming circular symmetry around the center of the bleached spot (solid line).
As shown below, using the angular averaged data Ir(r,t) instead of the two-dimensional data Ir(x,y,t) provides a significant advantage in terms of compensating for nonidealities in the images. Circular median averaging could also be used to eliminate strongly scattering objects or intensity spikes in the images.
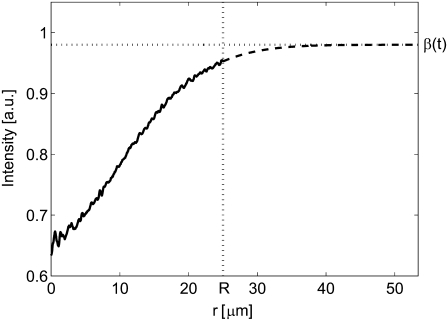
Since the Hankel transform is defined over the interval 0 < r < ∞ (see Eq. 3), the shape of Ir(r,t) outside the field of view must be estimated. The simplest method is to assume that cr,i equals unity for r > R, which is valid when there is no net influx of fluorescently labeled molecules at the edge of the image. This assumption is valid at short times after bleaching, but fails to represent the experimental conditions at longer times. A more generic approach is to fit the tail of Ir(r,t) to a Gaussian curve according to Eq. 8 (see also Fig. 3), where it is assumed that the net influx of molecules over the edge of the image is dominated by a single diffusing component,
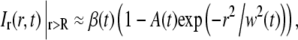 |
(8) |
where A(t) and w(t) are fitting parameters. The expression in Eq. 8 will be valid as long as the net influx of fluorescently labeled molecules into the field of view is dominated by a single diffusing component. Note that for a two-component system with a slow and a fast diffusing component this expression will be a reasonably good approximation for all times. The reason for this is that at short times the fast component dominates, whereas at longer times the fast component has reached equilibrium and the slow component will then dominate the influx of molecules into the field of view. The Gaussian fit to the concentration is a good approximation if the distance R is much greater than the width of the bleached region in the first frame after photobleaching. The solution to Eq. 1, for a single diffusing molecule, will then approach the solution obtained from a point source at t = 0, which has a Gaussian shape. Furthermore, the bleached spot in our experiments could, to a good approximation, be described by a Gaussian profile, which further motivates the use of the expression for Ir(r,t) at r > R described by Eq. 8.
FIGURE 3.
The intensity Ir(r,t) after a certain time, t, in the interval 0 < r < R (solid line). The dashed line at r > R is a Gaussian curve fit to the tail of Ir(r,t), according to Eq. 8.
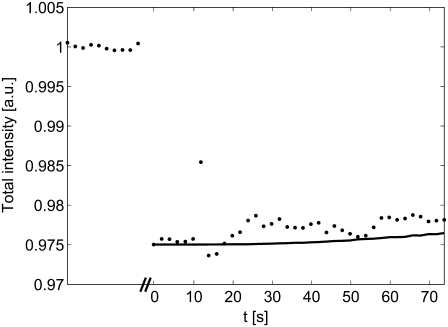
Note that both Eqs. 7 and 8 contain the term β(t). If there is no temporal intensity variation in the images, β(t) equals one. However, the temporal variations may, in reality, be nonnegligible compared to the recovery of the fluorescently labeled molecules (see Fig. 4). Since the total amount of fluorescent molecules in the sample must be constant, it can be shown that the term β(t) can be expressed as in Eq. 9 (see the Appendix for details),
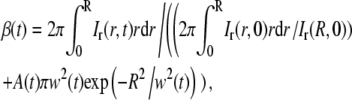 |
(9) |
where Ir(R,0) is the relative intensity at the edge of the field of view at t = 0 s. Furthermore, from Eq. 9 it is seen that β(t) can be described as a function of A(t) and w(t). It is thus sufficient to use the two parameters A(t) and w(t) when fitting the tail of Ir(r,t) to Eq. 8. The values of A(t) and w(t) can then be used, together with Eq. 9, to estimate β(t).
FIGURE 4.
Illustration of typical intensity variations before and after bleaching (t = 0 s), where the total intensity is the sum over the entire image (the data during bleaching have been omitted). The solid line is the total intensity versus time after compensating for temporal variations using Eq. 9. The slow increase in the total intensity is due to a net influx of molecules from outside of the field of view.
From Eq. 7, the relative concentration of fluorescently labeled molecules can be written as
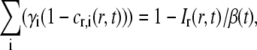 |
(10) |
where β(t) is given by Eq. 9. According to Eqs. 3 and 4, the Hankel transform of the left-hand side of Eq. 10 equals F(k,t). The Hankel transform of the right-hand side of Eq. 10 will then yield the relation between the measured quantity Ir(r,t) and F(k,t),
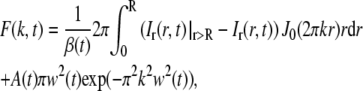 |
(11) |
where J0 is the 0th order Bessel function and Ir(r,t)|r>R is defined as in Eq. 8 (for all radial distances, 0 < r <∞). Ir(r,t)|r>R has been introduced into Eq. 11 since Ir(r,t) is only determined in the interval r < R, while the Hankel transform in Eq. 3 is defined over the entire interval 0 < r < ∞. However, since Ir(r,t)|r>R = Ir(r,t), when r > R, the right-hand side of Eq. 11 corresponds to the Hankel transform of the right-hand side of Eq. 10. The Hankel transform was determined using a numerical method developed by Guizar-Sicairos and Gutierrez-Vega (30).
Equation 4 relates the individual values of Di and γi to the measured value of F(k,t) defined in Eq. 11, but is given here for a system with two diffusing components with an immobile fraction of molecules, γ0:
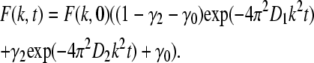 |
(12) |
In Eq. 12, it is assumed that fi(k,0) = F(k,0) for all components. However, if the different components also bleach differently, the latter condition is not necessarily valid. It can, however, be argued, that since the bleaching profile is expected to be proportional to the light intensity for moderate bleaching, then fi(k,0) ≈ AiF(k,0), where Ai is a constant describing the bleaching of the ith component. Hence, Eq. 4 will still be valid, but now with γi replaced by γiAi. However, this will only influence the determined values of γi, not Di.
From Eq. 12 it can be seen that for each spatial frequency, k, F(k,t) can be described as a double time-dependent exponential function with the unknown exponent 4π2k2Di and the prefactors γ2 (intensity fraction) and γ0 (immobile fraction). With γ2 and γ0 assumed independent of k, a fitting algorithm can be used to determine Di separately for each value of k, thus yielding the dependence of the diffusion coefficients on the spatial frequency, Di(k). The values of Di(k) may also depend on the fraction of immobile molecules, which acts as stationary obstacles in the path of the diffusing molecules. Thus Di(k) will, implicitly in this work, also depend on γ0. In reality, the range of k values must be restricted to k < kmax, where kmax is chosen such that the Hankel transform at k < kmax is not critically affected by noise. However, at larger values of k the magnitude of F(k,t) will be small and may therefore be dominated by noise. Note further, that for a system characterized by Brownian diffusion, Di(k) is expected to be independent of k. Hence, to further improve the accuracy of the fitted parameters, and to obtain an effective value of Di, curve fits of F(k,t) versus time were made simultaneously for all values of k < kmax. In these curve fits, the amplitude of F(k,0) was free to vary, while the values of D1, D2, γ2, and γ0 were chosen so as to be independent of k. All curves were fitted using a nonlinear curve fitting routine in MATLAB.
RESULTS AND DISCUSSION
We start with an illustration of how the various steps in the analysis (radial averaging, compensation for temporal variations, and a net influx of fluorescently labeled molecules into the field of view) influence the determined value of the diffusion coefficient. For this purpose we will investigate the recovery of a single diffusing component with a diffusion coefficient D = 2.5 μm2 s−1, which is a representative value for fluorescently labeled lipids in an egg PC bilayer (31–34). Furthermore, the concentration at t = 0 is assumed to be
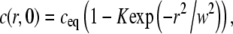 |
(13) |
where K = 0.5 and w = 10 μm. The concentration profile in Eq. 13 corresponds approximately to the experimental situation for moderate bleaching with a Gaussian light source. The solution to Fick's second law (see Eq. 1) for the initial concentration in Eq. 13 can be shown to be
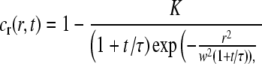 |
(14) |
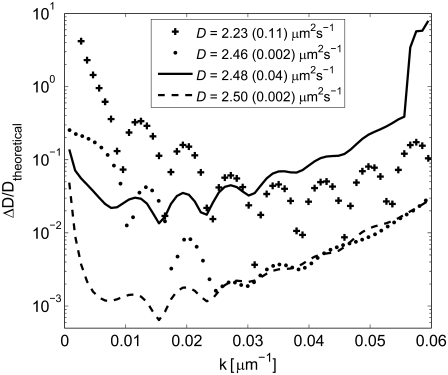
where τ = w2/(4D), cr(r,t) = c(r,t)/ceq, and D = Dtheoretical = 2.5 μm2 s−1. If there are no immobile molecules, then the intensity I(r,t) will be proportional to cr(r,t), with a proportionality constant q. To mimic a typical FRAP experiment, the data were transformed into a 512 × 512 pixel image, with a pixel size of 0.267 × 0.267 μm. The proportionality constant q was set to 5000, yielding an SNR of 71 in the data when Poisson-distributed noise was added to the pixels in each image. This is a representative value for a typical experimental situation (see below). The data in each image were also subjected to random temporal fluctuations with a magnitude of 1% of the total intensity. The fluorescently-labeled molecules were assumed to be bleached by 1% during the recovery process, which was monitored in the time interval from 0 to 198 s in steps of 2 s. The results obtained for the different steps are shown in Fig. 5, which displays the relative error in the determined diffusion coefficient, ΔD/Dtheoretical, at different spatial frequencies k. The inset in Fig. 5 shows the effective value of the diffusion coefficient, obtained by including all values of k up to kmax (= 0.06 μm−1) in the curve fits.
FIGURE 5.
Curves showing the relative error in D versus k after the different compensation steps in the FRAP analysis, determined from 50 separate simulations for each case. The values depicted by crosses (+) were obtained when only radial averaging of Ir(x,y,t) was performed. The values depicted by dots (•) have in addition been corrected for temporal variations in β(t), but not for a net influx of molecules into the field of view. The values depicted by solid and dashed lines were both corrected for temporal variations and a net influx of molecules into the field of view without and with radial averaging, respectively. The effective mean values of D for the different cases are presented in the inset, where the values in parenthesis are the standard deviations of D determined from 50 simulations.
As can be seen in Fig. 5, the relative error gradually decreases after each step in the analysis. When the data were not corrected for temporal variations in the intensity (crosses), the accuracy of the analysis was relatively poor, with an effective value of D being >20% lower than the theoretically correct value (see inset in Fig. 5). Also note that, in this case, the error dependence of D on k displays an oscillatory behavior. The error was observed to be smallest at values of k corresponding to 2πkR = α1, where J1(α1) = 0 and J1 is the first-order Bessel function. This corresponds to points where the Hankel transform of a constant value over the image is zero. The dominating error, when not compensating for temporal variations in the images, can thus be attributed to an erroneous offset in the data. This offset arises since the function 1-Ir(r,t)/β(t) is subjected to the Hankel transform in the generation of F(k,t) (see Eqs. 10 and 11). If the variations in β(t) are not taken into account, which means that 1-Ir(r,t) is used instead of 1-Ir(r,t)/β(t), then this quantity will have an offset equal to 1-β(t) over the entire field of view.
When compensating for the temporal variations in the intensity (dots), the accuracy of the analysis improves significantly for all values of k. The oscillatory behavior observed when not compensating for temporal variations is also significantly reduced. This confirms that the dominating error when temporal variations are present is mainly due to an erroneous offset in 1-Ir(r,t)/β(t)). However, the determined values of D may still be inaccurate due to a net influx of fluorescently labeled molecules from outside the field of view. This is best illustrated in Fig. 5 for small values of k, where the error after compensating for a net influx of molecules (dashed line) is significantly reduced compared to the situation without compensation (dots). The improvement at larger values of k, on the other hand, is negligible. This is attributed to the fact that the magnitude of the Hankel transform rapidly decreases with time at large values of k. Thus, the analysis for large values of k will solely depend on the first few frames, where the influence from a net influx of molecules from outside the field of view is negligible. Note, however, that the overall lower magnitude of the Hankel transform at large values of k makes this region less reliable (see below).
Data were also analyzed without performing radial averaging, but with compensation for temporal variations and a net influx of molecules (solid line). In this case, the radial profile was obtained from a line drawn from the center of the bleached spot to the edge of the field of view of the 512 × 512 pixel image. The resulting error was approximately a factor-of-20 larger than with radial averaging (dashed line) for all values of k. Note also that if temporal variations and a net influx of molecules are not compensated for, the effective value of D will be erroneous (see inset in Fig. 5).
In summary, the most important conclusion that can be drawn from this theoretical evaluation of the Hankel transform method is that the accuracy improves significantly after using radial averaging and the various compensation steps. Values of D(k) that were accurate to within 1% over the main part of the range in Fig. 5 were obtained with an effective value of D, accurate to within 0.1% of the theoretical value.
SIMULATED SYSTEMS
The influence of signal/noise ratio
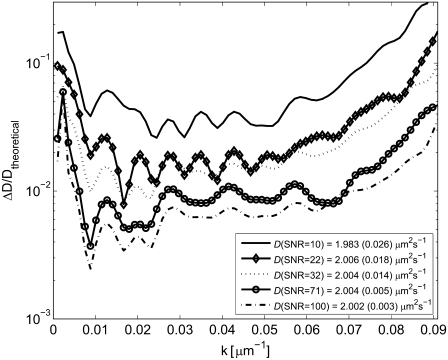
The influence of SNR on the outcome of the analysis was investigated by using finite element simulations of the recovery of fluorescently labeled molecules in a SPB with varying SNRs. The intensity, Ipre, in the simulated images was varied (Ipre = 100, 500, 1000, 5000, and 10,000 photon counts) after which Poisson-distributed noise was added. The SNR is then given by SNR = I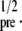 Fig. 6 shows the relative error in the determined diffusion coefficient, ΔD/Dtheoretical, versus k for various SNRs, where the errors are determined from 50 simulations for each SNR. These simulations were carried out for a single diffusing component, where Dtheoretical was chosen to be 2 μm2 s−1 and γ0 = 0. The effective diffusion coefficient was determined using all Hankel transforms with k < 0.06 μm−1.
Fig. 6 shows the relative error in the determined diffusion coefficient, ΔD/Dtheoretical, versus k for various SNRs, where the errors are determined from 50 simulations for each SNR. These simulations were carried out for a single diffusing component, where Dtheoretical was chosen to be 2 μm2 s−1 and γ0 = 0. The effective diffusion coefficient was determined using all Hankel transforms with k < 0.06 μm−1.
FIGURE 6.
The relative error in D, ΔD/Dtheoretical, versus k for different signal/noise ratios (SNRs). The analysis was made on a simulated system with Dtheoretical = 2 μm2 s−1, γ0 = 0, and added Poisson-distributed noise. The values in the inset are effective values of D calculated using all values of k < 0.06 μm−1. The standard deviations from 50 separate simulations are indicated within parentheses.
Fig. 6 shows that the standard error in the determined diffusion coefficient decreases with increasing SNR, but the Hankel transform method still yields accurate values of the diffusion coefficient at low signal levels. For example, the determined effective diffusion coefficient for a photon count of 100 (SNR = 10) is ∼10 times more accurate using the Hankel transform method than previously published results for similar systems using the traditional methods of FRAP analysis (12,14).
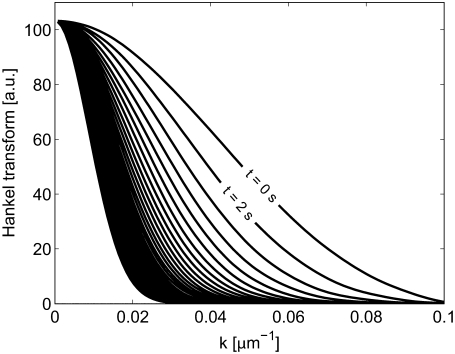
Furthermore, as can be seen in Fig. 6, the determined values of D are most accurate for an intermediate range of k values. The reason for the larger errors at small values of k is the slow temporal variation in the Hankel transform in this regime, thus making the fit more sensitive to noise in the signal (see Fig. 7). Conversely, for large values of k, the magnitude of the Hankel transform will be low and therefore also more susceptible to noise in the analysis (see Fig. 7). It is therefore important not to use data at too high values of k when fitting the Hankel transform to determine an effective diffusion coefficient. However, the slow temporal change in the magnitude of the Hankel transform at small values of k will not influence the accuracy of the analysis when using a range of k values to determine the effective diffusion coefficient. This is due to the fact that the accuracy in the analysis will only depend on the relative error in the Hankel transform at each value of k. The error in the Hankel transform at small values of k is limited due to the integral definition of β(t) in Eq. 9. Thus only an upper limit of k needs to be considered when determining the effective diffusion coefficient. The upper limit was set to kmax = 0.06 μm−1 when determining the effective diffusion coefficient from the simulated FRAP data. Although this limit will depend on the diffusion coefficients in the system, the noise in the images, the size of the bleached spot and the size and number of pixels in the images, it was seen to be a suitable limit for all simulated cases.
FIGURE 7.
The Hankel transform, F(k,t), as a function of k for different times after bleaching: t = 0, 2, 4,…, 78 s. The Hankel transforms are made on the system simulated in Fig. 6 with an SNR = 71.
Analysis of simulated two-component systems
Simulations were also performed for a system with two diffusing components, keeping the SNR fixed at a value corresponding to Ipre = 5000 photon counts (SNR = 71). The effective value of D and the standard deviation obtained from 50 simulations are presented for each simulated system in Table 1. The effective diffusion coefficients were determined using all Hankel transforms with k < 0.06 μm−1.
TABLE 1.
Determined values of the diffusion coefficient from simulated FRAP experiments for a two-component system with D1 = 2 μm2 s−1, γ0 = 0, and different values of D2
| D1,theoretical [μm2 s−1] | D2,theoretical [μm2 s−1] | γ2 | D1 [μm2 s−1] | D2 [μm2 s−1] | γ2 |
|---|---|---|---|---|---|
| 2 | 0 | 0.1* | 2.003 (0.005) | 0.001 (0.002) | 0.1006 (0.001)* |
| 2 | 0.2 | 0.1 | 2.001 (0.007) | 0.1998 (0.008) | 0.1004 (0.002) |
| 2 | 0.2 | 0.5 | 2.001 (0.011) | 0.1998 (0.002) | 0.5000 (0.002) |
| 2 | 0.5 | 0.3 | 1.999 (0.018) | 0.497 (0.011) | 0.2989 (0.008) |
| 2 | 1 | 0.3 | 1.993 (0.060) | 0.977 (0.074) | 0.289 (0.056) |
The intensity fraction between the two components, γ2, was also varied while the SNR was fixed at 71. The standard deviations from 50 separate simulations are indicated within parentheses.
For this case, γ2 corresponds to the fraction of immobile molecules.
The results in Table 1 show that the Hankel transform method can also be used to accurately analyze two-component systems under typical experimental conditions. However, the accuracy with which the diffusion coefficients of the two components can be determined depends on the SNR and on how close D1 is to D2. If the SNR is low and D1 is close to D2, then the ability to distinguish between the two diffusion coefficients decreases. However, note that the two diffusing components could be distinguished and determined to within a couple of percent even if the two diffusion coefficients did not differ from each other by more than a factor of two (D1 = 2 μm2 s−1 and D2 = 1 μm2 s−1). It should be noted that in these simulations it was assumed that the only source of error in the signal was from Poisson-distributed noise. In reality, other errors may also influence the analysis, as discussed previously in connection with Fig. 5 and in connection with the analysis of the experimental data (see below).
The effect of a noncircular bleaching profile
Another potential source of experimental error is variations in the shape of the bleached spot from the assumed circular symmetry. Fig. 8 shows three extreme situations that could occur in reality: 1), the center of mass is chosen erroneously; 2), the bleached spot has the shape of an ellipse, which may occur if the light source is not perpendicular to the sample surface; and 3), the bleached area has the shape of a square, thus representing a situation when a square aperture, rather than a focused laser, is used for photobleaching. The theoretical diffusion coefficient was D = 2 μm2 s−1 with no immobile molecules (γ0 = 0) and with an SNR = 71. The effective diffusion coefficient was determined using the Hankel transform method with k < 0.06 μm −1. The mean value and standard error, from 50 separate simulations, were determined for the different cases, yielding: 1), D = 2.005 μm2 s−1 (0.010 μm2 s−1); 2), D = 2.002 μm2 s−1 (0.002 μm2 s−1); and 3), D = 2.006 μm2 s−1 (0.015 μm2 s−1). Hence, the standard error when determining the diffusion coefficient was <1% of the theoretical value in all three cases and there was no significant difference in the accuracy of D(k) compared with the curve with an SNR = 71 in Fig. 6. This indicates that the error induced in the analysis by a bleached spot that lacks circular symmetry is essentially negligible. Furthermore, an error in the choice of the center of mass had no critical impact on the determined value of D. These results stem from the fact that circular averaging makes the error due to deviations from circular symmetry and an erroneous center of mass appear as a second-order effect in the analysis. If only data along a line through the center of the bleached spot were to be used, the error due to deviations from circular symmetry would appear as a first-order effect.
FIGURE 8.
(a) An erroneous choice of the center of mass (depicted by the open cross), where the error is x0 = w = 5 μm. (b) Bleached area in the shape of an ellipse with an aspect ratio of two: c(x,y,0) = ceqexp(−K × moH exp(−x2/w2 − y2/(2w)2)). (c) Bleached area in the shape of a square: c(x,y,0) = ceqexp(−K) when the absolute values of both x and y is <w, and c(x,y,0) = ceq otherwise. Parameters: ceq = 1, K = 2, and w = 5 μm for all simulations.
Experimental systems
The Hankel transform method for FRAP analysis was evaluated on a number of previously well-characterized and relevant experimental model systems:
Systems with a single diffusing component; either rhodamine-DHPE or NBD C12-HPC lipids in a supported lipid bilayer, or rhodamine-DHPE-modified lipid vesicles tethered to an unmodified supported lipid bilayer.
A two-component system consisting of rhodamine-DHPE-labeled lipid vesicles tethered to a rhodamine-DHPE-labeled supported lipid bilayer.
The Hankel transform method was also compared with the results obtained using two common traditional methods of FRAP analysis. The first method (the Fourier transform method) is based on the method introduced by Tsay and Jacobson (14) and Johnson et al. (35). In this method the relative intensity, 1-Ir(x,y,t), of each image is first subjected to a two-dimensional fast Fourier transform. The fast Fourier transform is calculated at the spatial frequencies (kx,ky) = (nx,ny)/L, where nx and ny are integer numbers different from zero and L is the length of the field of view. This choice of spatial frequencies has the effect that the Fourier transform, over the image, of a constant will be zero, thus minimizing the effects of temporal noise in the data (see the discussion in connection with Fig. 5). The Fourier-transformed data, F(k,t), are then plotted as a function of 4π2k2t and subsequently fitted to a double-exponential function (see Eq. 12), yielding D1, D2, γ2, and γ0.
The second traditional method (the integral method) is a slightly modified variant of the method introduced by Axelrod et al. (8) and Berk et al. (18), in which the total fluorescent intensity, Itot(t), inside a circle of radius w centered on the bleached area, is used to determine the diffusion coefficients of the system. Under the assumption that the intensity after bleaching can be described by a Gaussian function: I(r,0) = Ipre(1−Kexp(−r2/w2)), the following relation between Itot(t) and the diffusion coefficients in a two-component system applies (the derivation of this expression is shown in the Appendix),
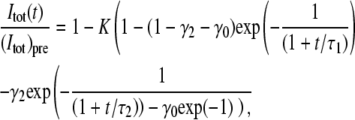 |
(15) |
where (Itot)pre is the value of Itot(t) before bleaching, τ1 = w2/(4D1), τ2 = w2/(4D2), γ2 is the intensity fraction of the second component and γ0 the fraction of immobile molecules. The width w is determined from a Gaussian fit to the first intensity profile after bleaching. Note that neither radial averaging nor compensation of temporal variations is used in the traditional methods of FRAP analysis, even though some of the compensation steps introduced in this study could also be used to improve the accuracy and robustness of these methods.
A single diffusing component
Fig. 9 a compares the frequency transform, F(k,t), of a series of FRAP images using either the Fourier transform or the Hankel transform method.
FIGURE 9.
(a) Comparison between the Fourier transform (shaded circles) and the Hankel transform method (solid dots) when analyzing the diffusion of rhodamine-DHPE in an SPB. Values at k = 0 have been omitted from the Fourier-transformed data due to excessive noise at these values. The inset shows a single-exponential curve fit to the Hankel-transformed data yielding D = 2.86 μm2 s−1 and γ0 = 0.065, where Eq. 12 with γ2 = 0 have been applied for the curve fit. The same fit to the Fourier-transformed data gave D = 2.84 μm2 s−1 and γ0 = 0.061 (fit not shown). (b) The total intensity Itot(t) versus time for the system in panel a, with values uncompensated for temporal variations (dots) and compensated for temporal variations (circles) (see inset for the total intensity on the CCD camera as a function of time). The shaded line is a curve fit to the compensated values of Itot(t) yielding D = 2.60 μm2 s−1 and γ0 = 0.068, where Eq. 15 with γ2 = 0 have been used for the curve fit.
The overall shapes of the curves are similar, yielding DHankel = 2.86 μm2 s−1 and (γ0)Hankel = 0.065 compared to DFourier = 2.84 μm2 s−1 and (γ0)Fourier = 0.061, where Eq. 12 with γ2 = 0 was used to fit the curves. This is comparable to the results obtained by other groups on similar systems (31,33,36), although lipid diffusivity is known to depend strongly on several external parameters such as temperature (34), the lipid composition (35), the fluorescent probe (35), and the ion concentration in the buffer (31). However, there is a clear reduction in noise when using the Hankel transform method, due primarily to the compensation for temporal variations in the data. In particular, the Hankel-transformed data in Fig. 9 a have a five-times-higher SNR than the corresponding Fourier-transformed data, obtained from the standard error in the fitted curves.
Fig. 9 b shows the total intensity Itot(t) used in the integral method, obtained from the same FRAP data as shown in Fig. 9 a. Using values for Itot(t) that were corrected for temporal variations (note that this is not performed in the traditional methods), such as drift and bleaching, the diffusion coefficient obtained and the fraction of immobile molecules were: D = 2.60 μm2 s−1 and γ0 = 0.068, where Eq. 15 with γ2 = 0 was used to fit the curves. Note that although the fit is good (R2 = 0.9997), the values obtained with the integral method are ∼10% lower than the values obtained using the Fourier transform and the Hankel transform methods. This difference is attributed to the fact that the concentration of fluorescent molecules in the first frame after bleaching did not have an exact Gaussian profile. Since Eq. 15 assumes an initial Gaussian concentration of fluorescent molecules, the deviation from a Gaussian profile in the experimental data is expected to influence the outcome of the analysis. As stated by others (8), the concentration profile of unbleached molecules will not have a perfect Gaussian profile after bleaching with a light source with a Gaussian intensity profile, but the approximation of a Gaussian profile will be good for moderate bleaching. However, fitting the first concentration profile after bleaching with the full expression in Axelrod et al. (8) did not yield a noticeably better description of the bleach profile than using a Gaussian curve fit for the experimental data.
Furthermore, it was observed that the integral method was more sensitive to errors in the data when fitting Itot(t), compared to the Fourier and the Hankel transform methods. The total intensity, Itot(t), decays slowly with time, which makes the determination of the fraction of immobile molecules uncertain if there is noise in the data. If γ0 is erroneously determined this will then influence the value of D obtained from the analysis, since these values are related according to Eq. 15. Using a range of k values for the curve fits causes both the Fourier- and the Hankel-transformed data to change more rapidly with time, which means that the uncertainty in the determination of γ0 is reduced with both these methods. To improve the accuracy of the integral method, the measurement time should be prolonged. However, when measurements are performed over longer times another problem arises, namely temporal fluctuations such as drifts in the overall intensity and bleaching during recovery.
Bleaching and drifts in intensity during recovery (see the inset in Fig. 9 b) can have a detrimental effect on the accuracy of the analysis. When the noncompensated values in Fig. 9 b (dots) were used to determine D and γ0, as is generally the case in traditional FRAP analysis (8,18), the values obtained were D = 3.42 μm2 s−1 and γ0 = 0.20. Thus, if not corrected for, temporal variations in the images may lead to an erroneous estimation of both the diffusion coefficient and the fraction of immobile molecules in the analyzed system. Both the Fourier transform and the Hankel transform methods are also sensitive to temporal fluctuations in the images, mainly because these fluctuations can give rise to an erroneous offset in the signal that is to be transformed. However, if the spatial frequencies are chosen such that the transform of a constant will be zero, then the sensitivity to temporal fluctuations is reduced (see the discussion in connection with Fig. 5). For a more thorough analysis of D(k), it may be desirable to use a finer interval of k, and in such cases, correction for temporal variations will be essential.
The error in D(k) for three different one-component systems
To evaluate the error in the value of the diffusion coefficient, D was determined in a wide interval of k values for three different single diffusing components (Fig. 10): rhodamine-DHPE; NBD C12-HPC; and 35-nm lipid vesicles tethered to an unlabeled SPB. For all cases γ2 was set to zero when analyzing the one-component data. The fluidity of the SPB used for the FRAP analysis of tethered vesicles was confirmed by labeling and monitoring the recovery of NBD C12-HPC lipids in the bilayer. The error bars show standard deviations obtained from multiple measurements on different areas of the sample and from measurements on different samples. The effective diffusion coefficient, D, and the fraction of immobile molecules, γ0, for the different systems are presented in the inset in Fig. 10.
FIGURE 10.
D vs. k for different types of single molecule diffusion in a SPB: rhodamine-DHPE (crosses), NBD C12-HPC (circles), and 35-nm tethered vesicles (dots). The error bars indicate the standard error at each value of k. The inset shows measured values of the diffusion coefficient for the different types of molecules, with the standard deviations indicated within parentheses. Mean values and standard deviations are calculated from multiple measurements at different locations on each sample and from different samples. To eliminate the influence from a faster diffusing component observed for the tethered vesicles, the first 50 frames after bleaching were omitted in the analysis for this specific case.
The variation in D versus k is low: 4, 6, and 8% for rhodamine-DHPE, NBD C12-HPC, and 35-nm lipid vesicles, respectively, where the variation is defined as the standard deviation of D divided by the mean value of D in the interval shown in Fig. 10. This behavior is expected for molecules undergoing Brownian diffusion. This is especially evident in the interval 0.005 μm−1 < k < 0.015 μm−1 where the standard deviation in the determined value of D is smallest. This observation is consistent with the results obtained from the analysis of simulated data, which showed that the accuracy when determining D using a single value of k was highest at intermediate values of k (see Figs. 5 and 6). As discussed above, the reason for this observation is that at small values of k the Hankel transform changes slowly with time, while at large values of k the magnitude of the Hankel transform is small, thus making the change in the Hankel transform with time largest for intermediate values of k.
The effective diffusion coefficient of NBD C12-HPC (DNBD = 2.21 μm2 s−1) was observed to be slightly lower than that of rhodamine-DHPE (Drhod = 2.79 μm2 s−1). This is in agreement with previously published data for similar systems (32,34). The diffusion coefficient determined for the 35-nm tethered vesicles (Dves = 0.11 μm2 s−1) was significantly smaller than that of both NBD C12-HPC and rhodamine-DHPE, but this value is also in good agreement with previously published data on similar systems (23,37). When studying the recovery of the fluorescently labeled vesicles it was observed that there was also a fast component, although with a significantly lower magnitude than that of the slow component. One possible explanation of this observation is that some of the NBD-labeled lipids in the bilayer were excited when studying the recovery. An alternative explanation is that a small fraction of the rhodamine-labeled lipids in the vesicles were transferred to the SPB. However, the influence of this faster component was eliminated by omitting the first 50 frames after bleaching when analyzing the data.
It is also worthwhile to note that the NBD-labeled lipids bleached much faster than the rhodamine-labeled lipids, at a certain light intensity. Thus, to avoid bleaching, the intensity of the light illuminating the sample must be kept low. However, this results in a lower SNR, since the light emitted by the fluorophores will also be low. There is thus a compromise between the amount of light emitted by the fluorophores and the degree of bleaching during recovery. This must be taken into consideration when analyzing the data using traditional methods of FRAP analysis (8,14). However, the Hankel transform method uses the spatial information from the acquired images to compensate for bleaching. Thus, since bleaching can be compensated for in the analysis, a higher light intensity can be used when illuminating samples yielding a higher SNR in the acquired images.
Multiple diffusing components
To test the applicability of the FRAP analysis on a more challenging system, a model system consisting of two identically labeled diffusing components, rhodamine-DHPE in the SPB and rhodamine-labeled tethered vesicles, was analyzed. In addition to being a suitable test for the Hankel transform method, the ability to analyze systems with more than one diffusing component may also be of practical interest when investigating the mobility of other biological model systems (12,38). Fig. 11 a shows typical recovery curves obtained with the Hankel transform method (solid dots) together with the corresponding values obtained with the Fourier transform method (shaded circles). Fig. 11 b shows the results for Itot(t), obtained using the integral method, together with a single-component (see inset) and a double-component curve fit according to Eq. 15.
FIGURE 11.
(a) All transformed values, F(k,t), versus 4π2k2t for a system consisting of two diffusing components: rhodamine-DHPE and 35-nm tethered lipid vesicles. The solid dots and shaded circles are data obtained by using the Hankel and the Fourier transform methods, respectively. The shaded line, through the solid data points, is a curve fit to the Hankel-transformed data. (b) Itot(t) for the two-component system, with a double-component curve fit (R2 = 0.9981) to the data according to Eq. 15 (shaded line). The inset shows a single-component curve fit with an immobile fraction of molecules to the same data (shaded line), yielding R2 = 0.9976.
The analysis of the two-component system using the Hankel transform method yielded one fast, D1 = 2.49 μm2 s−1 (0.21 μm2 s−1), and one slow component, D2 = 0.13 μm2 s−1 (0.005 μm2 s−1), where the values in parentheses are standard deviations calculated from six different locations on the sample. The intensity fraction of the slow component was γ2 = 0.41 (0.03), and the fraction of immobile molecules was negligible. These values are in good agreement with the values measured for the single-component systems of rhodamine-DHPE-labeled lipids in the SPB (Drhod = 2.79 μm2 s−1) and tethered lipid vesicles (Dves = 0.11 μm2 s−1), as shown in Fig. 10. Note also that, from the results shown in Fig. 11 a, an accurate analysis of a two-component system requires measurements over both a wide range of k values and long times to reveal the slow-diffusing component.
Strikingly, the two traditionally used methods were significantly less reliable in determining the diffusivity of the two-component system. One problem associated with the integral method is that both a single- and a double-component fit to Eq. 15 could describe the experimental data essentially equally well: 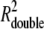 = 0.9981 and
= 0.9981 and 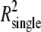 = 0.9976 (see inset in Fig. 11 b). Furthermore, the double-component curve fits yielded D1 = 0.74 μm2 s−1, D2 = 0.08 μm2 s−1, and γ2 = 0.19, which deviate significantly from the values obtained using the Hankel transform method and from the values obtained from the analysis of single-diffusing components. One reason for this is that temporal variations were not compensated for in the case illustrated in Fig. 11 b. However, even after compensating for temporal variations, the value determined for D1 was still a factor-of-2 lower than expected, while the values of D2 and γ2 were significantly improved. The reason for this is attributed to one of the assumptions behind Eq. 15, namely that the fluorescent intensity from the two diffusing components can be described by a single Gaussian curve immediately after bleaching. A careful analysis of the FRAP images revealed that this was not the case for the two-component experiments. Ir(r,t) had two discernible components already in the first frame after bleaching, where the faster component had a wider concentration profile than the slow component. This, in turn, resulted in the underestimation of the faster component by at least a factor of two, even after compensating for temporal variations. The observation of a wider concentration profile for the fast molecules is attributed to the fact that this component had time to diffuse a significant distance from the start of bleaching to the first frame in the recovery images. This effect may also influence the outcome of the Fourier and Hankel transform methods. However, the Hankel transform method was not critically affected by the duration of the bleaching time in the current experiments, as seen from the analysis of the two-component system. Despite this, it is worth noting that, for optimal accuracy when analyzing two-component systems, the bleaching time should be kept as short as possible.
= 0.9976 (see inset in Fig. 11 b). Furthermore, the double-component curve fits yielded D1 = 0.74 μm2 s−1, D2 = 0.08 μm2 s−1, and γ2 = 0.19, which deviate significantly from the values obtained using the Hankel transform method and from the values obtained from the analysis of single-diffusing components. One reason for this is that temporal variations were not compensated for in the case illustrated in Fig. 11 b. However, even after compensating for temporal variations, the value determined for D1 was still a factor-of-2 lower than expected, while the values of D2 and γ2 were significantly improved. The reason for this is attributed to one of the assumptions behind Eq. 15, namely that the fluorescent intensity from the two diffusing components can be described by a single Gaussian curve immediately after bleaching. A careful analysis of the FRAP images revealed that this was not the case for the two-component experiments. Ir(r,t) had two discernible components already in the first frame after bleaching, where the faster component had a wider concentration profile than the slow component. This, in turn, resulted in the underestimation of the faster component by at least a factor of two, even after compensating for temporal variations. The observation of a wider concentration profile for the fast molecules is attributed to the fact that this component had time to diffuse a significant distance from the start of bleaching to the first frame in the recovery images. This effect may also influence the outcome of the Fourier and Hankel transform methods. However, the Hankel transform method was not critically affected by the duration of the bleaching time in the current experiments, as seen from the analysis of the two-component system. Despite this, it is worth noting that, for optimal accuracy when analyzing two-component systems, the bleaching time should be kept as short as possible.
When making a two-component fit to the data using the Fourier transform method, regardless of whether this was a statistically significant improvement from a one-component fit or not, the determined values were of the same order of magnitude as those obtained with the Hankel transform method. However, the spread in D1 was twice as high and that in D2 four times higher compared to the values obtained with the Hankel transform method. The existence of two diffusing components was also more difficult to assess using the Fourier transform method than with the Hankel transform method. The reason for this is that the noise in the Fourier-transformed data (see Fig. 11 a) generally made it harder to evaluate whether a two-component fit described the system significantly better than a one-component fit with an immobile fraction of molecules. The extra noise in the Fourier-transformed data compared to the data from the new method is primarily attributed to temporal variations in the images, but the effect of a net influx of fluorescently labeled molecules into the field of view is also expected to influence the outcome of the analysis. For the Fourier transform method, the latter complication is related to the fact that the measurement time that yields accurate results is limited by the time it takes for the fastest molecules to diffuse out of the field of view (18,39). However, the Hankel transform method does not suffer from this limitation, since a net influx of molecules into the field of view is efficiently compensated for in the analysis, thus eliminating the constraint imposed on the measurement time.
CONCLUSIONS
Fluorescence recovery after photobleaching is a well-known method of determining the diffusive properties of molecular species in two-dimensional systems. This article presents a new method (the Hankel transform method) that compensates for temporal variations, reduces the effect of noise, and is independent of the shape of the bleached area. The method, which was implemented in MATLAB, was demonstrated to accurately be able to determine the diffusion coefficient from simulated, as well as experimental, data over a wide range of conditions and SNRs. In comparison to traditional methods of FRAP analysis (8,14), the Hankel transform method was shown to be less sensitive to noise and temporal drifts in the images. Drifts in the illumination and bleaching during the recovery of a FRAP experiment can have especially detrimental effects on the outcome of the analysis, if not compensated for. Thus, the Hankel transform method can improve the analysis of systems under nonideal experimental conditions, where traditional methods of FRAP analysis encounter difficulties. Furthermore, its insensitivity to noise makes the Hankel transform method especially suited for the analysis of systems with multiple diffusion coefficients, shown by the accurate analysis of both simulated and experimental data in this article.
The Hankel transform method utilizes circular averaging to reduce the spatial noise in the analyzed data as well as to make the data easier to handle. This requires that the analyzed images possess circular symmetry, which might be considered a serious disadvantage of the method. However, many FRAP setups have light sources with circular symmetry and deviations from circular symmetry were shown, for most practical cases, to have a negligible effect on the determination of the diffusion coefficient. Another potential limitation of circular averaging is that it will only measure isotropic diffusion. However, many of the lateral transport modes in, for example, cell membranes, as well as in other systems, are isotropic (2). If specifically anisotropic behavior is being investigated then other techniques could be used (14,16). Note that the compensation steps introduced in the current work could also be used, with some adaptation, to improve the accuracy of these techniques.
The new method was named the Hankel transform method since the FRAP data are first subjected to a Hankel transform before the diffusion coefficients of the studied system are determined. Hankel transformation of the data has the advantage that no prior knowledge of the initial bleaching profile is necessary. Furthermore, there is a clear advantage in using a Hankel transform when analyzing the mobility of a heterogeneous system where the diffusion is anomalous. Measuring the total fluorescent intensity, as in conventional FRAP analysis, will only give an effective value of the diffusion coefficient in the system, assuming Brownian diffusion. In contrast, by analyzing the spatial frequency dependence of the diffusion, information about the mobility over different length scales can be obtained, as currently being investigated by our group. The presented method of FRAP analysis thus opens up exciting possibilities in situations where the investigated system displays non-Brownian diffusion. Furthermore, the shown reliability of the method when analyzing systems under nonideal conditions points to a general applicability of the method for improved analysis of diffusion in complex systems.
APPENDIX
The total intensity for r < w
The concentration of the ith component, ci(r,t), in the studied system is assumed to obey Fick's second law, as presented in Eq. 16 (28),
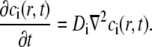 |
(16) |
The solution to Eq. 16 can be shown to be (8,11)
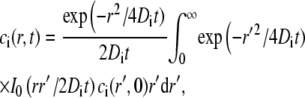 |
(17) |
where I0 is the first-order modified Bessel function. For an initially Gaussian concentration profile ci(r,0) = ci,eq(1−Kiexp(−r2/w2)), and the value of ci(r,t) in Eq. 17 is given by
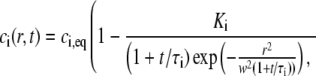 |
(18) |
where τi = w2/(4Di). In Eq. 18 it is assumed that the initial width of the bleached profile is the same for all components. The relative concentration of unbleached molecules of the ith type, cr,i(r,t) = ci(r,t)/ci,eq, is assumed to be proportional to the light intensity detected by the CCD camera, Ii(r,t), with the proportionality constant qi. The total intensity, Itot(t), within a circle of radius R is then
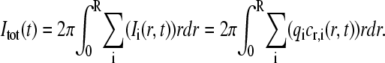 |
(19) |
Inserting the expression for ci(r,t) given in Eq. 18 into Eq. 19 yields
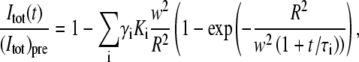 |
(20) |
where (Itot)pre is the value of Itot(t) before bleaching, 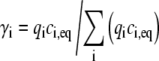 and τi = w2/(4Di). For two diffusing components and an immobile fraction of molecules, γ0, Eq. 20 equals the expression in Eq. 21, where Ki = K and R = w have been used,
and τi = w2/(4Di). For two diffusing components and an immobile fraction of molecules, γ0, Eq. 20 equals the expression in Eq. 21, where Ki = K and R = w have been used,
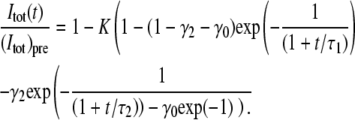 |
(21) |
Derivation of the expression for β(t)
The relative intensity, Ir(r,t), is given by
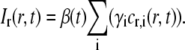 |
(22) |
The function Ir(r,t)|r>R is defined according to Eq. 23, where A(t) and w(t) are chosen such that Ir(r,t)|r>R = Ir(r,t) when r > R,
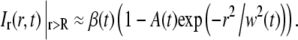 |
(23) |
To determine β(t), one can utilize the fact that the total amount of fluorescently labeled molecules in the system must be constant. The integral of Ir(r,t)−Ir(r,t)|r>R will then yield
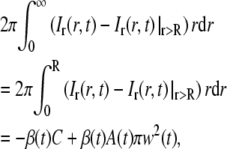 |
(24a) |
where
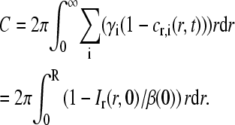 |
(24b) |
In Eq. 24b it is assumed that A(0) = 0. This is equivalent to ci(R,0) = ci,eq, which will be an accurate assumption if the bleached spot in the first frame after photobleaching is smaller than the field of view. Furthermore, since Ir(r,t) = Ir(r,t)|r>R when r > R, the integral on the left-hand side of Eq. 24a can be restricted to the interval 0 < r < R. Inserting the expression for Ir(r,t)|r>R in Eq. 23 into Eq. 24a then yields
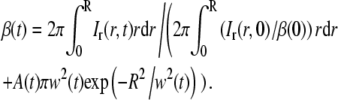 |
(25) |
The value of β(0) can be estimated from the relative intensity immediately after bleaching at t = 0. Hence, the concentration at r = R is equal to the equilibrium value ci,eq, which yields
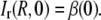 |
(26) |
Inserting this relation for β(0) into Eq. 25 finally gives
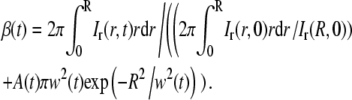 |
(27) |
This work was financially supported by the Swedish Research Council for Engineering Sciences, contract No. 2005-3140, and the Ingvar grant from the Strategic Research Foundation.
Magnus P. Jonsson's and Fredrik Höök's present address is Dept. of Applied Physics, Chalmers University of Technology, SE-41296 Gothenburg, Sweden.
Editor: Michael Edidin.
References
- 1.Saffman, P. G., and M. Delbruck. 1975. Brownian-motion in biological-membranes. Proc. Natl. Acad. Sci. USA. 72:3111–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobson, K., A. Ishihara, and R. Inman. 1987. Lateral diffusion of proteins in membranes. Annu. Rev. Physiol. 49:163–175. [DOI] [PubMed] [Google Scholar]
- 3.Saxton, M. J., and K. Jacobson. 1997. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. 26:373–399. [DOI] [PubMed] [Google Scholar]
- 4.Meyvis, T. K. L., S. C. De Smedt, P. Van Oostveldt, and J. Demeester. 1999. Fluorescence recovery after photobleaching: a versatile tool for mobility and interaction measurements in pharmaceutical research. Pharm. Res. 16:1153–1162. [DOI] [PubMed] [Google Scholar]
- 5.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444–456. [DOI] [PubMed] [Google Scholar]
- 6.Klonis, N., M. Rug, I. Harper, M. Wickham, A. Cowman, and L. Tilley. 2002. Fluorescence photobleaching analysis for the study of cellular dynamics. Eur. Biophys. J. Biophys. 31:36–51. [DOI] [PubMed] [Google Scholar]
- 7.Reits, E. A. J., and J. J. Neefjes. 2001. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3:E145–E147. [DOI] [PubMed] [Google Scholar]
- 8.Axelrod, D., D. E. Koppel, J. Schlessinger, E. Elson, and W. W. Webb. 1976. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16:1055–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edidin, M., Y. Zagyansky, and T. J. Lardner. 1976. Measurement of membrane protein lateral diffusion in single cells. Science. 191:466–468. [DOI] [PubMed] [Google Scholar]
- 10.Peters, R., J. Peters, K. H. Tews, and W. Bahr. 1974. Microfluorimetric study of translational diffusion in erythrocyte-membranes. Biochim. Biophys. Acta. 367:282–294. [DOI] [PubMed] [Google Scholar]
- 11.Soumpasis, D. M. 1983. Theoretical-analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41:95–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon, G. W., B. Chazotte, X. F. Wang, and B. Herman. 1995. Analysis of simulated and experimental fluorescence recovery after photobleaching—data for two diffusing components. Biophys. J. 68:766–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubitscheck, U., P. Wedekind, and R. Peters. 1994. Lateral diffusion measurement at high-spatial-resolution by scanning microphotolysis in a confocal microscope. Biophys. J. 67:948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsay, T. T., and K. A. Jacobson. 1991. Spatial Fourier-analysis of video photobleaching measurements—principles and optimization. Biophys. J. 60:360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain, R. K., R. J. Stock, S. R. Chary, and M. Rueter. 1990. Convection and diffusion measurements using fluorescence recovery after photobleaching and video image-analysis—in vitro calibration and assessment. Microvasc. Res. 39:77–93. [DOI] [PubMed] [Google Scholar]
- 16.Kapitza, H. G., G. McGregor, and K. A. Jacobson. 1985. Direct measurement of lateral transport in membranes by using time-resolved spatial photometry. Proc. Natl. Acad. Sci. USA. 82:4122–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmon, E. D., R. J. Leslie, W. M. Saxton, M. L. Karow, and J. R. McIntosh. 1984. Spindle microtubule dynamics in sea-urchin embryos—analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 99:2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berk, D. A., F. Yuan, M. Leunig, and R. K. Jain. 1993. Fluorescence photobleaching with spatial Fourier-analysis—measurement of diffusion in light-scattering media. Biophys. J. 65:2428–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss, M. 2004. Challenges and artifacts in quantitative photobleaching experiments. Traffic. 5:662–671. [DOI] [PubMed] [Google Scholar]
- 20.Smith, B. A., W. R. Clark, and H. M. McConnell. 1979. Anisotropic molecular-motion on cell-surfaces. Proc. Natl. Acad. Sci. USA. 76:5641–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sackmann, E. 1996. Supported membranes: scientific and practical applications. Science. 271:43–48. [DOI] [PubMed] [Google Scholar]
- 22.McConnell, H. M., T. H. Watts, R. M. Weis, and A. A. Brian. 1986. Supported planar membranes in studies of cell-cell recognition in the immune-system. Biochim. Biophys. Acta. 864:95–106. [DOI] [PubMed] [Google Scholar]
- 23.Benkoski, J. J., and F. Hook. 2005. Lateral mobility of tethered vesicle—DNA assemblies. J. Phys. Chem. B. 109:9773–9779. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson, M. P., P. Jonsson, A. B. Dahlin, and F. Hook. 2007. Supported lipid bilayer formation and lipid-membrane-mediated biorecognition reactions studied with a new nanoplasmonic sensor template. Nano Lett. 7:3462–3468. [DOI] [PubMed] [Google Scholar]
- 25.Tsai, J., E. Sun, Y. Gao, J. C. Hone, and L. C. Kam. 2008. Non-Brownian diffusion of membrane molecules in nanopatterned supported lipid bilayers. Nano Lett. 8:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan, Y. H. M., P. Lenz, and S. G. Boxer. 2007. Kinetics of DNA-mediated docking reactions between vesicles tethered to supported lipid bilayers. Proc. Natl. Acad. Sci. USA. 104:18913–18918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunnarsson, A., P. Jonsson, R. Marie, J. O. Tegenfeldt, and F. Hook. 2008. Single-molecule detection and mismatch discrimination of unlabeled DNA targets. Nano Lett. 8:183–188. [DOI] [PubMed] [Google Scholar]
- 28.Cussler, E. L. 1997. Diffusion: Mass Transfer in Fluid Systems. Cambridge University Press, Cambridge, UK.
- 29.Arfken, G. B., and H. J. Weber. 2001. Mathematical Methods for Physicists. Academic Press, San Diego, CA.
- 30.Guizar-Sicairos, M., and J. C. Gutierrez-Vega. 2004. Computation of quasi-discrete Hankel transforms of integer order for propagating optical wave fields. J. Opt. Soc. Am. A. 21:53–58. [DOI] [PubMed] [Google Scholar]
- 31.Bockmann, R. A., A. Hac, T. Heimburg, and H. Grubmuller. 2003. Effect of sodium chloride on a lipid bilayer. Biophys. J. 85:1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilmanshin, R., C. E. Creutz, and L. K. Tamm. 1994. Annexin-IV reduces the rate of lateral lipid diffusion and changes the fluid-phase structure of the lipid bilayer when it binds to negatively charged membranes in the presence of calcium. Biochemistry. 33:8225–8232. [DOI] [PubMed] [Google Scholar]
- 33.Patel, A. R., and C. W. Frank. 2006. Quantitative analysis of tethered vesicle assemblies by quartz crystal microbalance with dissipation monitoring: binding dynamics and bound water content. Langmuir. 22:7587–7599. [DOI] [PubMed] [Google Scholar]
- 34.Tamm, L. K. 1988. Lateral Diffusion and fluorescence microscope studies on a monoclonal-antibody specifically bound to supported phospholipid bilayers. Biochemistry. 27:1450–1457. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, M. E., D. A. Berk, D. Blankschtein, D. E. Golan, R. K. Jain, and R. S. Langer. 1996. Lateral diffusion of small compounds in human stratum corneum and model lipid bilayer systems. Biophys. J. 71:2656–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schutz, G. J., H. Schindler, and T. Schmidt. 1997. Single-molecule microscopy on model membranes reveals anomalous diffusion. Biophys. J. 73:1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshina-Ishii, C., Y. H. M. Chan, J. M. Johnson, L. A. Kung, P. Lenz, and S. G. Boxer. 2006. Diffusive dynamics of vesicles tethered to a fluid supported bilayer by single-particle tracking. Langmuir. 22:5682–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg, M. L., and D. Axelrod. 1993. Anomalously slow mobility of fluorescent lipid probes in the plasma-membrane of the yeast Saccharomyces cerevisiae. J. Membr. Biol. 131:115–127. [DOI] [PubMed] [Google Scholar]
- 39.Johnson, E. M., D. A. Berk, R. K. Jain, and W. M. Deen. 1995. Diffusion and partitioning of proteins in charged agarose gels. Biophys. J. 68:1561–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]