Abstract
The three-dimensional organization of nuclear compartments within living cells determines genome function and yet their underlying self-organizing principles are unclear. We visualize in real-time transcriptionally active compartments (TCs) by the transient enrichment of fluorescently-labeled uridine 5′-triphosphate molecules within living cells. These TCs partially colocalize with active RNA-Pol II in the cell nucleus. Fluorescence anisotropy maps of chromatin compaction evidences a more open chromatin structure at the TCs. Using live-cell timelapse imaging, heterogeneity in the dynamic behavior of TCs has been revealed which falls into three distinct classes: subdiffusive, super-diffusive, and normal diffusive behavior. In contrast, the mobility of a candidate gene locus, either in the repressed or activated state, undergoes a differential restricted motion that is coupled to TC movement. Further TC dynamics is directly affected by small molecule chromatin structure modulators and adenosine triphosphate depletion. This heterogeneous behavior in TC dynamics within living cells could provide an interesting paradigm to explore the spatiotemporal dimension to gene transcription control.
INTRODUCTION
Recent experiments have revealed that the eukaryotic nucleus is spatially, temporally, and functionally compartmentalized within living cells (1,2). Chromosomes, DNA complexed with histone, and other nuclear proteins, are nonrandomly organized and occupy distinct territories within the nucleus. Further chromosome positions are found to be maintained through mitosis and are organized into gene-rich euchromatin and compact heterochromatin regions within the crowded three-dimensional architecture of the nucleus (3,4). In addition, the nuclear lamina, that forms the structural scaffold, is found to play an important role in maintaining nuclear organization and global transcription levels (5,6). Functional compartments, such as the nuclear speckles, transcription and replication factories, Cajal bodies, and other nuclear organelles are found to be interspersed between chromosome territories (7,8). Current studies have uncovered that some of these nuclear compartments and their components are mobile within the nucleus and their dynamic mobility is found to be linked with transient interactions with chromatin regions to elicit functional responses (9–13). Despite this, the links between their dynamic organizations in determining genome function remain unclear.
Conversely compartments with RNA polymerases are found to be spatially distributed in distinct immobile submicron regions of the nucleus called “transcription factories” (14). Genes are found to actively reposition into immobile transcription factories, enriched in nuclear proteins and nucleoside triphosphates required for the transcription control of a fully functional mRNA (15). In this article, we visualize in real-time transcriptionally active compartments (TCs) via the transient enrichment of fluorescently-labeled uridine 5′-triphosphate (UTP) molecules within living cell nuclei, which partially colocalize with active RNA-Pol II. We probe the status of chromatin compaction at TCs using fluorescence anisotropy imaging. Using live-cell timelapse imaging, we observe that these TCs are dynamic within the cell nucleus, exhibiting heterogeneity in their dynamics. TC mobility can be grouped into three distinct classes: subdiffusive, superdiffusive, and normal diffusive behavior. In contrast, the mobility of a candidate gene locus undergoes a restricted motion that is coupled to TC dynamics. Further TC mobility is critically dependent on the spatiotemporal organization of chromatin, temperature, and adenosine triphosphate (ATP). This heterogenous behavior in TC dynamics within living cells could provide an interesting paradigm to explore the spatiotemporal dimension to gene transcription control within a dynamic cell nucleus.
METHODS
Cell culture, inhibitors, and antibody stain
Wild-type HeLa cells and cells stably expressing H2B-EGFP were grown in Dulbecco's Eagle Medium (DMEM) containing 5% fetal bovine serum (FBS) at 37°C in 5% CO2. Trichostatin-A (TSA) treatment involved addition of 100 ng/ml TSA (Sigma, St. Louis, MO) and incubation for various time periods. Adenosine triphosphate (ATP) depletion was done by 1 h incubation with 10 mM sodium azide and 6 mM 2-deoxy-d-glucose in M1 buffer without glucose. Cells were synchronized at early S phase by incubation with 10 μM Aphidicolin (Sigma) for 16 h. Antibody labeling of active RNA-Pol II (Abcam, Cambridge, UK) was carried out as described in Sproul et al. (3).
Fluorescent-labeled UTP incorporation
The protocol of uridine 5′-triphosphate (UTP) incorporation is functionally similar to the incorporation of 5-bromouridine 5′-triphosphate (BR-UTP) to mark transcription sites in fixed cell preparations (16). Here Texas-Red (TR) labeled UTP (Molecular Probes, Eugene, OR) or Cy5(Cyanine5)-UTP was used to mark TC. UTP molecules were incorporated into cells by a brief hypotonic shock with Krebs-Henseleit-bicarbonate buffer (10 mM HEPES at pH 7.4, 30 mM KCl) containing 10 μM UTP for 5–10 min. Cells are then washed with DMEM and incubated with 5% FBS at 37°C in 5% CO2 for 5–10 min before imaging. The imaging medium used throughout is 5% FBS in DMEM without phenol red.
Gene locus marking
For labeling gene loci, 96 lacO repeats were introduced upstream of the Tet responsive element sequence preceding the PCMVmin promoter (BD Biosciences, San Jose, CA) driving the enhanced green fluorescent proteins (EGFP) gene. This plasmid was transiently cotransfected in HeLa cells with another plasmid coding for mRFP-LacI-NLS that localizes to the nucleus and binds specifically to lacO sequence. To regulate gene expression, tetracycline controlled transcriptional silencer (tTS) was transiently expressed using pTet-tTS vector (Clontech). Forty micromolar of 5,6-dichloro-1-ß-D-ribobenzimidazole (DRB) (Sigma) were added to cells for 1 h for inhibiting transcription elongation. For de-repression, the medium was changed and cells incubated in the fresh non-DRB medium for 16 h.
Live-cell imaging
Timelapse single-plane images or Z-stacks of TC and gene locus were acquired on a Zeiss LSM 510 scanning confocal microscope using 63×, 1.4 NA objective. To avoid the cross-talk between chromatin (marked with H2B-EGFP), TC (marked with Texas Red (TR)-UTP or Cy5-UTP), or gene locus (marked with lacI-mRFP), they were imaged in multitrack mode.
Single-particle tracking analysis
To obtain the trajectory 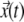 for individual TC or gene locus, a LabVIEW program (National Instruments, Austin, TX) was written to track their center of mass. Using
for individual TC or gene locus, a LabVIEW program (National Instruments, Austin, TX) was written to track their center of mass. Using 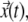 the average velocity of TC was calculated by windowing displacements of the TC in different time intervals ranging from the ∼1.5 s to 400 s. The trajectories of a minimum of 30 TC were pooled from different cells to estimate their average velocity distribution. Using trajectories
the average velocity of TC was calculated by windowing displacements of the TC in different time intervals ranging from the ∼1.5 s to 400 s. The trajectories of a minimum of 30 TC were pooled from different cells to estimate their average velocity distribution. Using trajectories 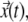 the mean-square displacement for each TC was calculated using
the mean-square displacement for each TC was calculated using 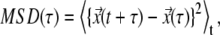 where
where 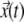 represents position of particle at time t, and τ is arbitrary interval of time. The parameters determining the mean-squared displacement, D the diffusion constant, and α are then used to characterize the nature of TC dynamics (as described in Fig. S2 in Supplementary Material, Data S1).
represents position of particle at time t, and τ is arbitrary interval of time. The parameters determining the mean-squared displacement, D the diffusion constant, and α are then used to characterize the nature of TC dynamics (as described in Fig. S2 in Supplementary Material, Data S1).
Fluorescence anisotropy imaging
The fluorescence image of the H2B-EGFP cell nuclei is split into two components based on its polarization. These two (I‖ and I⊥) images are acquired simultaneously/sequentially on different or same charge-coupled device cameras (Roper Scientific, Trenton, NJ). The fluorescence anisotropy image r is constructed by computing the anisotropy for each pixel using the relation 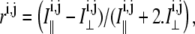 where i, j are the indices of a pixel in the respective image. The anisotropy values depend on the rotational correlation time of the fluorescent probe (H2B-EGFP) in comparison to its fluorescence lifetime, hence allowing us to assay the relative compaction of the chromatin assembly.
where i, j are the indices of a pixel in the respective image. The anisotropy values depend on the rotational correlation time of the fluorescent probe (H2B-EGFP) in comparison to its fluorescence lifetime, hence allowing us to assay the relative compaction of the chromatin assembly.
RESULTS
Marking UTP-enriched transcription compartments (TC) within living cells
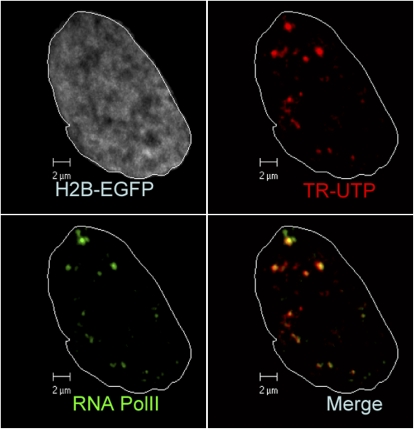
To visualize TC, fluorescently-labeled Texas Red UTP (TR-UTP) molecules were incorporated into the nucleus within living HeLa cells stably expressing histone H2B fused with EGFP (H2B-EGFP) as described in Methods. UTP molecules localize via transient enrichment in transcriptionally active compartments (TC) both in the nucleus and the cytoplasm. Fig. 1 shows the enrichment of TR-UTP at certain pockets inside the nucleus. The three-dimensional reconstruction of the marked foci is shown in Fig. S1 a in Data S1. Antibody staining of active RNA-Pol II showed partial colocalization with TC (Fig. 1), establishing that a fraction of them in the nucleus are transcription factories. In addition, those TCs in the cytoplasm are colocalized with mitochondria marked by MitoTracker Red (Fig. S1 b in Data S1).
FIGURE 1.
Fluorescent labeling of TC in living HeLa cell nucleus: TCs (red) are shown to colocalize with RNA-Pol II antibody (green) in cell nucleus where H2B-EGFP is shown in gray. White line marks out the nuclear boundary Scale bar: 2 μm.
Chromatin compaction at transcription compartments
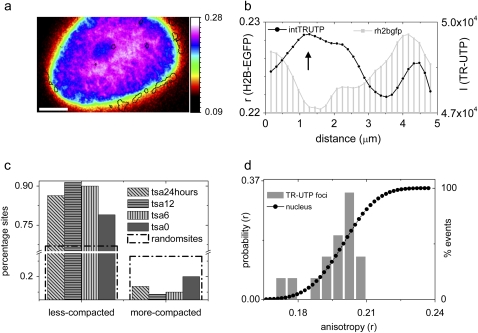
To gain insight into the state of chromatin architecture at TCs, we next investigated chromatin compaction proximal to TC in untreated HeLa cells using fluorescence anisotropy imaging method as described before (17). For this, TR-UTP molecules were incorporated in HeLa cells stably expressing the core histone H2B-EGFP. The anisotropy profile of H2B-EGFP as well as the intensity profile of TR-UTP incorporated within TC was acquired to estimate the chromatin compaction around the TC. Fig. 2 a is a color-coded anisotropy profile of H2B-EGFP reflecting the spatiotemporal heterogeneity in chromatin compaction within the nucleus where the location of TCs is indicated by a black outline. TCs are seen to localize more often in regions of lowered chromatin compaction. A typical line scan across a TC (Fig. 2 b) shows lowering of H2B-EGFP anisotropy values at the regions of maximal TR-UTP intensity signifying transcriptionally active regions indicated by UTP enrichment. Detailed analysis was performed on multiple TC sites (∼100) to estimate the frequency of lower chromatin decompaction at TC. For quantification, the analysis involved finding the ratios of H2B-EGFP anisotropy values averaged for the circular region under the TR-UTP peaks to that averaged for a neighboring annular region. A ratio > 1 suggests a more compacted state of the chromatin at TC whereas a ratio <1 implies a less-compacted state. A plot of the percentage of sites having decompacted chromatin in comparison to a neighboring concentric region in normal cells as well as TSA-treated cells for different durations is shown in Fig. 2 c. A significant fraction (∼80%) of TCs exhibited similar chromatin decompaction in control as well as TSA-treated cells in comparison to 66% as expected if the sites were to be randomly arranged in the nucleus (Fig. 2 c). The percentage of TC sites above the dotted line in Fig. 2 c was statistically significant. Fig. 2 d plots the cumulative anisotropy distribution of all pixels inside the nucleus and the anisotropy distribution of pixels under TC. TCs localize at regions of varied chromatin compaction and it is evident that regions of relatively high compaction (r > 0.22) are avoided as shown in Fig. 2 d.
FIGURE 2.
Analysis of TC and chromatin compaction: (a) Color-coded anisotropy profile of H2B-EGFP with solid lines marking TCs. (b) A typical line scan across a TC showing the TR-UTP intensity profile (solid, right axis) and the underlying H2B-EGFP anisotropy (r) profile (shaded, left axis). (c) Percentage sites which have underlying chromatin relatively more or less compacted then the neighborhood for TCs sites in HeLa cells under various hours of TSA treatment and for randomly chosen points inside the nucleus. (d) Cumulative distribution of fluorescence anisotropy of the nucleus (solid circles). The shaded bars represent anisotropy distribution below TC foci.
ATP-dependent dynamic organization of TCs
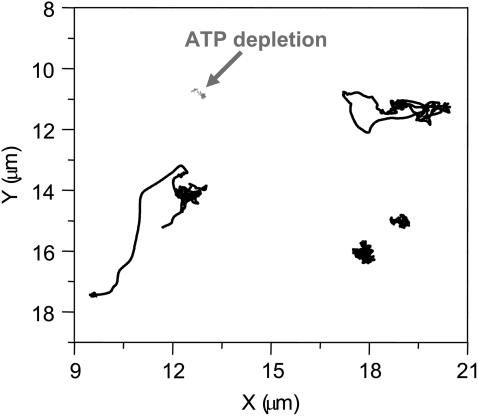
Importantly, dual-track, timelapse confocal microscopy images of TC marked H2B-EGFP cells, acquired at 1.6-s intervals, revealed their dynamics within the cell nucleus (Fig. 3, and Movie S1 in Data S1). A plot of trajectories, deduced from single-particle tracking analysis (Methods), exhibited by different TC within the same nucleus is shown in Fig. 3. From these trajectories, a fraction of TC can directionally move up to a few microns while others are much more restricted to less than micron-scale mobility. Depletion of adenosine triphosphate (ATP) in these cells reduces the mobility of TCs considerably (Fig. 3). The individual XY trajectories of TCs were further analyzed to obtain their mean-squared displacement MSD(τ) ≈ D × τα (18), where α was used as a parameter to characterize the nature of TC diffusion (Fig. S2, a and b, in Data S1). Mean α-values was used to classify the mobility of TC that diffuse freely (normal diffusion, 0.8 < α < 1.2), in a physically constrained environment (subdiffusion α < 0.8) or in the presence of external forces (super-diffusion α > 1.2). The mean α ∼ 0.6 (see Methods) and velocities Vavg ∼ 71 nm/s of TCs were computed from their trajectories (Fig. 3). We find that a small but statistically significant fraction of TCs (∼18%) undergo super diffusion or normal diffusion (∼23%), while a larger number of TCs exhibit subdiffusive (∼59%) transport (Fig. S3 a in Data S1). Interestingly, the mean velocity and velocity distributions of TCs (Fig. S2 c in Data S1) were similar to that of cytoskeletal motors obtained from single-molecule in vitro mobility assays (19). In addition, the nature of superdiffusive transport of TCs was found to be independent of its size (Fig. S2 d in Data S1).
FIGURE 3.
Nature of TC dynamics: XY trajectories of various TCs from the same nucleus are shown in solid representation, and in the ATP-depleted cell, are shown in shaded representation.
Role of chromatin assembly on TC dynamics
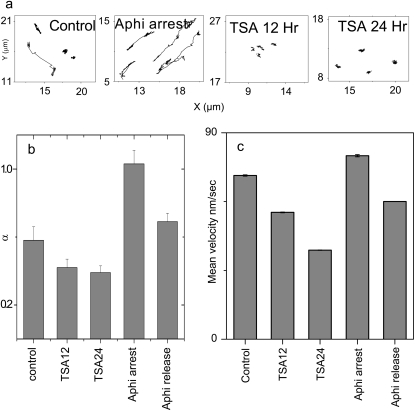
To investigate the effect of chromatin organization on TC dynamics, we repeated the TC mobility experiments in cells treated with a Trichostatin-A (TSA); a histone deacetylase inhibitor (20) (HDAC) that decondenses chromatin or with Aphidicolin; and a cell-stage specific inhibitor (21). Fig. 4 a shows a representative XY trajectory of TC under each of the above perturbations while Fig. 4, b and c, are averages of >30 TCs between different experiments. Upon treatment with TSA for 12 or 24 h, the super-diffusive component of TC was abolished, resulting in a significant reduction of both mean α to 0.4 and Vavg to 40 nm/s at 24 h (Fig. 4, b and c; and Fig. S3, a and b and Movie S2 in Data S1). TSA treatment also caused an increase in the number of TCs per cell (data not shown). Notably, in Aphidicolin-treated cells with early-S phase arrest, a significant fraction of TCs exhibit superdiffusive transport resulting in mean α increasing to 1 and the Vavg to 80 nm/s (Fig. 4, b and c; and Fig. S3 c and Movie S3 in Data S1). However, 7 h beyond the withdrawal of early-S phase arrest, the characteristics of TC dynamics qualitatively revert to that of control cells. TSA and Aphidicolin modulate chromatin assembly and concomitant with this, only the nuclear fraction of TC dynamics is affected (Fig. S3 d in Data S1).
FIGURE 4.
(a) Typical XY trajectories of TCs in Aphidicolin-arrested and TSA-treated cells. (b) Mean α-values for TCs in cells treated with TSA (HDAC inhibitor) or with Aphidicolin (cell-stage specific inhibitor). (c) Corresponding mean velocity for TC. Error bars correspond to standard error.
Contrast between TC and gene locus dynamics
The decondensed state of chromatin at TCs suggests a functional coupling between chromatin (gene) loci and TC. To probe this coupling, we monitored gene locus mobility in relation to its expression state and contrasted that with the mobility of TC. For this, a gene locus, with a Tet responsive element as described in Methods, is made visible by the addition of 96 lacO sites upstream of the gene (22), to which binding of transiently expressed mRFP-lacI results in a bright punctate structure in the nucleus (Fig. 5 a; and Fig. S4, a and b, in Data S1). The center panel in Fig. 5 a shows EGFP expression from the transfected gene locus. The right panel of Fig. 5 a shows a representative image of the gene locus (shown in red) colocalizing with a Cy5-UTP marked TC (shown in green). Dual track timelapse confocal images of the gene locus marked with mRFP and TC marked with Cy5-UTP was acquired at 1 s intervals for 50–150 s. Calculation of locus positions and mean α-values from the locus trajectories were obtained as described previously for TC. The trajectories of gene loci in expressing state undergo constrained or subdiffusion within a radius of ∼0.5 μm and under repression show reduced mobility (Fig. 5 b). Inset to Fig. 5 b depicts a representative plot of the trajectories of both TF and gene locus (see also Fig. S4 and Movie S4 in Data S1). Mean α for the locus varied from ∼0.6 in the expressing state to 0.23 in the nonexpressing state induced by repressor protein tTS. Interestingly, adding DRB (a transcription elongation inhibitor (23)) resulted in a reduction of mean α to levels as observed for repression by tTS. However, addition of TSA did not significantly alter the mean α-values in the expressing or the repressed state of gene locus (Fig. 5 c). Table S1 in Data S1 summarizes the mean α and Vavg for both TC and gene locus mobility under all the experimental conditions.
FIGURE 5.
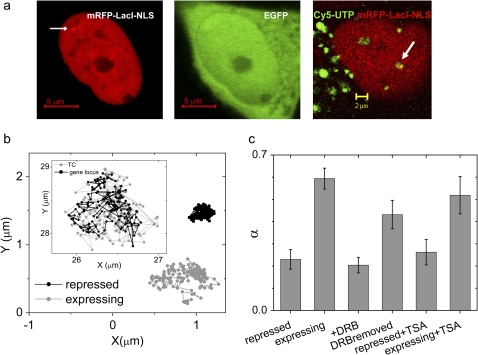
Nature of gene loci mobility: (a) Image of HeLa cell expressing mRFP-lacI-NLS (left panel) and EGFP (middle panel). Scale bar: 5 μm. Arrow indicates position of the EGFP gene locus. Right panel shows a cell where gene loci and TCs are labeled in red and green, respectively. Arrow indicates a colocalization of a gene locus and TCs. Scale bar: 2 μm (b) typical trajectories measured for cells expressing EGFP and under repression. (Inset) Trajectories of a gene locus and TC. (c) Plot of mean α-values of gene loci under various conditions. Error bars correspond to standard error.
DISCUSSIONS
Thus, using quantitative live-cell imaging and single-particle tracking analysis we report a heterogeneous behavior in the dynamics of transcriptionally active compartments within the cell nucleus. UTP incorporation in living cells marks distinct compartments both in the nucleus as well as in the cytoplasm. The nuclear fractions partially colocalize with antibodies to active RNA-Pol II, suggesting that a fraction of these compartments are transcription factories. Quantitative fluorescence anisotropy imaging evidenced a physical coupling between chromatin decondensation and TC positioning. Consistent with earlier observations of nuclear compartments (7,8,16,24), TCs are enriched in interchromatin territories with decondensed chromatin structure.
Timelapse confocal imaging of TC dynamics revealed that TCs exist in three distinct states: those exhibiting confined diffusion, normal diffusion, and super-diffusion with active and directed motion across the nucleus in an ATP and temperature-dependent manner. Single-particle tracking analysis of TC trajectories reveals that, while a large fraction of TCs exhibit confined diffusion, a statistically significant small fraction of TC undergoes super-diffusive transport within interspersed chromatin territories, suggesting a role for molecular motors in their active transport. Concomitant with this observation, perturbations to chromatin assembly alters the dynamic nature of TC. The enhancement of directional transport of TC in Aphidicolin-arrested cells implies that cell-stage specific functional alterations of chromatin structure have a strong influence on TC dynamics. In contrast, active movement of TC was inhibited by TSA-induced modulation of global chromatin packing, perhaps due to enhanced association between chromatin and TCs. The stalling of TC mobility with ATP depletion suggests a direct correlation with ATP-dependent chromatin remodeling motors in their dynamic organization.
In contrast, single-particle tracking of a labeled candidate gene locus evidences confined mobility. Notably for both TCs and the transcribing gene locus, the mean α was found to be ∼0.6, which reduced to ∼0.2 for either a stalled TC or a repressed gene locus suggesting a strong correlation with the underlying nuclear architecture. Gene locus mobility, either in the repressed or transcribing state, remained unaltered with TSA. In contrast, addition of transcription inhibitor DRB or induction of tTS repressor protein resulted in reduced mobility of gene locus. Taken together, our results suggest a novel spatiotemporal organization of TCs that shows heterogeneous behavior in terms of their dynamics and its correlation with chromatin architecture and gene locus mobility.
The functional organization of the cell nucleus has been conjectured to have profound impact on genome regulation (1,2). Though cytoskeletal and nuclear architectural proteins and motors (25–27) have been implicated in gene regulation, the molecular components involved in organization of transcription control in three dimensions remain largely unknown. Early in development and cellular differentiation, genes are repositioned within the nucleus and are often clustered around active sites of transcription (28–32). Regulatory genes are increasingly found to reposition during transcription, and the mobility of TC, reported here, could provide a physical basis for such reorganization. Perhaps the plasticity in the organization of TCs may establish such specific enriched functional compartments. Recent studies have also revealed a dynamic nuclear organization where gene locus, RNA-Pol II, transcription factors, histones, and other nuclear proteins are found to undergo distinct diffusive transport (33–39). Further, the spatial positioning of gene locus has been found to critically determine its function (40,41). In this context, the dynamic organization of TC implies new and more subtle design principles governing eukaryotic gene transcription within the three-dimensional architecture of living cell nuclei. Eliciting the underlying mechanisms will require an understanding of the molecular components that participate in controlling both TC and gene locus mobility, the self-organization rules that govern the dynamic assembly and disassembly of enriched compartments within the cell nucleus and its functional implications in diverse developmental contexts.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Satyajit Mayor, Yamuna Krishnan, Veronica Rodrigues, Apurva Sarin, and Ron Vale for critical reading of the manuscript.
We thank the imaging facility at The National Centre for Biological Sciences and Department of Science and Technology Nanoscience Initiative of the Government of India for funding. D.K.S. and B.B. were awarded the Kanwal Rekhi doctoral fellowship for career development and S.M. a Council of Scientific and Industrial Research doctoral fellowship.
Editor: Stuart M. Lindsay.
References
- 1.Misteli, T. 2007. Beyond the sequence: cellular organization of genome function. Cell. 128:787–800. [DOI] [PubMed] [Google Scholar]
- 2.Lanctot, C., T. Cheutin, M. Cremer, G. Cavalli, and T. Cremer. 2007. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 8:104–115. [DOI] [PubMed] [Google Scholar]
- 3.Sproul, D., N. Gilbert, and W. A. Bickmore. 2005. The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 6:775–781. [DOI] [PubMed] [Google Scholar]
- 4.Chakalova, L., E. Debrand, J. A. Mitchell, C. S. Osborne, and P. Fraser. 2005. Replication and transcription: shaping the landscape of the genome. Nat. Rev. Genet. 6:669–677. [DOI] [PubMed] [Google Scholar]
- 5.Gruenbaum, Y., A. Margalit, R. D. Goldman, D. K. Shumaker, and K. L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6:21–31. [DOI] [PubMed] [Google Scholar]
- 6.Tang, C. W., A. Maya-Mendoza, C. Martin, K. Zeng, S. Chen, D. Feret, S. A. Wilson, and D. A. Jackson. 2008. The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J. Cell Sci. 121:1014–1024. [DOI] [PubMed] [Google Scholar]
- 7.Lamond, A. I., and D. L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4:605–612. [DOI] [PubMed] [Google Scholar]
- 8.Cook, P. R. 1999. The organization of replication and transcription. Science. 284:1790–1795. [DOI] [PubMed] [Google Scholar]
- 9.Gorski, S. A., M. Dundr, and T. Misteli. 2006. The road much traveled: trafficking in the cell nucleus. Curr. Opin. Cell Biol. 18:284–290. [DOI] [PubMed] [Google Scholar]
- 10.Platani, M., I. Goldberg, A. I. Lamond, and J. R. Swedlow. 2002. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 4:502–508. [DOI] [PubMed] [Google Scholar]
- 11.Muratani, M., D. Gerlich, S. M. Janicki, M. Gebhard, R. Eils, and D. L. Spector. 2002. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 4:106–110. [DOI] [PubMed] [Google Scholar]
- 12.Misteli, T., J. F. Caceres, and D. L. Spector. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 387:523–527. [DOI] [PubMed] [Google Scholar]
- 13.Vargas, D. Y., A. Raj, S. A. Marras, F. R. Kramer, and S. Tyagi. 2005. Mechanism of mRNA transport in the nucleus. Proc. Natl. Acad. Sci. USA. 102:17008–17013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iborra, F. J., A. Pombo, D. A. Jackson, and P. R. Cook. 1996. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J. Cell Sci. 109:1427–1436. [DOI] [PubMed] [Google Scholar]
- 15.Marenduzzo, D., I. Faro-Trindade, and P. R. Cook. 2007. What are the molecular ties that maintain genomic loops? Trends Genet. 23:126–133. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, D. A., A. B. Hassan, R. J. Errington, and P. R. Cook. 1993. Visualization of focal sites of transcription within human nuclei. EMBO J. 12:1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee, B., D. Bhattacharya, and G. V. Shivashankar. 2006. Chromatin structure exhibits spatio-temporal heterogeneity within the cell nucleus. Biophys. J. 91:2297–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian, H., M. P. Sheetz, and E. L. Elson. 1991. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 60:910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Svoboda, K., and S. M. Block. 1994. Force and velocity measured for single kinesin molecules. Cell. 77:773–784. [DOI] [PubMed] [Google Scholar]
- 20.Toth, K. F., T. A. Knoch, M. Wachsmuth, M. Frank-Stohr, M. Stohr, C. P. Bacher, G. Muller, and K. Rippe. 2004. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 117:4277–4287. [DOI] [PubMed] [Google Scholar]
- 21.Pedrali-Noy, G., S. Spadari, A. Miller-Faures, A. O. Miller, J. Kruppa, and G. Koch. 1980. Synchronization of HeLa cell cultures by inhibition of DNA polymerase alpha with Aphidicolin. Nucleic Acids Res. 8:377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasser, S. M. 2002. Visualizing chromatin dynamics in interphase nuclei. Science. 296:1412–1416. [DOI] [PubMed] [Google Scholar]
- 23.Chodosh, L. A., A. Fire, M. Samuels, and P. A. Sharp. 1989. 5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 264:2250–2257. [PubMed] [Google Scholar]
- 24.Mitchell, J. A., and P. Fraser. 2008. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 22:20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vreugde, S., C. Ferrai, A. Miluzio, E. Hauben, P. C. Marchisio, M. P. Crippa, M. Bussi, and S. Biffo. 2006. Nuclear myosin VI enhances RNA polymerase II-dependent transcription. Mol. Cell. 23:749–755. [DOI] [PubMed] [Google Scholar]
- 26.Nunez, E., Y. S. Kwon, K. R. Hutt, Q. Hu, M. D. Cardamone, K. A. Ohgi, I. Garcia-Bassets, D. W. Rose, C. K. Glass, M. G. Rosenfeld, and X. D. Fu. 2008. Nuclear receptor-enhanced transcription requires motor- and LSD1-dependent gene networking in interchromatin granules. Cell. 132:996–1010. [DOI] [PubMed] [Google Scholar]
- 27.Miralles, F., and N. Visa. 2006. Actin in transcription and transcription regulation. Curr. Opin. Cell Biol. 18:261–266. [DOI] [PubMed] [Google Scholar]
- 28.Brown, J. M., J. Leach, J. E. Reittie, A. Atzberger, J. Lee-Prudhoe, W. G. Wood, D. R. Higgs, F. J. Iborra, and V. J. Buckle. 2006. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 172:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambeyron, S., N. R. Da Silva, K. A. Lawson, and W. A. Bickmore. 2005. Nuclear re-organization of the HOXB complex during mouse embryonic development. Development. 132:2215–2223. [DOI] [PubMed] [Google Scholar]
- 30.Chuang, C. H., A. E. Carpenter, B. Fuchsova, T. Johnson, P. de Lanerolle, and A. S. Belmont. 2006. Long-range directional movement of an interphase chromosome site. Curr. Biol. 16:825–831. [DOI] [PubMed] [Google Scholar]
- 31.Osborne, C. S., L. Chakalova, K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065–1071. [DOI] [PubMed] [Google Scholar]
- 32.Kooren, J., R. J. Palstra, P. Klous, E. Splinter, M. von Lindern, F. Grosveld, and W. de Laat. 2007. Beta-globin active chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J. Biol. Chem. 282:16544–16552. [DOI] [PubMed] [Google Scholar]
- 33.Taddei, A., G. Van Houwe, F. Hediger, V. Kalck, F. Cubizolles, H. Schober, and S. M. Gasser. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 441:774–778. [DOI] [PubMed] [Google Scholar]
- 34.Cabal, G. G., A. Genovesio, S. Rodriguez-Navarro, C. Zimmer, O. Gadal, A. Lesne, H. Buc, F. Feuerbach-Fournier, J. C. Olivo-Marin, E. C. Hurt, and U. Nehrbass. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773. [DOI] [PubMed] [Google Scholar]
- 35.Yao, J., K. M. Munson, W. W. Webb, and J. T. Lis. 2006. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 442:1050–1053. [DOI] [PubMed] [Google Scholar]
- 36.Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. I. Rothblum, R. D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 298:1623–1626. [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharya, D., A. Mazumder, S. A. Miriam, and G. V. Shivashankar. 2006. EGFP-tagged core and linker histones diffuse via distinct mechanisms within living cells. Biophys. J. 91:2326–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elf, J., G. W. Li, and X. S. Xie. 2007. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 316:1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darzacq, X., Y. Shav-Tal, V. de Turris, Y. Brody, S. M. Shenoy, R. D. Phair, and R. H. Singer. 2007. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14:796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumaran, R. I., and D. L. Spector. 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 180:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddy, K. L., J. M. Zullo, E. Bertolino, and H. Singh. 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 452:243–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.