Abstract
ABC transporters constitute one of the most abundant membrane transporter families. The most common feature shared in the family is the highly conserved nucleotide binding domains (NBDs) that drive the transport process through binding and hydrolysis of ATP. Molecular dynamics simulations are used to investigate the effect of ATP hydrolysis in the NBDs. Starting with the ATP-bound, closed dimer of MalK, four simulation systems with all possible combinations of ATP or ADP-Pi bound to the two nucleotide binding sites are constructed and simulated with equilibrium molecular dynamics for ∼70 ns each. The results suggest that the closed form of the NBD dimer can only be maintained with two bound ATP molecules; in other words, hydrolysis of one ATP can lead to the opening of the dimer interface of the NBD dimer. Furthermore, we observed that the opening is an immediate effect of hydrolysis of ATP into ADP and Pi rather than the dissociation of hydrolysis products. In addition, the opening is mechanistically triggered by the dissociation of the LSGGQ motif from the bound nucleotide. A metastable ADP-Pi bound conformational state is consistently observed before the dimer opening in all the simulation systems.
INTRODUCTION
ATP-binding cassette (ABC) transporters are integral membrane proteins that use the chemical energy of ATP hydrolysis to promote active transport across the cellular membrane (1). Numerous ABC transporters have been found in all three kingdoms of life, mediating translocation of various substrates ranging from ions and small molecules to large biopolymers. In the human genome, there are 48 genes encoding ABC family proteins; a large number of these genes have been functionally characterized and associated with genetic diseases or have presented multidrug resistance in cancer cells (2,3).
Whereas eukaryotic ABC transporters are exclusively exporters, the majority of prokaryotic ABC transporters have evolved as importers (4,5). With the aid of an extracellular/periplasmic protein for specific substrate acquisition, ABC importers play important roles in nutrient uptake among prokaryotes. ABC exporters also exist in prokaryotic cells, playing crucial roles for survival and virulence due to their involvement in lipid and toxin secretion (6,7) and drug resistance (8). Owing to these essential roles, ABC transporters constitute an important target for drug and antibiotic development.
Four basic building blocks are present in all ABC transporters regardless of the direction of the transport: two transmembrane permease domains (TMDs) providing the substrate translocation pathway and two nucleotide binding domains (NBDs) that bind and hydrolyze ATP and provide energy to drive the transport. In contrast to the TMDs, which share little or no sequence similarity among ABC transporters, the NBDs are highly conserved, suggesting a common mechanism of activation in different ABC transporters. NBD crystal structures have been determined for a number of ABC transporters as functional dimers (9–13) and in complex with TMDs in several complete ABC transporter structures (14–22). A consistent mechanistic view regarding the role of the NBDs in the transport has emerged from these dimeric NBD crystal structures, which seems to be applicable to both ABC importers and exporters; namely, the NBDs appear as a closed dimer in the ATP-bound form, in which two ATP molecules are sandwiched between the conserved Walker A motif of one monomer and the ABC signature motif (the LSGGQ motif) of the other (Fig. 1 A). In contrast, most ADP-bound or nucleotide-free states of NBDs are crystallized as monomers or as open dimers. Therefore, it seems that ATP binding and hydrolysis controls the opening and closure of the NBDs, which in turn can modulate the conformational states of the TMDs where substrate translocation takes place.
FIGURE 1.
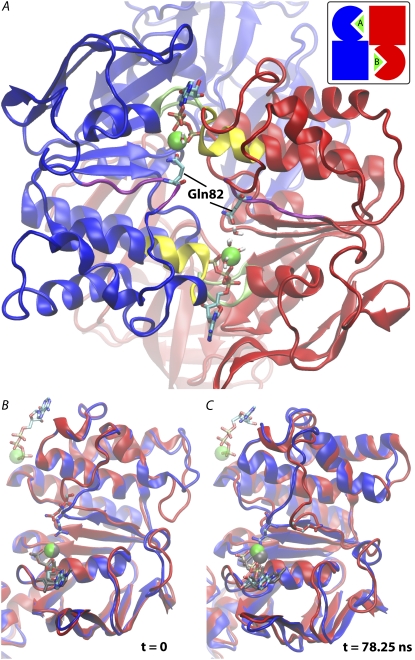
Initial structure of MalK dimer used for MD simulations. The following color scheme is used in all figures: blue ribbon for chain C and red ribbon for chain D. Atoms are colored as follows: carbon in cyan, oxygen in red, nitrogen in blue, phosphorus in brown, and magnesium in green. The ATP binding motifs are colored as Walker A in green, Q-loop in purple, and the LSGGQ motif in yellow. (A) The initial structure for all the MD simulations in this study with a schematic representation of the dimer arrangement in the inset. Gln-82 and water molecules bound to Mg2+ are drawn as sticks. In the schematic presentation, each MalK monomer is represented by an oval (RecA-like subdomain) and a square (helical subdomain) and is colored accordingly. Triangles represent the bound nucleotides within the two binding sites, named sites A and B, respectively. The regulatory domains, which are included in the simulations, are shown in the back with faded colors. (B and C) Overlaid structures of chains C and D at the beginning and at the end of the 78.25 ns simulation of the ATP/ATP system. ATP and Mg2+ associated with each monomer are shown as sticks and van der Waals spheres, respectively. To enhance the contrast, ATP-Mg2+ bound to chain C is colored gray, and its location relative to the LSGGQ motif of chain D is shown as a transparent model. The two Gln-82 residues are highlighted as ball-and-stick models, whereas the sticks are colored with respect to the protein monomer, and the balls are colored per element. For clarity, the regulatory domains are not shown.
Most of our knowledge about the mechanism of NBDs originates from extensive structural and biochemical studies on the Escherichia coli maltose transporter MalK. Specifically, the NBD of MalK, which is an ABC transporter, has been crystallized as a dimer in three different nucleotide-bound and conformational states: 1), the closed dimer with ATP (10); 2), the open dimer, which is either cocrystallized with ADP or allows ATP hydrolysis during crystallization (11) in the nucleotide-free, open state (10); and 3), the semi-open, nucleotide-free form (10). MalK is also present as a closed dimer in the recent crystal structure of the intact maltose transporter complex trapped in an ATP-bound, substrate-occluded state (19).
The nature of all these structures supports the hypothesis that the opening and closure of the NBD dimers, which are key events in the transport mechanism, are governed by ATP binding and hydrolysis in NBDs. However, some details of this mechanism are still largely unknown. Specifically, the stoichiometry of ATP hydrolysis per transport cycle and whether the two active sites work cooperatively or alternatively has been the subject of long debate (23–27).
Molecular dynamics (MD) simulations can be used to investigate the detailed mechanisms and to capture structural transitions at high temporal and spatial resolutions. Previous simulations of ABC transporters have revealed several features that are in agreement with experimental observations (28–33). MD simulations of the open and semiopen forms of MalK dimers with ATP docked into their nucleotide binding sites resulted in a closing motion of the two monomers (30). The same effect has also been observed in simulations with ATP docked into the nucleotide-free form of the vitamin B12 transporter BtuCD (28). Simulation of the nucleotide-free, semiopen form of MalK resulted in a greater degree of dimer opening and separation of the NBDs (30). Moreover, independent simulation studies carried out on the ATP-docked BtuCD led to a proposed model involving alternate ATP binding and hydrolysis at the two active sites, which was based on the structural asymmetry of the NBDs observed during the simulations (28,31). Asymmetric behaviors have also been observed when one of the two bound ATP molecules was replaced by ADP in MD simulations of an archaeal ABC transporter (29,33). Interestingly, an asymmetric behavior has also been reported for a simulation in which both ATP molecules remained bound to their respective active sites of this archaeal ABC transporter (29).
The opening of NBD dimers during the transition from the ATP-bound state to the ADP-bound or nucleotide-free state is evident from crystal structure (11) and MD simulations (30). It is unclear, however, whether the opening motion is a direct consequence of ATP hydrolysis (that is, conversion of ATP to ADP-Pi) or a result of the subsequent dissociation of the phosphate anion. Furthermore, it is not known whether one ATP hydrolysis event can initiate the opening or whether both catalytic sites need to work cooperatively to induce the process. In this study, we use long, atomistic MD simulations to investigate the effect of ATP hydrolysis in the closed NBD dimer of MalK and to study the detailed mechanism involved in dimer opening of the NBDs in ABC transporters immediately after the hydrolysis of ATP. All four possible nucleotide binding configurations are simulated in a MalK dimer. The results suggest that the closed dimer is stable only when both nucleotide binding sites are occupied by ATP. In other words, a single ATP hydrolysis event seems to be sufficient to induce the opening of the dimer. The observed opening appears to be an immediate consequence of ATP hydrolysis and to be independent of phosphate dissociation.
COMPUTATIONAL METHODS
The structure of the ATP-bound dimeric MalK taken from Protein Data Bank (PDB) entry 1Q12 (10), including the C-terminal regulatory domains, was used as the initial structure for all the simulations. Between the two biological units in the asymmetric unit, the dimer composed of chains C and D was chosen for model building because it includes a larger number of crystal water molecules. Two Mg2+ ions missing from the crystal structure were manually placed between the β- and γ-phosphates of the bound ATP molecules based on the position of Na+ ions in the crystal structure of the ATP-bound NBD dimer of MJ0796 (PDB entry 1L2T (9)). The complex was then solvated in a 90 × 100 × 90 Å3 periodic box; it also was neutralized and ionized with 200 mM NaCl. The structures were energy minimized for 3000 steps before the MD simulations. Mg2+ locations were monitored during the minimization to ensure proper and stable positioning.
MD simulations were carried out with NAMD 2.6 (34) using the CHARMM27 force field with φ/ψ cross-term map corrections (35). Water molecules were simulated with the TIP3P model (36). Simulation conditions were maintained at 1.0 atm constant pressure by the Nosé-Hoover Langevin piston method (37,38) and at 310 K constant temperature by Langevin dynamics with a damping coefficient at 0.5 ps−1. The time step used for the simulations was 1 fs, and the coordinates were recorded every 1 ps. A cutoff of 12 Å was used for short-range, nonbonded interaction, and long-range electrostatic interactions were computed using the particle mesh Ewald method (39).
Equilibrium simulations were conducted on the doubly ATP-bound (ATP/ATP) MalK dimer for 8.25 ns, the last frame of which was used as the starting structure for the simulations of the dimer in different nucleotide-bound forms. To simulate the effect of ATP hydrolysis, ATP was converted to ADP and Pi by manually replacing the γ-phosphate with a dibasic phosphoric acid molecule 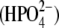 (see Fig. S1 in Supplementary Material, Data S1). Four different systems with different nucleotide/phosphate combinations were generated; thus, all ATP/ATP, ATP/ADP-Pi, ADP-Pi/ATP, and ADP-Pi/ADP-Pi active site configurations were generated and minimized. One simulation of 70 ns was performed for each of the four systems. We note that, in this work, we have not simulated the process of ATP hydrolysis itself, and we have only investigated the effect of conversion of ATP to ADP and Pi. We also note that long MD simulations (at least ∼50 ns in this research) proved essential to draw relevant conclusions regarding the mechanism and process of dimer opening, because the initial 20 to 30 ns of the simulation did not provide a consistent picture of the relationship between ATP hydrolysis and dimer opening.
(see Fig. S1 in Supplementary Material, Data S1). Four different systems with different nucleotide/phosphate combinations were generated; thus, all ATP/ATP, ATP/ADP-Pi, ADP-Pi/ATP, and ADP-Pi/ADP-Pi active site configurations were generated and minimized. One simulation of 70 ns was performed for each of the four systems. We note that, in this work, we have not simulated the process of ATP hydrolysis itself, and we have only investigated the effect of conversion of ATP to ADP and Pi. We also note that long MD simulations (at least ∼50 ns in this research) proved essential to draw relevant conclusions regarding the mechanism and process of dimer opening, because the initial 20 to 30 ns of the simulation did not provide a consistent picture of the relationship between ATP hydrolysis and dimer opening.
Protein subdomains in the system are referred to as the helical subdomains (residues 88–151) and RecA-like subdomains (residues 4–87 and 152–235) in the two monomers. Key nucleotide binding motifs relevant to dimer opening are the Walker A motif (residues 36–43) and the LSGGQ motif (residues 134–141). The distances between the centers of masses of these subdomains and motifs, in combination with the hydrogen bond distances between ATP (or the replacing ADP-Pi) and binding site residues, were used to monitor the destabilization of the active sites and the opening of the dimer. The average value of these distances in the ATP/ATP simulation system (see Figs. 3 E and 4 D) are used as a reference. Opening/dissociation events in the posthydrolysis systems are defined as significant deviations of these geometrical measures (beyond three standard deviations) from the ATP/ATP state. Data analysis and molecular images were made with visual molecular dynamics (VMD) software (40).
FIGURE 3.
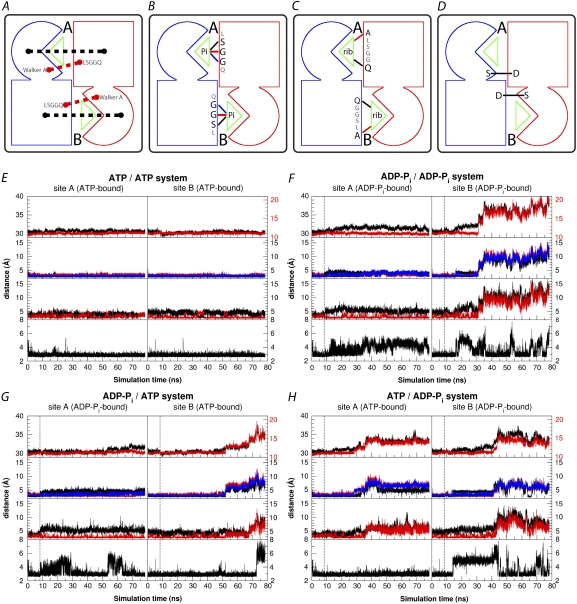
Dimer opening and dynamics of the binding sites. (A–D) Schematic representations of the distances shown in panels E–H. (A) The dimer opening was measured in two ways: 1), the distance between the centers of masses of the RecA-like subdomain of the cis monomer and that of the helical subdomain of the trans monomer (black traces); and 2), the distance between the centers of masses of the Walker A motif (residue 36–43) and the LSGGQ motif (residue 134–141) flanking the nucleotide binding sites (red traces). (B) The distances between the key γ-phosphate binding residues of the LSGGQ motif and the closest oxygen atom from either ATP or Pi: side-chain hydroxy of Ser-135 (black), and backbone amides of Gly-136 (red) and Gly-137 (blue). (C) Distances between the ribose ring and the helical subdomain: between the side-chain carbonyl oxygen of Gln-138 (the conserved Q of the LSGGQ motif) and the 3′ oxygen of the bound nucleotide (black), and between the backbone carbonyl oxygen of Ala-133 and the 2′ oxygen of the bound nucleotide (red). (D) The distance of a direct hydrogen bond contact between the two monomers between the conserved Ser-38 (Walker A motif) and the conserved Asp-165 (D-loop) residues. (E–H) Time series of the distances shown in A–D. Distances pertaining to binding site A are plotted on the left side and those for site B on the right side of each panel. The top graph in each panel shows the intersubdomain distances (black) and the intermotif distances (red). The second graph shows the distances between phosphate (ATP γ-phosphate or Pi) and LSGGQ motif residues (Ser-135 in black, Gly1-36 in red, and Gly-137 in blue). The third graph of each panel shows the distance between the ribose ring of the nucleotide and the helical subdomain residues (Gln-138 in black and Ala-133 in red). The last graph plots the distance between Ser-38 and Asp-165. The vertical dashed lines at 8.25 ns of ADP-Pi including systems indicate the time point that one or both ATP are converted to ADP-Pi from the ATP/ATP system.
FIGURE 4.
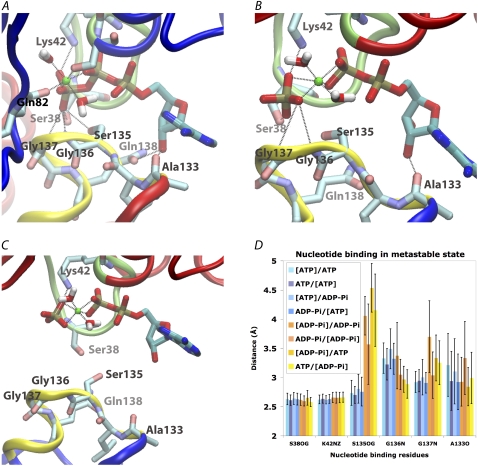
Nucleotide binding site configurations. (A–C) Representative nucleotide binding site configurations in different systems. Dashed lines represent hydrogen bond or Mg2+-oxygen coordination. (A) The ATP-bound state of site A of the ATP/ATP system at the end of the simulation (t = 78.25 ns). (B) The ADP-Pi state of site B of ATP/ADP-Pi system at t = 1 ns after hydrolysis. (C) The ADP-Pi state of site B of ATP/ADP-Pi system at the end of simulation (t = 70 ns after hydrolysis). The LSGGQ motif is already completely dissociated from the nucleotide. (D) Comparison of the nucleotide binding sites in the ATP-bound state and metastable ADP-Pi state. Average distances between hydrogen bond partners from protein and nucleotides at ATP-bound sites (blue bars) or at ADP-Pi-bound sites (yellow/orange bars). The brackets in the legends indicate the nucleotide binding sites used in the plot. All distances are sampled every 5 ps and averaged over the simulation time before dimer opening started (Fig. 3): 78.25 ns for both sites A and B in the ATP/ATP systems; 70 ns for site A of the ADP-Pi/ADP-Pi system; 50 ns for site A of the ADP-Pi/ATP system and 40 ns for site B of this system; 30 ns for site B of the ATP/ADP-Pi system; 20 ns for site B of the ADP-Pi/ADP-Pi system and site A of the ATP-/ADP-Pi system.
RESULTS AND DISCUSSION
To study the effect of ATP hydrolysis and to investigate the stoichiometry of NBDs in ABC transporters, we used the crystal structure of the ATP-bound, closed form of MalK (PDB entry 1Q12 (10)) for MD simulations (Fig. 1 A). The two nucleotide binding sites in ABC transporters are located between the two monomers, and the sites are mainly formed by the Walker A motif of the cis monomer and the LSGGQ motif of the trans monomer. For clarity, we named the binding site between the Walker A motif of chain C and the LSGGQ motif of chain D as site A and the other binding site as site B (Fig. 1 A, inset).
Four simulation systems were built based on the crystal structure with different active site configurations: 1), ATP/ATP to represent the original crystal structure and to use as a control; 2), ADP-Pi/ADP-Pi to simulate the effect of simultaneous ATP hydrolysis at both active sites; and 3), two hybrid ATP/ADP-Pi systems with the ADP-Pi at either site A or site B to study the effect of single ATP hydrolysis. With 70 ns of MD simulations on each of the four systems, our aim was to cover all possible patterns of ATP hydrolysis in this dimeric system and to study the mechanism of conformational changes induced by the hydrolysis event.
Dimer opening is a direct effect of ATP hydrolysis
The most apparent consequence of ATP hydrolysis in all three ADP-Pi containing systems is the opening of the dimer interface. Regardless of the number of hydrolyzed ATP molecules (either in site A or B or both), the closed dimer appears to be destabilized by ATP hydrolysis, which eventually results in the dissociation of the LSGGQ motif from one or both of the bound nucleotides (Fig. 2). Because the interactions between the two NBDs are dominated by a hydrogen bond network around ATP, especially with the γ-phosphate, the dissociation of the LSGGQ motif from the nucleotide can lead to the separation of the two monomers.
FIGURE 2.
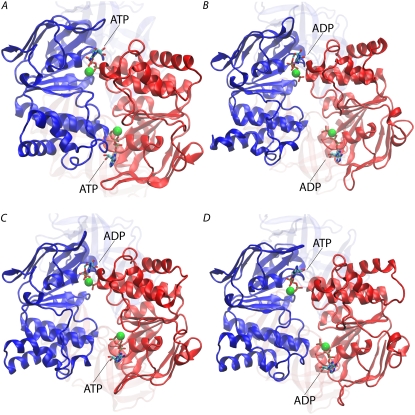
Dimer opening induced by ATP hydrolysis. The four panels are the final structure of the four simulation systems; (A) ATP/ATP, (B) ADP-Pi/ADP-Pi, (C) ADP-Pi/ATP, and (D) ATP/ADP-Pi. Whereas ATP/ATP dimer shows a stable, closed structure throughout the simulation, all the other systems exhibit clear opening of one or both active sites. The regulatory domains are shown in faded colors.
To quantitate the dimer separation, the distance between the RecA-like subdomain of the cis monomer and the helical subdomain of the trans monomer, as well as the distances between the two major nucleotide binding motifs (Walker A and LSGGQ) across the binding site, were monitored during the simulations (Fig. 3 A and E–H, top panels). To characterize the interactions within the binding site, the contacts between the γ-phosphate and the key binding residues of the LSGGQ motif (Ser-135, Gly-136, and Gly-137 (Fig. 3 B)), the residues between the ribose ring and the helical subdomain (Ala-133 and Gln-138 (Fig. 3 C)), and a direct (nucleotide-independent) contact between conserved residues from the two monomers (Ser-38 and Asp-165 (Fig. 3 D; see Fig. S2 in Data S1)) were also measured as the distance between respective hydrogen bonding partners (Fig. 3, E–H, lower three panels).
These distances in the ATP/ATP system clearly indicate a highly stable dimeric structure stabilized by tightly bound ATP (Fig. 3 E). In contrast, dimer separation at one or both nucleotide binding sites are observed in all three of the other systems after ATP hydrolysis (Fig. 2, B–D). The increasing intersubdomain distances in the three posthydrolysis systems closely correlate with the increased distances between the LSGGQ motif and the substrate/product (Fig. 3, F–H), as well as the unbinding of ribose from the helical subdomain, especially from the backbone carbonyl of Ala-133. In other words, the dimer separation is accompanied by the dissociation of the LSGGQ motif from the nucleotide, which requires the release of the nucleotide from the LSGGQ motif region at both the phosphate and ribose ends. The nucleotide binding site B is disrupted in the ADP-Pi/ADP-Pi and ADP-Pi/ATP systems due to the separation of the two flanking subdomains (Figs. 2, B and C, and 3, F and G), whereas both active sites are disrupted in the ATP/ADP-Pi system by the same mechanism (Figs. 2 D and 3 H).
During the dimer opening, the intersubdomain distance increases from ∼30.5 Å to >33 Å once all three hydrogen bonds between the LSGGQ motif and the Pi, as well as the hydrogen bond between the ribose ring and the helical subdomain, rupture at the same time (Fig. 3, F–H). The open states captured in our simulations resemble that of the MalK dimer in the semiopen crystal structure (PDB entry 1Q1B (10)). In the latter, the intersubdomain distance is ∼33.5 Å and the only connection between the two NBDs is provided by a hydrogen bond between Ser-38 of the Walker A motif and the conserved Asp-165 of the D-loop of the other monomer (this hydrogen bond is partially disrupted in our ADP-Pi-bound sites, as discussed later). The rotation of the helical subdomains, which appears to constitute a key structural difference observed in the ADP-bound or nucleotide-free crystal structures, is not observed in any of our posthydrolysis simulations (see Fig. S3 in Data S1). The helical subdomain rotation might require the dissociation of the inorganic phosphate from the ADP and Mg2+.
As the dimer opens between the Walker A motif of the cis monomer and the LSGGQ motif of the trans monomer, ADP remains bound to the Walker A motif and with Pi tightly associated with the nucleotide through the Mg2+ ion. Moreover, the connection of the side chains of Ser-38 and Lys-42 in the Walker A motif to the ATP γ-phosphate is also preserved in the ADP-Pi-bound form, even after the LSGGQ motif is completely dissociated (breaking all hydrogen bonds between the Pi and the LSGGQ motif, as illustrated in Fig. 4 C). Because the Pi in all of the ADP-Pi containing systems stays bound to the nucleotide binding site and yet dimer opening is consistently observed in all three posthydrolysis systems, the opening appears to be a direct consequence of ATP hydrolysis, which does not require the dissociation of the hydrolysis product.
The Hill coefficient of ATP hydrolysis is 1.7 or 2 for the NBDs measured (41–44), suggesting that ATP hydrolysis is carried out by a dimeric form of the NBDs. Furthermore, crystal structures of ATP-bound, monomeric forms of the NBDs are available for many ABC transporters (45–50), indicating that the dimer formation is required only for ATP hydrolysis and not for its binding. Notably, in both hybrid systems (ATP/ADP-Pi and ADP-Pi/ATP; Fig. 2, C–D), the LSGGQ motif is dissociated from the ATP-bound site once the dimer opening has been initiated shortly after ATP hydrolysis in the other binding site. This dissociation leaves the ATP-bound site nonhydrolytic. Therefore, if the second ATP hydrolysis does not take place within a short time after the first one, it will not take place at all during the remainder of the transport cycle.
Mechanism of hydrolysis-induced opening
Examining the details of structural changes of the nucleotide binding sites induced by ATP hydrolysis in all three ADP-Pi containing systems, the first immediate consequence of ATP hydrolysis is the rearrangement of the nascent Pi and the Mg2+ ion. Upon the conversion of γ-phosphate to Pi, the latter quickly departs from the remaining ADP moiety and changes its orientation such that the Mg2+ ion moves in between the β-phosphate of ADP and the newly formed Pi. The Mg2+ ion is now coordinated by four oxygen atoms from the two phosphates (Fig. 4 B) instead of two oxygen atoms in the ATP-bound state (Fig. 4 A). When the hydrolysis occurs in the nucleotide binding site A, where the Gln-82 is connected to the Mg2+ ion, the two coordination ligands of the Mg2+ ion provided by the protein in the ATP state (Ser-43 of the Walker A motif and Gln-82 of the Q-loop) are replaced by the two flanking phosphate species. The other two coordination ligands for Mg2+ are contributed by two water molecules, which remain tightly bound and do not exchange throughout the 70 ns simulation time in all the ADP-Pi containing systems (the two water molecules in the ATP-bound site of all the ATP containing systems are also tightly bound to the Mg2+ ion associated with ATP). The origin of the Pi and Mg2+ rearrangement in our simulations appears to be a repulsive electrostatic interaction between ADP and Pi, which are both negatively charged.
The rearrangement has two major effects on the protein structure. First, in site A, it alters the Q-loop conformations of chain C where the Mg2+-Gln-82 coordination is lost. This alteration can be observed in both the ADP-Pi/ADP-Pi and the ADP-Pi/ATP system where the ATP molecule in site A is hydrolyzed. In both simulation systems, once ATP is hydrolyzed, the side chain of Gln-82 of chain C is immediately unleashed from the active site due to Mg2+ and Pi rearrangement. The unleashed Gln-82 eventually results in a disordered conformation of the Q-loop (see Fig. S4 in Data S1). Also, the Q-loop is shown to be coupled to the TMDs in the intact maltose transporter (19), as well as in other ABC transporters (14–18,20–22). The Q-loop in an intact ABC transporter, therefore, may not be fluctuating as much as we observed in our simulations.
The second local effect of ATP hydrolysis is the rupture of the hydrogen bond between the ATP γ-phosphate and the side chain of Ser-135 of the LSGGQ motif. On examination of the detailed interactions between the Pi and the nucleotide binding residues at the ADP-Pi-bound sites, it is noticeable that the distance between Ser-135 and Pi always increases significantly before the complete dissociation of the LSGGQ motif from Pi (Fig. 3, E–H, black traces in second panels). The rupture of the Ser-135-Pi hydrogen bond is due to the displacement of Pi. After the Mg2+-Pi rearrangement, the Pi moves slightly closer to Gly-136 (Fig. 4 B). The rupture of the Ser-135-Pi hydrogen bond seems to be a key step in destabilizing the nucleotide binding at the dimer interface, because it is the strongest hydrogen bond between the helical subdomain and the nucleotide (that is, it shows the shortest distance and the least fluctuation among the hydrogen bonds provided by the helical subdomains (Fig. 4 D)). Meanwhile, the dimer interface in the posthydrolysis sites is further destabilized by the partial rupture of the hydrogen bond between Ser-38 and Asp-165 (Fig. 3, E–H, bottom panels). Although this contact between the monomers is not directly mediated by nucleotide binding, Ser-38 is involved in γ-phosphate binding and its side chain remains bound to Pi after hydrolysis. Therefore, it is possible that the Mg2+ and Pi rearrangement changes the backbone configuration of Ser-38 slightly to maintain its side-chain binding, thus destabilizing the hydrogen bond across the dimer interface.
The opening of the dimer interface does not always start with the binding site at which ATP is hydrolyzed. In the ADP-Pi/ATP system, for instance, separation of the two monomers is more evident at the ATP-bound site. We attribute this behavior to the stochastic nature of the process. Hydrolysis of ATP at one or both binding sites results in destabilization of the dimer interface, increasing the probability of opening at both sites and allowing either one to open in a stochastic manner. One would still predict a higher probability of opening at the hydrolysis site. However, having only a single simulation per configuration does not allow us to provide sufficient statistics to examine this prediction.
Metastable ADP-Pi state of the nucleotide binding site
In a comparison of the sequence of events involved in dimer opening in three ADP-Pi containing systems (Fig. 3, F–H), no clear retention time can be identified between the hydrolysis and the departure of the LSGGQ motif from the Pi, reflecting the stochastic nature of the hydrolysis-induced dimer opening. However, a specific distance pattern among γ-phosphate binding residues can be found in all binding sites with ADP-Pi before the dimer opening. We can define this state in which local effects of ATP hydrolysis (within the binding site) have taken place, but the dimer opening has not been initiated as a metastable state. These local changes include an increased Ser-135-Pi distance and fluctuating Gly-137-Pi distance that lasts until the complete dissociation of the Pi from all three hydrogen bonds of the LSGGQ motif or until the end of simulations. These changes suggests a metastable ADP-Pi-bound state, which is shown in detail in Fig. 4 B.
A detailed comparison of this metastable ADP-Pi state and the ATP-bound state is shown in Fig. 4 D. Among the γ-phosphate binding residues, the connection between Ser-135 and Pi is affected most by the hydrolysis event, whereas the binding of Walker A residues (Ser-38 and Lys-42) is not affected at all. The tight connection of Walker A residues to phosphate lasts even after all the hydrogen bonds between the LSGGQ motif and the nucleotide are disrupted. In contrast, the Gly-136-Pi interaction is slightly strengthened in this metastable state, whereas the Gly-137-Pi connection is destabilized (Fig. 4 D). In contrast to the strong hydrogen bond between the Pi and Walker A residues or Ser-135 in the ATP-bound form, the hydrogen bond distances between the Pi and Gly-136 or Gly-137, as well as the hydrogen bond between ribose and Ala-133, indicate a weak binding in both the ATP-bound and ADP-Pi metastable states. However, the dimer opening requires the rupture of all four hydrogen bonds between the helical subdomain and the nucleotide, as shown in Fig. 3, F–H.
Conformation of Q-loop glutamine is independent of dimer opening
The conformations of the two Q-loops differ slightly in the MalK crystal structure. The differences between the two monomers is likely due to the absence of Mg2+ in the original crystal structure, which appears to be important in stabilizing the Q-loop conformation in the simulations. Apart from the Q-loops, the structures are mostly identical between the two MalK monomers (Fig. 1 B). With the Mg2+ ions placed in the two binding sites between the ATP β- and γ-phosphates, the local environments for the two Mg2+ ions are different in the initial structures of sites A and B. Both Mg2+ ions are hexacoordinated with five coordinating ligands identical at both sites, namely, the oxygen atoms of the ATP β- and γ-phosphates, the side chain of Ser-43, and two water molecules all contribute to the coordination of Mg2+ ion. The last coordination ligand for the Mg2+ ion at site A is the side-chain carbonyl of Gln-82 (Q-loop glutamine) of chain C, whereas the other Mg2+ ion at site B is coordinated by an additional water molecule (Fig. 1 A).
The two different starting structures affect the dynamics of the Q-loops in the two monomers, as revealed by the MD simulation of the ATP/ATP system. Lacking the anchorage point provided by the Gln-82-Mg2+ coordination, the Q-loop of chain D is more flexible and shows a large fluctuation and displacement during the simulation (Fig. 1 C; see Fig. S4 in Data S1). Water molecules, including the hydration shell of the bound nucleotides, freely diffuse at the dimer interface, with the exception of those water molecules directly coordinated by the two Mg2+ cations. The disordered Gln-82 of chain D probes various conformations over time, frequently approaching the Mg2+ ion. At some instances, Gln-82 indeed establishes a hydrogen bond with an immobile, Mg2+-bound water molecule. However, the linkage is too transient to stabilize the Q-loop conformation and the Gln-82 side chain is unable to replace the Mg2+-bound water during the 78.25 ns simulation time span. As mentioned above, the Q-loop is closely coupled to the TMD, and so its fluctuations could be significantly reduced in an intact ABC transporter.
It has been suggested that the Q-loop glutamine orients nucleophilic water needed for ATP hydrolysis through the binding of Mg2+ cofactor, as shown in the crystal structure of MJ0796 (9). Lacking the Gln-82-Mg2+ link in chain D can possibly render site B nonhydrolytic. However, the ATP binding between the two monomers and the stability of the dimer interface were not affected during our simulation of the ATP/ATP system. As shown in Fig. 3 B, the tight binding between the LSGGQ motif and the ATP γ-phosphate was maintained throughout the simulation, and the intersubdomain distances across the two nucleotide binding sites were essentially unaltered. The MalK dimer maintained its dimeric ATP-bound conformation throughout the entire simulation time (78.25 ns (Fig. 2 A)).
In contrast, even though the dimer is largely open in the ATP/ADP-Pi system at the end of the simulation, the Gln-82 of chain C remains tightly bound to the Mg2+ cofactor at site A (Fig. 2 D), and the Q-loop of chain C preserves its conformation as in the ATP-bound crystal structure (see Fig. S4 in Data S1). It can be concluded, therefore, that the dimer opening and the dissociation of the LSGGQ motif from the bound nucleotide are both independent of Gln-82-Mg2+ anchorage of the Q-loop.
Mechanistic implications
The stoichiometry of ATP hydrolysis and substrate transport in ABC transporters has long been a disputable subject (23,24,27). This stoichiometry was reported to be 1.4:1 in reconstituted maltose transporter (51), albeit much higher numbers were obtained for some of the experiments in the same study (51), possibly due to repetitive futile hydrolytic runs. Our results suggest that the closed dimeric MalK can only exist when both nucleotide binding sites are occupied by ATP, that is, ATP hydrolysis in either or both active sites can induce the opening of the MalK dimer. It is still unclear how ATP binding and hydrolysis trigger conformational changes of the TMDs, specifically whether one or more open-closed cycles of NBDs are required for substrate transport. However, assuming that the transport mechanism relies on a single conversion between the open and the closed states, our simulations suggest that one ATP hydrolysis event is sufficient to complete one transport cycle.
Several ABC transporters with only one hydrolytic nucleotide binding site have now been identified, for example, the well-known cystic fibrosis transmembrane conductance regulator (CFTR; reviewed by Gadsby and colleagues (52)) and the recently characterized yeast multidrug resistant exporter PDR5 (53). Both proteins contain nonconserved residues at key positions of one of the two nucleotide binding sites that abolish the ATP hydrolysis activity but preserve ATP binding. Yet, both proteins are fully functional ABC transporters (53–55). We note that ATP binding at both nucleotide binding sites is still required for NBD dimer formation and, hence, for ATP hydrolysis at the hydrolytic nucleotide binding site.
A mutant form of NBDs of Salmonella typhimurium histidine permease (HisP) including a hybrid dimer with one deactivated nucleotide binding site is also shown to be functional (56). The mutation at the corresponding location in MalK (H192R) severely impairs the functionality in hybrid MalK dimers (57). However, this impairment could result from a tighter arrangement of the NBD pair in MalK than in HisP, in which a lengthy side chain such as arginine would sterically hinder dimer association.
Other than HisP, a hybrid NBD dimer constructed in mouse multidrug resistant protein P-glycoprotein (MDR3) also shows that one mutant active site does not prevent transport (58). Furthermore, results of mixing catalysis-deficient and wild-type E. coli hemolysin A exporter HlyB-NBDs also agree with the mutation studies mentioned above (59). All these hybrid NBD dimers in ABC transporters demonstrate that single ATP hydrolysis can drive the transport but requires ATP binding at both binding sites.
Even though one ATP hydrolysis seems to be sufficient to trigger the opening of NBD dimers, we do not exclude the possibility of simultaneous or consecutive ATP hydrolysis events taking place in a single closed to open cycle for a symmetric NBD dimer such as MalK. Given that the two active sites share, on average, an equal probability of hydrolysis and that both the formation and the full disruption of the active sites take a certain amount of time to complete, a second ATP hydrolysis event could take place before the dimer opening is complete. Especially in an intact ABC transporter, where the opening of NBD dimers are coupled to conformational changes of massive TMDs (19,60), the rate of dimer opening might be much slower than the one in our simulations, thus providing more opportunity for a second hydrolysis event. Therefore, the average ATP hydrolysis per cycle is predicted to be between one and two for symmetric NBD dimers, which is consistent with the reported stoichiometry for the maltose transporter of E. coli (51).
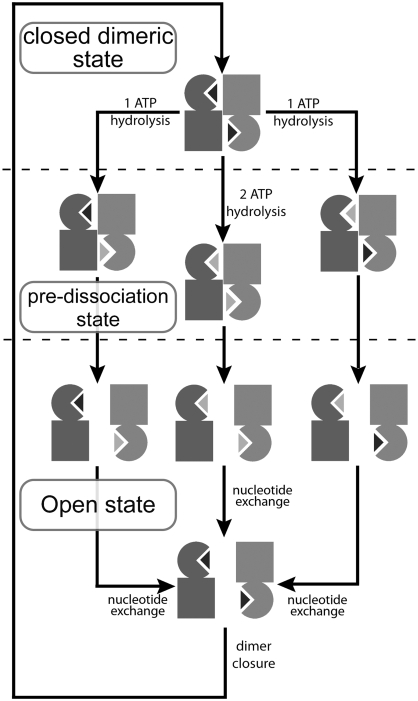
Another open question regarding the transport mechanism is whether the two active sites work in a synchronized manner or alternatively (24,26). Based on our simulation results, ATP hydrolysis events in the two active sites do not have to be simultaneous nor in a certain order to open a MalK dimer. Because ATP hydrolysis in either or both active sites can trigger domain opening, it is possible that the two sites work in a stochastic manner and that the transport cycle can be initiated with either or both ATP molecules hydrolyzed (Fig. 5). However, if the substrate transport is more frequently coupled to a single ATP hydrolysis cycle, it is more likely for symmetric NBD dimers to have a better chance to operate alternatively. This likelihood can be attributed to the conservation of Mg2+ binding from the Q-loop glutamine in the ATP occluded active site, after ATP hydrolysis in the trans active site and the following dimer separation. Because the Q-loop glutamine at the posthydrolytic binding site is unleashed from the nucleotide, it requires more time to probe its conformational space so that it can settle appropriately close to the newly associated ATP-Mg2+ complex and orient the nucleophilic water for the next round of hydrolysis. In contrast, because of the preservation of the Gln-Mg2+ bond, the ATP-bound site can quickly reach the hydrolysis-ready state upon dimer association. This agility could be a possible origin for what Higgins and Linton call the “memory of previous nucleotide state” (24), a requirement of the NBD monomer if the dimeric NBDs hydrolyze ATP in an alternating manner.
FIGURE 5.
Proposed mechanism for ATP binding and hydrolysis. MalK dimers are shown in the inset of Fig. 1 A. The bound nucleotides are shown as dark (ATP) or light (ADP) gray triangles. The transport cycle is composed of alternations between an open and a closed dimer in which the closing transition can only be achieved by ATP binding in both nucleotide binding sites. The opening motion can be accomplished by ATP hydrolysis in either or both nucleotide binding sites.
CONCLUSIONS
We have performed extended MD simulations on the dimeric form of the NBDs of MalK in all possible combinations of the protein, ATP, and ADP-Pi, to study the immediate effect of ATP hydrolysis on the structure and dynamics of the dimer. Within the time scale of our simulations, we found the dimer to be very stable when bound to two ATP molecules, whereas ATP hydrolysis in either or both binding sites was found to induce the opening of the dimer. Notably, to capture the process of opening, simulations on the order of at least tens of nanosecond are required. We conclude, therefore, that a single ATP hydrolysis is sufficient to induce the opening of the dimer during the transport cycle. The opening of the dimer seems to be directly induced by ATP hydrolysis in either one or both of the two nucleotide binding sites rather than being an effect of the dissociation of hydrolysis products from the active sites. The separation of the LSGGQ motif from the nucleotide, which is induced by rearrangement of the nascent Pi and the bound Mg2+ ion in the active site after hydrolysis, appears to be a key step in the opening of the dimer.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
The authors thank Dr. Amy L. Davidson for careful reading of our manuscript and insightful comments.
Simulations in this study were performed at the Bigred cluster of Indiana University and the Abe cluster at National Center for Supercomputing Applications (Teragrid grant number MCA06N060), along with Turing cluster in University of Illinois at Urbana-Champaign.
Editor: Edward H. Egelman.
References
- 1.Holland, I. B., S. P. C. Cole, K. Kuchler, and C. F. Higgins. 2003. ABC Proteins: From Bacteria to Man. Academic Press, London.
- 2.Dean, M., and T. Annilo. 2005. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 6:123–142. [DOI] [PubMed] [Google Scholar]
- 3.Gillet, J.-P., T. Efferth, and J. Remacle. 2007. Chemotherapy-induced resistance by ATP-binding cassette transporter genes. Biochim. Biophys. Acta. 1775:237–262. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241–268. [DOI] [PubMed] [Google Scholar]
- 5.Albers, S.-V., S. M. Koning, W. N. Konings, and A. J. M. Driessen. 2004. Insights into ABC transport in archaea. J. Bioenerg. Biomembr. 36:5–15. [DOI] [PubMed] [Google Scholar]
- 6.Holland, I. B., L. Schmitt, and J. Young. 2005. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway. Mol. Membr. Biol. 22:29–39 [review]. [DOI] [PubMed] [Google Scholar]
- 7.Doerrler, W. T. 2006. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol. Microbiol. 60:542–552. [DOI] [PubMed] [Google Scholar]
- 8.Lubelski, J., W. N. Konings, and A. J. M. Driessen. 2007. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 71:463–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith, P. C., N. Karpowich, L. Millen, J. E. Moody, J. Rosen, P. J. Thomas, and J. F. Hunt. 2002. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell. 10:139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., G. Lu, J. Lin, A. L. Davidson, and F. A. Quiocho. 2003. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell. 12:651–661. [DOI] [PubMed] [Google Scholar]
- 11.Lu, G., J. M. Westbrooks, A. L. Davidson, and J. Chen. 2005. ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation. Proc. Natl. Acad. Sci. USA. 102:17969–17974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaitseva, J., S. Jenewein, T. Jumpertz, I. B. Holland, and L. Schmitt. 2005. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 24:1901–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaitseva, J., C. Oswald, T. Jumpertz, S. Jenewein, A. Wiedenmann, I. B. Holland, and L. Schmitt. 2006. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. EMBO J. 25:3432–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 296:1091–1098. [DOI] [PubMed] [Google Scholar]
- 15.Pinkett, H. W., A. T. Lee, P. Lum, K. P. Locher, and D. C. Rees. 2007. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science. 315:373–377. [DOI] [PubMed] [Google Scholar]
- 16.Dawson, R. J. P., and K. P. Locher. 2007. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 581:935–938. [DOI] [PubMed] [Google Scholar]
- 17.Hollenstein, K., D. C. Frei, and K. P. Locher. 2007. Structure of an ABC transporter in complex with its binding protein. Nature. 446:213–216. [DOI] [PubMed] [Google Scholar]
- 18.Hvorup, R. N., B. A. Goetz, M. Niederer, K. Hollenstein, E. Perozo, and K. P. Locher. 2007. Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science. 317:1387–1390. [DOI] [PubMed] [Google Scholar]
- 19.Oldham, M. L., D. Khare, F. A. Quiocho, A. L. Davidson, and J. Chen. 2007. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 450:515–521. [DOI] [PubMed] [Google Scholar]
- 20.Ward, A., C. L. Reyes, J. Yu, C. B. Roth, and G. Chang. 2007. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl. Acad. Sci. USA. 104:19005–19010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber, S., M. Comellas-Bigler, B. A. Goetz, and K. P. Locher. 2008. Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science. 321:246–250. [DOI] [PubMed] [Google Scholar]
- 22.Kadaba, N. S., J. T. Kaiser, E. Johnson, A. Lee, and D. C. Rees. 2008. The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science. 321:250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patzlaff, J. S., T. van der Heide, and B. Poolman. 2003. The ATP/substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 278:29546–29551. [DOI] [PubMed] [Google Scholar]
- 24.Higgins, C. F., and K. J. Linton. 2004. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11:918–926. [DOI] [PubMed] [Google Scholar]
- 25.van der Does, C., and R. Tampé. 2004. How do ABC transporters drive transport? Biol. Chem. 385:927–933. [DOI] [PubMed] [Google Scholar]
- 26.Oswald, C., I. B. Holland, and L. Schmitt. 2006. The motor domains of ABC-transporters. What can structures tell us? Naunyn Schmiedebergs Arch. Pharmacol. 372:385–399. [DOI] [PubMed] [Google Scholar]
- 27.Dawson, R. J. P., K. Hollenstein, and K. P. Locher. 2007. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Mol. Microbiol. 65:250–257. [DOI] [PubMed] [Google Scholar]
- 28.Oloo, E. O., and D. P. Tieleman. 2004. Conformational transitions induced by the binding of MgATP to the vitamin B12 ATP-binding cassette (ABC) transporter BtuCD. J. Biol. Chem. 279:45013–45019. [DOI] [PubMed] [Google Scholar]
- 29.Campbell, J. D., and M. S. P. Sansom. 2005. Nucleotide binding to the homodimeric MJ0796 protein: a computational study of a prokaryotic ABC transporter NBD dimer. FEBS Lett. 579:4193–4199. [DOI] [PubMed] [Google Scholar]
- 30.Oloo, E. O., E. Y. Fung, and D. P. Tieleman. 2006. The dynamics of the MgATP-driven closure of MalK, the energy-transducing subunit of the maltose ABC transporter. J. Biol. Chem. 281:28397–28407. [DOI] [PubMed] [Google Scholar]
- 31.Ivetac, A., J. D. Campbell, and M. S. P. Sansom. 2007. Dynamics and function in a bacterial ABC transporter: simulation studies of the BtuCDF system and its components. Biochemistry. 46:2767–2778. [DOI] [PubMed] [Google Scholar]
- 32.Sonne, J., C. Kandt, G. H. Peters, F. Y. Hansen, M. Ø. Jensen, and D. P. Tieleman. 2007. Simulation of the coupling between nucleotide binding and transmembrane domains in the ATP binding cassette transporter BtuCD. Biophys. J. 92:2727–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, P. M., and A. M. George. 2007. Nucleotide-dependent allostery within the ABC transporter ATP-binding cassette. J. Biol. Chem. 282:22793–22803. [DOI] [PubMed] [Google Scholar]
- 34.Phillips, J. C., R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kale, and K. Schulten. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKerell, A. D., Jr., M. Feig, and C. L. Brooks III. 2004. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25:1400–1415. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 37.Martyna, G. J., D. J. Tobias, and M. L. Klein. 1994. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101:4177–4189. [Google Scholar]
- 38.Feller, S. E., Y. H. Zhang, R. W. Pastor, and B. R. Brooks. 1995. Constant pressure molecular dynamics simulation — the Langevin piston method. J. Chem. Phys. 103:4613–4621. [Google Scholar]
- 39.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald. An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 40.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD : visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 41.Liu, C. E., P.-Q. Liu, and G. F.-L. Ames. 1997. Characterization of the adenosine triphosphatase activity of the periplasmic histidine permease, a traffic ATPase (ABC transporter). J. Biol. Chem. 272:21883–21891. [DOI] [PubMed] [Google Scholar]
- 42.Moody, J. E., L. Millen, D. Binns, J. F. Hunt, and P. J. Thomas. 2002. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J. Biol. Chem. 277:21111–21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janas, E., M. Hofacker, M. Chen, S. Gompf, C. van der Does, and R. Tampé. 2003. The ATP hydrolysis cycle of the nucleotide-binding domain of the mitochondrial ATP-binding cassette transporter Mdl1p. J. Biol. Chem. 278:26862–26869. [DOI] [PubMed] [Google Scholar]
- 44.Benabdelhak, H., L. Schmitt, C. Horn, K. Jumel, M. A. Blight, and I. B. Holland. 2005. Positive co-operative activity and dimerization of the isolated ABC ATPase domain of HlyB from Escherichia coli. Biochem. J. 386:489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung, L.-W., I. X. Wang, K. Nikaido, P.-Q. Liu, G. F.-L. Ames, and S.-H. Kim. 1998. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 396:703–707. [DOI] [PubMed] [Google Scholar]
- 46.Verdon, G., S. V. Albers, B. W. Dijkstra, A. J. Driessen, and A.-M. W. Thunnissen. 2003. Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations. J. Mol. Biol. 330:343–358. [DOI] [PubMed] [Google Scholar]
- 47.Verdon, G., S. V. Albers, N. van Oosterwijk, B. W. Dijkstra, A. J. Driessen, and A.-M. W. Thunnissen. 2003. Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus. J. Mol. Biol. 334:255–267. [DOI] [PubMed] [Google Scholar]
- 48.Ose, T., T. Fujie, M. Yao, N. Watanabe, and I. Tanaka. 2004. Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3. Proteins. 57:635–638. [DOI] [PubMed] [Google Scholar]
- 49.Lewis, H. A., S. G. Buchanan, S. K. Burley, K. Conners, M. Dickey, M. Dorwart, R. Fowler, X. Gao, W. B. Guggino, W. A. Hendrickson, J. F. Hunt, M. C. Kearins, D. Lorimer, P. C. Maloney, K. W. Post, K. R. Rajashankar, M. E. Rutter, J. M. Sauder, S. Shriver, P. H. Thibodeau, P. J. Thomas, M. Zhang, X. Zhao, and S. Emtage. 2004. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 23:282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis, H. A., X. Zhao, C. Wang, J. M. Sauder, I. Rooney, B. W. Noland, D. Lorimer, M. C. Kearins, K. Conners, B. Condon, P. C. Maloney, W. B. Guggino, J. F. Hunt, and S. Emtage. 2005. Impact of the ΔF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J. Biol. Chem. 280:1346–1353. [DOI] [PubMed] [Google Scholar]
- 51.Davidson, A. L., and H. Nikaido. 1990. Overproduction, solubilization, and reconstitution of the maltose transport system from Escherichia coli. J. Biol. Chem. 265:4254–4260. [PubMed] [Google Scholar]
- 52.Gadsby, D. C., P. Vergani, and L. Csanady. 2006. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 440:477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ernst, R., P. Kueppers, C. M. Klein, T. Schwarzmueller, K. Kuchler, and L. Schmitt. 2008. A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proc. Natl. Acad. Sci. USA. 105:5069–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aleksandrov, L., A. A. Aleksandrov, X. B. Chang, and J. R. Riordan. 2002. The first nucleotide binding domain of cystic fibrosis transmembrane conductance regulator is a site of stable nucleotide interaction, whereas the second is a site of rapid turnover. J. Biol. Chem. 277:15419–15425. [DOI] [PubMed] [Google Scholar]
- 55.Basso, C., P. Vergani, A. C. Nairn, and D. C. Gadsby. 2003. Prolonged nonhydrolytic interaction of nucleotide with CFTR's NH2-terminal nucleotide binding domain and its role in channel gating. J. Gen. Physiol. 122:333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikaido, K., and G. F.-L. Ames. 1999. One intact ATP-binding subunit is sufficient to support ATP hydrolysis and translocation in an ABC transporter, the histidine permease. J. Biol. Chem. 274:26727–26735. [DOI] [PubMed] [Google Scholar]
- 57.Davidson, A. L., and S. Sharma. 1997. Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J. Biol. Chem. 179:5458–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tombline, G., L. A. Bartholomew, G. A. Tyndall, K. Gimi, I. L. Urbatsch, and A. E. Senior. 2004. Properties of P-glycoprotein with mutations in the “catalytic carboxylate” glutamate residues. J. Biol. Chem. 279:46518–46526. [DOI] [PubMed] [Google Scholar]
- 59.Zaitseva, J., S. Jenewein, A. Weidenmann, H. Benabdelhak, I. B. Holland, and L. Schmitt. 2005. Functional characterization and ATP-induced dimerization of the isolated ABC-domain of the haemolysin B transporter. Biochemistry. 44:9680–9690. [DOI] [PubMed] [Google Scholar]
- 60.Hollenstein, K., R. J. Dawson, and K. P. Locher. 2007. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17:412–418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.