Abstract
Oxidation of methionine residues in calmodulin (CaM) lowers the affinity for calcium and results in an inability to activate target proteins fully. To evaluate the structural consequences of CaM oxidation, we used infrared difference spectroscopy to identify oxidation-dependent effects on protein conformation and calcium liganding. Oxidation-induced changes include an increase in hydration of α-helices, as indicated in the downshift of the amide I′ band of both apo-CaM and Ca2+-CaM, and a modification of calcium liganding by carboxylate side chains, reflected in antisymmetric carboxylate band shifts. Changes in carboxylate ligands are consistent with the model we propose: an Asp at position 1 of the EF-loop experiences diminished hydrogen bonding with the polypeptide backbone, an Asp at position 3 forms a bidentate coordination of calcium, and an Asp at position 5 forms a pseudobridging coordination with a calcium-bound water molecule. The bidentate coordination of calcium by conserved glutamates is unaffected by oxidation. The observed changes in calcium ligation are discussed in terms of the placement of methionine side chains relative to the calcium-binding sites, suggesting that varying sensitivities of binding sites to oxidation may underlie the loss of CaM function upon oxidation.
INTRODUCTION
Calmodulin (CaM), the ubiquitous Ca2+-binding regulatory protein of eukaryotes, is the chief mediator of Ca2+-dependent signaling events in the cell (1,2). In response to rises in intracellular Ca2+ levels, CaM binds up to four Ca2+ ions and undergoes a marked conformational change, enabling the protein to recognize and bind a multitude of target proteins and to reorganize its distribution among Ca2+-dependent versus Ca2+-independent binding partners (1–3). Among the targets activated by CaM are metabolic enzymes, kinases and phosphatases, ion channels, and transcription factors, which places CaM in a pivotal “switch” role in such cellular processes as inflammatory response, transcription, and synaptic potentiation (2,3).
Calmodulin shares many structural features with other small Ca2+-binding proteins such as troponin C, calbindin, and recoverin (4). Calmodulin contains four canonical EF-hand Ca2+ binding motifs, which exist as pairs in the N-terminal and C-terminal lobes separated by a metastable linker sequence (5–10). EF-hands are helix-loop-helix structures that were named using traditional helix nomenclature for the protein albumin and represent a common binding motif for calcium (86). Cooperative binding of four Ca2+ ions to the four EF-hands stabilizes an elongated CaM structure, in which the hydrophobic target-binding surfaces are exposed on either lobe (7). The utility of CaM as a signaling protein, therefore, depends on its ability to structurally couple Ca2+ binding with target protein recognition. Disrupting either of these events, or their physical linkage, can compromise or abolish CaM function.
Although pathological CaM mutations are rare (CaM has an identical sequence in all vertebrates), covalent posttranslational modifications of CaM are common, and modulate the Ca2+ binding affinity and extent of fractional activation of target proteins (11–15). One such modification is the oxidation of methionine residues by reactive oxygen species, which generate methionine sulfoxide (MetSO) to form oxidized CaM (CaMox). Oxidation of the critical Met residues, Met144 and Met145, to MetSO greatly reduces the ability of CaM to activate target proteins (11,15–20), and decreases the protein's affinity for Ca2+ (21–24). Oxidation of CaM methionines in vivo was suggested to serve as a means of dampening Ca2+ signaling in response to prolonged stress, aging, or physical insults such as ionizing radiation (11,25,26). In this respect, ionizing radiation generates large amounts of reactive oxygen species in cells (27) and mobilizes Ca2+-dependent responses, of which CaM is an integral part (25,28). Oxidation of CaM methionines, therefore, may provide a means of “cross-talk” between the oxidative and cell-signaling effects of radiation exposure.
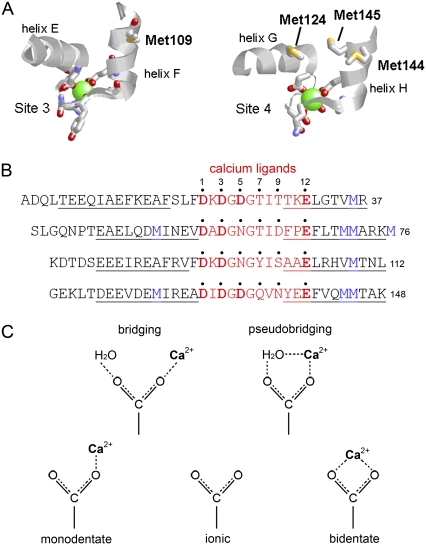
The structural mechanism underlying the effects of Met oxidation on CaM function, however, remains uncertain. It was suggested that Met oxidation results in global conformational changes that disrupt the helical structure of apo-CaM, interfering with the ability of CaMox to bind all four Ca2+ ions and activate target proteins (23,29). Alternatively, oxidation may result in a more subtle conformational change that allows complete Ca2+ binding, but selectively alters Ca2+ liganding to diminish the binding affinity. This possibility is supported by the proximity of methionine residues to Ca2+ ligands within the CaM structure (Fig. 1). Ca2+ binds to an EF-hand domain, which consists of a helix-loop-helix structure, where the majority of Ca2+ coordination residues are located in the EF-loop, and an additional residue is in the subsequent N-terminal helix (Fig. 1 B). Within the 12-residue binding sequence, highly conserved Asp and Glu side chains coordinate Ca2+ at Asp1, Asp3, Asp5 (or Asn5), and Glu12 (4). All of these side chains coordinate Ca2+ through a single oxygen atom (i.e., monodentate coordination, Fig. 1 C), except for Glu12, which binds in a bidentate mode. The coordination sphere is completed by a backbone amide carbonyl at position 7 and a water molecule stabilized by the side chain at position 9. The CaM sequence shows that all Ca2+-binding sites have at least one Met residue in the C-terminal EF-hand α-helix, whereas Ca2+-binding sites 2 and 4 have two, with an additional Met residue in the N-terminal helix (Fig. 1). A disruption of the EF-hand helices by the presence of MetSO has the potential to alter Ca2+ liganding.
FIGURE 1.
The EF-hand coordination of Ca2+ in CaM. (A) Structures of the third (left) and fourth (right) calcium-binding sites of CaM (7) (Protein Data Bank code 1cll), indicating the position and orientation of Met residues with respect to calcium (green)-liganding groups. (B) Sequence alignment of 12 amino-acid Ca2+ binding sequences (red) in EF-hands of vertebrate CaM. Residues providing a calcium ligand are numbered and marked with a dot. Ligands at positions 7 and 9 in each EF-loop coordinate calcium through a backbone carbonyl or through a water molecule bound to the side chain, respectively. All other residues coordinate calcium directly via the side chain. Acidic residues (Asp or Glu), in which coordination occurs via the side-chain carboxylate group, are shown in boldface. The proximity of methionines (blue) is indicated, showing that all binding sites have at least one Met residue in equivalent positions in one of the helices that flank the EF-loop (helical sequences are underlined). (C) Nomenclature for carboxylate side-chain coordination geometries.
To investigate the structural effects of Met oxidation on the conformation of CaM and how the liganding of Ca2+ is altered, we examined H2O2-oxidized and nonoxidized CaM in solution, using Fourier-transform infrared (FTIR) spectroscopy. An established technique for the determination of protein conformation (30–35), FTIR spectroscopy also exhibits exquisite sensitivity and an ability to monitor the structure of both the protein backbone and amino-acid side chains, making it ideal for rapid and simultaneous measurements of protein conformation and ligand binding (30,36–38). Particularly useful to our investigations is the demonstrated ability of FTIR spectroscopy to discriminate the different coordination modes (Fig. 1 C) of Glu or Asp side chains bound to metal ions (39–44). Our spectroscopic data provide evidence that Met oxidation increases solvent exposure of the α-helices of CaM, and modifies the coordination of Ca2+ by carboxylate side chains in the EF-loop. These findings support the view that the oxidation of CaM modifies its function primarily through disruption of tertiary contacts, and suggests that the loss of Ca2+ affinity upon oxidation is a direct result of a perturbation of Ca2+ binding sites.
EXPERIMENTAL PROCEDURES
Materials
Deuterium oxide (D2O, 99.9 atom % D) was purchased from Aldrich Chemical Co. (Milwaukee, WI). Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) was purchased from Fluka (Basel, Switzerland). All other buffers and chemicals were from Sigma (St. Louis, MO), were of reagent grade, and were used without additional purification.
Expression and purification of CaM
Recombinant vertebrate CaM was produced by expression in Escherichia coli from the pET15b expression vector, as described previously (17). Protein was purified by hydrophobic interaction chromatography on a phenyl-sepharose CL4B column, as described previously (45). Purity of all proteins was at least 95%, as determined from polyacrylamide gel electrophoresis (data not shown). Calmodulin was stored as a lyophilized powder at −20°C until use. In all experiments, the concentration of CaM was determined spectrophotometrically, using an extinction coefficient of 2980 M−1 cm−1 at 280 nm (calculated with the ProtParam tool on the ExPASy website, http://ca.expasy.org/tools/).
Oxidation of CaM methionine residues
Unless otherwise stated, the oxidation of CaM was performed as described previously (15). Briefly, lyophilized protein was dissolved to ∼1 mg/mL in oxidation buffer (50 mM Tris-HCl, 120 mM KCl, 1 mM MgCl2, 100 μM CaCl2, pH 7.4), to which concentrated hydrogen peroxide (H2O2) was added to a final concentration of 50 mM. Oxidation was allowed to proceed at room temperature (23°C ± 1°C) for 24 h, and was stopped by passing the reaction volume through a 5-mL HiTrap desalting column (GE Healthcare, Uppsala, Sweden) containing Sephadex G25 and equilibrated with water. The void volume (containing CaMox) was immediately frozen and lyophilized, and was used within 2 weeks of preparation. Under these reaction conditions, complete oxidation of all nine methionine residues of CaM occurs (15,17), as verified by observing the characteristic decrease in electrophoretic mobility of CaMox in the presence of 10 mM CaCl2 (17,21) (data not shown). Infrared spectra showed that one cycle of freezing-lyophilization did not appreciably alter the secondary structure and conformation of CaMox (data not shown). The oxidation of CaM was further verified by the presence of a very weak band at 1035 cm−1 in transmission-mode spectra in D2O (data not shown); sharp bands at 1030–1050 cm−1 in proteins were assigned to the S=O stretch vibration of MetSO (46).
Infrared spectroscopy
The CaM samples for FTIR spectroscopy were prepared by dissolving lyophilized protein in buffer at ∼10 mg/mL (∼600 μM). Samples in D2O were allowed to equilibrate at room temperature for at least 18 h before use, to allow for complete exchange of amide protons. Unless specified otherwise, all buffers contained 20 mM Tris-HCl (pH/pD 7.5) and 120 mM KCl. Buffers for Ca2+-CaM contained 10 mM CaCl2. Samples of Mg2+-CaM contained 150 mM MgCl2 and no calcium salts. Before the addition of buffer, apo-CaM and Mg2+-CaM samples were depleted of metals as follows: protein was diluted to ∼2 mg/mL in a solution of 2 mM EGTA and 2 mM ethylenediaminetetraacetic acid (EDTA), and allowed to sit for 3 h at room temperature. The protein solution was then dialyzed exhaustively against MilliQ deionized water for 24 h at 4°C, and then frozen, lyophilized, and reconstituted in the appropriate buffer. Immediately before use, samples were passed through a sterile 0.2-μm polyvinylidene fluoride filter, or in some cases, centrifuged for 5 min at 5000 × g to remove insoluble aggregate.
Infrared spectra were recorded on a Bruker Vector 22 FTIR spectrometer (Bruker Optics, Billerica, MA), equipped with a liquid N2-cooled photovoltaic mercury-cadmium-telluride detector, and continuously purged with dry, CO2-free air. Unless otherwise stated, solution spectra were obtained in attenuated total reflectance (ATR) mode, using the BioATRcell II (Harrick Scientific, Pleasantville, NY) with a 4-mm-diameter multireflection silicon ATR crystal (sample volume, 7.5 μL). All spectra were obtained at room temperature. To prevent cross-contamination, after each measurement, the ATR crystal was washed with 10 sample volumes of 2 mM EDTA/2 mM EGTA, followed by buffer. All spectra are the accumulated average of 256 scans at 2-cm−1 resolution, with a zero-filling factor of 2 (final spectra are 1 point per cm−1). Spectra were corrected for absorption of buffer and atmospheric water vapor by manually subtracting blank spectra, to obtain a flat baseline in the 1750–1850 cm−1 region. Because ATR spectra are known to exhibit altered peak intensity (particularly at high frequency) relative to absorption spectra (47), ATR spectra were converted to “simulated” absorbance-mode spectra by normalizing, using a linear function in the Opus 4.0 software program (Bruker Optics). Comparison of spectra collected in this manner with those obtained in transmission mode (using barium fluoride windows) showed no difference in band positions, indicating that anomalous dispersion effects were negligible in our experiments (data not shown). Overlaid spectra were normalized to a constant peak area from 1800–1600 cm−1. Subtraction, normalization, and differentiation of spectra were performed in Opus 4.0, and graphing and fine-structure enhancement (see below) were performed using IgorPro 6 (WaveMetrics, Inc., Lake Oswego, OR). Averaged data from at least 10 separate experiments are shown. Difference spectra are the average of individual subtractions obtained from at least six experiments. As a control, error spectra were determined by separating the unaveraged difference spectra into two datasets, which were subtracted and divided by 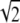 to compensate for differences in average and error spectral counts (48).
to compensate for differences in average and error spectral counts (48).
Fine-structure enhancement of FTIR spectra
The principle of fine-structure enhancement (FSE), a method of identifying component peaks of broad spectral bands, was described by Barth (49). Briefly, the FSE of a spectrum S involves generating a smoothed version of S (Ssm), and subtracting the smoothed spectrum from the original:
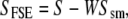 |
where SFSE is the final (fine structure-enhanced) spectrum, and W is a “weighting factor” (0 < W < 1). Difference spectra were smoothed over a 13-cm−1 window (25 data points), using a Savitsky-Golay algorithm, and the smoothed spectra were subtracted from the original spectra using a weighting factor W = 0.985, according to Barth (49). To verify that this procedure faithfully enhanced features present in the original absorbance spectra, the FSE procedure was also performed on the absorbance spectra (Supplementary Material, Data S1). The resulting enhanced spectra were qualitatively similar to those obtained using Fourier self-deconvolution, with a Lorentzian line and a bandwidth of 16 cm−1 (data not shown).
RESULTS
Oxidation-induced changes in ATR-FTIR absorbance spectra of CaM
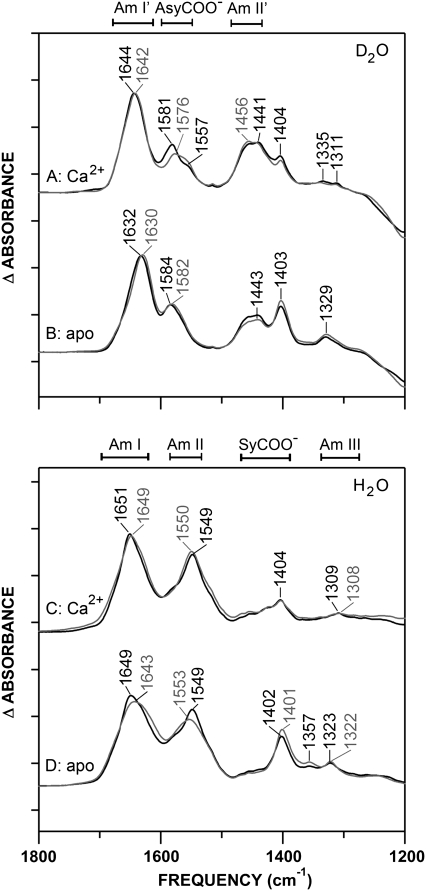
Fourier-transform infrared spectroscopy provides a powerful tool to study the effects of Met oxidation on the backbone fold and Ca2+-binding ligands of CaM. For comprehensive analysis, spectra were acquired in both D2O and H2O buffers, allowing greater visualization of amide and carboxylate (COO−) stretch regions, which are resolved differently in the two solvents. The amide regions (I, II, and III, as indicated in Fig. 2, C and D) reflect peptide backbone conformations in H2O, and these band positions are similar in D2O (Fig. 2, A and B) with the exception of the amide II′ (deuterated amide II) peak, which downshifts to reveal antisymmetric (Asy) COO− stretching bands near 1580 cm−1 (Fig. 2, A and B). Visualization of the Asy COO− bands in D2O provides information regarding Ca2+ liganding by Asp and Glu carboxylate side chains, as complemented by absorbance bands in the symmetric (Sy) COO− stretching region (∼1404 cm−1) that are more readily observed in H2O (30,36,42). The sensitivity of FTIR is apparent in the significant oxidation-induced spectral changes that reflect the effects of oxidation on the helical environment of CaM (Fig. 2, A–D, amide I/I′) and Ca2+ liganding (Fig. 2 A, Asy COO−).
FIGURE 2.
The FTIR absorbance spectra of CaM (black) and CaMox (gray) (10 mg/mL). Positions of major bands in D2O (A and B) and H2O (C and D) and their maxima (cm−1) are indicated. Spectra were acquired in Tris buffer, pH/pD 7.5, at room temperature. Sample buffer contained 10 mM CaCl2 (A and C), or samples were depleted of metals using EGTA/EDTA (B and D). Spectra were buffer-subtracted and normalized to a constant amide I/I′ band area, as described in Experimental Procedures. Tick marks on y axis represent Δ0.2 absorbance units.
Upon oxidation, the greatest perturbation occurs in the Asy-COO− region of the Ca2+-bound state (Fig. 2 A), where the peak at 1581 cm−1 (CaM) downshifts to 1576 cm−1 (CaMox), suggesting alterations in the Asp and Glu coordination of Ca2+ (42,50,51). In comparison, the Asy-COO− band of apo-CaM (Fig. 2 B) experiences very little shift, and retains intensity upon oxidation, indicating that the oxidation-dependent shift of Asp/Glu side-chain absorbances is specific for the Ca2+-bound state. An effect by oxidation on secondary structure can be seen in a downshift of the amide I/I′ bands (∼2 to 6 cm−1), yet overall, retention of the α-helical protein conformation is indicated by the nominal shift in the amide III band resolved in H2O (52–55). It is therefore evident that Met oxidation produces minimal changes in the conformation of CaM, but marked changes in the signal of side-chain carboxylate groups, suggesting an effect of oxidation on Ca2+-coordinating ligands.
Identification of FTIR bands associated with calcium liganding, using difference spectra
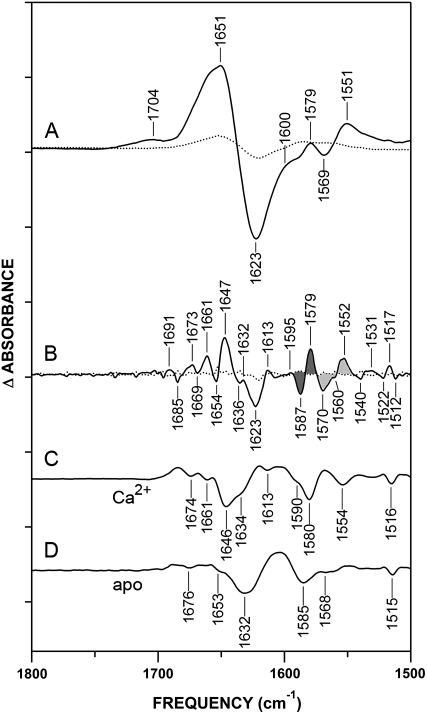
To aid in the interpretation of the oxidation effects observed in Fig. 2, we examined the Ca2+-CaM minus apo-CaM difference spectrum (Fig. 3 A), which reflects known structural changes upon Ca2+ binding to CaM. To reveal further component peaks, FSE (49) and second-derivative techniques (56) were used (Fig. 3, B–D). Importantly, the majority of peaks observed in the FSE difference spectrum correspond to minima in second-derivative spectra, providing strong evidence that the FSE bands reflect resolved structural features experiencing changes between the apo and Ca2+-bound states of CaM. Assignments of these bands are summarized in Table 1.
FIGURE 3.
Identification of D2O absorbance bands associated with Ca2+ ligands in nonoxidized CaM. (A) Ca2+-CaM-minus-apo-CaM difference spectrum (solid line) and error spectrum (averaged subtractions of spectra of like samples, dotted line). Positive peaks reflect structures in Ca2+-bound state, and negative peaks reflect structures in apo-state. (B) Fine structure-enhanced difference spectrum (solid) and error (dotted line), generated from A, as described in Experimental Procedures. Resulting spectra were multiplied by a factor of 10. Band shifts assigned to Ca2+-coordinating Asp and Glu carboxylates are shaded in dark gray and light gray, respectively (see also Table 1). (C) Second derivative of nonoxidized Ca2+-CaM and (D) apo-CaM absorbance spectra in Fig. 2, A and B. Resulting spectra were multiplied by a factor of 25. Minima correspond to positions of component bands in original absorbance spectra. Conditions are as described in legend of Fig. 2. Tick marks on y axis represent Δ0.05 absorbance units.
TABLE 1.
Assignment of bands in fine structure-enhanced FTIR difference spectra
| Turns/β | Bent/disordered helix | α-helix | Solvated α-helix | β-sheet | α-helix/β-sheet interaction | Asy-COO−, monodentate Asp | Asy-COO−, ionic Asp | Asy-COO−, ionic Glu | Asy-COO−, pseudobridging Asp | Asy-COO−, bidentate Glu | Asy-COO−, bidentate Asp | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ca2+-CaM | 1673 | 1661 | – | 1647 | 1632 | – | 1707, 1593 | – | – | 1580 | 1552 | – |
| Apo-CaM | 1685 | 1669 | 1654 | 1636 | – | – | 1587 | 1570 | – | – | – | |
| Ca2+-CaMox | 1674 | 1660 | – | 1645 | 1631 | 1623 | 1743, 1602 | – | – | 1570 | 1552 | 1549 |
| Apo-CaMox | 1683 | 1667 | 1653 | – | 1635 | 1622 | – | 1586 | 1571 | – | – | – |
| Reference for assignment | (31) | (31,58) | (31) | (58,61,63) | (31) | (65–67) | (40,41,43,78) | (43,58,78) | (43,58,78) | (78) | (41–43,78) | (78) |
Positions in cm−1 of component bands in amide I′ and Asy-COO− regions of fine structure-enhanced FTIR difference spectra. Assignments for Ca2+-CaM and apo-CaM were based, respectively, on positive and negative peaks in FSE difference spectra of Fig. 3 B and Fig. S2 B in Data S1, except for the monodentate and pseudobridging Asp bands that were based on Fig. 4.
The difference spectrum in Fig. 3 A shows a Ca2+-activated change in protein conformation in the amide I′ region and liganding in the Asy-COO− region. The appearance of positive peaks reflects changes associated with Ca2+ ligation, whereas negative peaks reflect the apo structure. Increased resolution of absorbance shifts are evident in both the FSE and second-derivative spectra, indicating that the broad features in Fig. 3 A are a compilation of overlapping bands (Fig. 3, B–D). In the FSE spectra, the 1570(−)/1552(+) cm−1 band shift upon Ca2+ binding is characteristic of EF-hand proteins, and reflects the formation of a bidentate Ca2+ coordination by Glu12 in the Ca2+ binding sequence (41–44,57) (Fig. 3 B). Carboxylate bands of Asp residues involved in calcium liganding are resolved in the FSE difference spectra at 1595(+)/1587(−)/1579(+) cm−1 (Fig. 3 B). These same features are well-resolved in the FSE absorbance spectra, where a single broad feature centered at 1587 cm−1 in apo-CaM is split into two peaks at 1593 cm−1 and 1580 cm−1 after Ca2+ activation (see Fig. S1 in Data S1). The large frequency shifts after Ca2+ activation, apparent in both the absorbance and difference spectra, reflect the fact that 11 of 17 Asp residues in CaM function as Ca2+ ligands (Fig. 1).
The 1587-cm−1 band apparent in apo-CaM is characteristic of Asp carboxylates in the ionic (nonliganded) state (36,42,43,58). The shift of Asp bands to both higher-frequency and lower-frequency positions upon calcium binding is indicative of two different liganding environments. The higher-frequency 1595(+) cm−1 peak is indicative of Asp carboxylates in a monodentate or bridging ligand structure, whereas the lower-frequency 1579(+) cm−1 band absorbs in a region characteristic of a weak pseudobridging interaction involving the Asp side chain, Ca2+, and solvent (36,39–43). Although they were not specifically commented on, these shifts can be seen in previously reported FTIR spectra of CaM (42,51). Examination of the relative orientation and surface accessibility of the Asp side chains in the EF-hands of CaM supports the assignments of the 1595(+) cm−1 and 1579(+) cm−1 peaks (4,59) (see Discussion).
The conserved Asp1 of the EF-loop has a lower surface accessibility than Asp3 and Asp/Asn5, and therefore has a greater probability of binding in either a monodentate or bridging fashion (1595(+) cm−1), where the peptide backbone is the hydrogen-bonding partner (60). On the other hand, Asp3 and Asp/Asn5 are in close proximity to the Ca2+-bound H2O (itself bound to the side chain at position 9), creating the potential for pseudobridging interactions (1579(+) cm−1) between Ca2+ and this water molecule. In CaMox, similar frequency shifts are observed in Ca2+-minus-apo difference spectra (see Fig. S2 in Data S1), although the absorbance frequencies of the Ca2+-bound Asp residues differ slightly from those in the nonoxidized protein (see below). Band shifts upon Ca2+ binding also occur in the Sy-COO− region in both CaM and CaMox (see Fig. S3 in Data S1). Although we do not assign these shifts to specific binding geometries, the appearance of multiple bands at 1451, 1428, and 1411 cm−1 upon Ca2+ binding supports the presence of two populations of Ca2+-bound aspartates in Ca2+-CaM.
Resolution of calcium-dependent increases in helix stability
Ca2+ binding also produces absorbance changes in the conformation-sensitive amide I′ band of CaM. Based on high-resolution structures of CaM, it is known that Ca2+ binding “opens” the target-binding pockets (located between the central linker and the helices flanking the EF-hands) and stabilizes helices in the linker region, elongating the molecule (1). This conformational change is reflected in the gain of intensity in the α-helical region of the amide I′ band and a loss of intensity at lower frequencies (unordered and β-sheet structures). In the FSE spectrum (Fig. 3 B), a marked band shift upon Ca2+ binding can be seen at 1654(−)/1647(+) cm−1. The 1654(−) cm−1 peak is a “classic” α-helical absorbance frequency, whereas the 1647(+) cm−1 band was previously assigned to highly solvent-exposed α-helices (61–63). This band pair therefore reflects helices that are shielded from solvent in the apo-state, and become exposed upon Ca2+ activation. Additional bands between 1691 cm−1 and 1661 cm−1 probably arise from the reorganization of turns and bends upon Ca2+ binding, whereas the 1636(−)/1632(+) cm−1 band pair suggests a reorientation of β-structure, which is responsible for allosteric communication between neighboring EF-hands of the protein (4,64). Consistent with this assignment, a comparison of the structures of apo-CaM and Ca2+-CaM reveals that the strands of these β-sheets become reoriented on Ca2+ binding (5,7,9,10). The broad 1623(−) cm−1 band, reflecting apo-CaM, occurs at a position consistent with previously described absorbances of extended chains (65,66), side chains of Arg, Tyr, or Gln residues (36), interactions between α-helices and β-sheets (67), and the intermolecular β-sheet (33,65). It is intriguing to note that evidence of interactions between the unstructured N-terminus and uncoiled central linker of apo-CaM was reported (68). The 1623(−) cm−1 band may reflect an intramolecular interaction by this structure that is similar in absorbance to a β-sheet/α-helix interaction. In addition, a Ca2+-induced gain in intensity at 1613 cm−1 could be associated with changes in Arg or Tyr hydrogen bonding (36,69), suggesting that side chains also contribute to the 1623(−) cm−1 band.
CaMox maintains native fold, whereas Met oxidation promotes helical destabilization
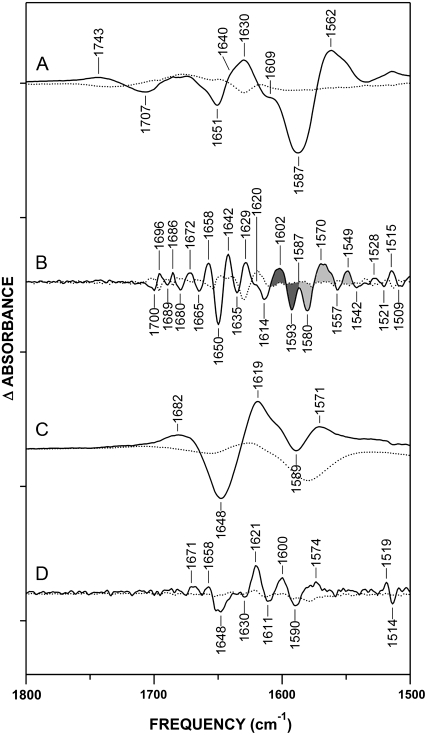
In the Ca2+-bound state, oxidation of CaM produces shifts in the amide I′ band, indicating that some degree of conformational change occurs upon Met oxidation (Fig. 4). The most notable of these changes is the 1650(−)/1642(+) cm−1 band pair, which reflects a shift of α-helical absorbance (Fig. 4 B). This 8-cm−1 downshift is comparable to that observed upon binding of Ca2+ (Fig. 3 B), strongly suggesting that it reflects an alteration in the hydration of α-helices. In fact, the oxidation of methionine residues was shown by NMR spectroscopy (16) and oxidative surface mapping (70) to induce disordering or transient unwinding of α-helices in the neighborhood of the oxidized residue, which would cause a downshift in the amide I′ absorbance frequency (61–63). We therefore attribute this band shift to the destabilization of helices induced by the oxidation of methionines. The shift to 1642 cm−1 could also be interpreted as a replacement of helices with an unordered structure. However, the retention of the amide III maximum at 1308/1309 cm−1 upon oxidation (Fig. 2 C, see Fig. S3 in Data S1) indicates that CaMox remains mostly helical, and that the 1650(−)/1642(+) cm−1 shift reflects a destabilization or transient unfolding, rather than a loss, of helices. This observation is also consistent with previous data showing that Ca2+ binding restores the native fold of CaMox (22,23). Other band shifts in the 1700–1658 cm−1 range suggest subtle rearrangements in turns and bends, whereas the 1635(−)/1629(+) cm−1 shift likely reflects a change in the β-sheets of CaM, possibly suggesting a different conformational preference in the EF-hands of CaMox.
FIGURE 4.
Conformational effects of oxidative modifications of CaM observed in D2O spectra: CaMox-minus-CaM difference spectrum of Ca2+ activated CaM (A and B) and apo-CaM (C and D) in D2O. (B) Fine-structure enhancement of A. Two populations of Asp residues are indicated by shading (see Results and Discussion): monodentate (dark gray) and pseudobridging and bidentate (light gray). (C) CaMox-minus-CaM difference spectrum of apo-CaM. (D) Fine-structure enhancement of C. In all traces, difference spectra are represented by solid lines, and error spectra by dotted lines. Fine-structure enhancement was performed as described in Experimental Procedures, and was multiplied by a factor of 25. Conditions are as described in legend of Fig. 2. Tick marks on y axis represent Δ0.05 absorbance units.
In apo-CaM, the overall shift of the amide I′ region suggests some loss of helical structure upon oxidation. However, as in Ca2+-CaM, apo-CaM retains an amide III maximum at 1322 cm−1, consistent with the retention of helical structure (Fig. 2 D). In the FSE spectrum (Fig. 4 D), the band pair 1648(−)/1621(+) cm−1 may reflect the loss of α-helical structure by oxidation-induced unraveling of the central linker, coupled with a potential increase in interactions between the linker and the N-terminus, as proposed by Faga et al. (68). The 1621(+) cm−1 band may also contain contributions from the intermolecular β-sheet because of protein aggregation (22), although precautions were taken to remove insoluble aggregate from samples before data collection (see Experimental Procedures). Further band shifts at 1671(+) cm−1 and 1658(+) cm−1 likely reflect a slight increase in turns or bent/disordered helices, either globally or in the metastable central linker helix of CaM (31,33,58).
Met oxidation alters calcium coordination by carboxylate side chains
The absorbance spectrum of Ca2+-CaMox differs most noticeably from that of Ca2+-CaM in the Asy-COO− stretching region (Fig. 2 A). This observation suggests that Met oxidation alters the coordination of Ca2+ by CaMox, because the Asy-COO− peak is sensitive to cation binding. To explore this possibility, we examined CaMox-minus-CaM difference spectra, in both the presence and absence of Ca2+ (Fig. 4), to investigate oxidation-dependent shifts in carboxylate absorbance. Comparison of the FSE spectra for Ca2+-CaM and apo-CaM shows an increased number of oxidation-induced spectral perturbations in the Asy-COO− region of the more rigid Ca2+-bound form (Fig. 4, B and D). This finding suggests that most of the change in Asy-COO− absorbance occurring upon Met oxidation results from perturbations of Ca2+-liganding Asp/Glu residues. The Asy-COO− shifts seen upon oxidation of apo-CaM, in contrast, likely reflect changes in solvent exposure or intramolecular interactions (e.g., salt bridging) of carboxylates not involved in Ca2+ binding (see below).
The prominent shifts observed in the Asy-COO− region of the Ca2+-CaM FSE difference spectra (Fig. 4 B) reveal two oxidation-induced band shifts at 1602(+)/1593(−)cm−1 and 1580(−)/1570(+) cm−1, which are consistent with changes in the absorbance of Asp residues. The negative bands at 1593 cm−1 and 1580 cm−1 reflect contributions by nonoxidized CaM, and correlate with bands assigned to the two Ca2+ liganding populations of Asp observed in the apo-CaM spectrum (Fig. 3 B). Therefore, we propose that the 1580(−)/1570(+) cm−1 downshift results from increased stabilization of the weak pseudobridging interaction between Asp, Ca2+, and solvent water formed upon Ca2+ binding. In addition, the 1602(+)/1593(−) cm−1 upshift correlates with a shift toward a stronger or more “monodentate-like” interaction with Ca2+. This scenario likely results from a loss or weakening of hydrogen bonding between the EF-loop Asp1 (1593(−) cm−1) and the polypeptide backbone, leading to a “pure” monodentate coordination (1602(+) cm−1) of Ca2+ by this Asp residue. Indeed, the FTIR spectrum of CaM saturated with Mg2+, which is known to complex with EF-hands solely through monodentate coordination by aspartates (4,71), has a primary Asy-COO− peak at 1600 cm−1 (data not shown), very close to the 1602-cm−1 peak observed in Fig. 4 B.
A critical ligand for maintaining Ca2+ coordination is the conserved glutamate residue at the twelfth position of the Ca2+ binding sequence (Fig. 1 B). In the Ca2+-bound state, this residue coordinates Ca2+ in the bidentate mode (4,5,7,8,10), and forms hydrogen bonds with the polypeptide backbone at positions 2 and 9 of the EF-loop (72,73). These interactions are believed to be responsible for both the Ca2+ specificity of the EF-hands of CaM, and for the allosteric coupling between Ca2+ binding sites (4,71–74). According to the data in Fig. 4, the oxidation of methionines in Ca2+-CaM has no apparent effect on the 1552-cm−1 peak associated with the Glu coordination of Ca2+ (Fig. 4 B), indicating that the bidentate coordination between Glu12 and Ca2+ is retained upon Met oxidation. This result is consistent with previous measurements indicating that the oxidation of all nine methionines to their sulfoxides does not affect the stoichiometry of Ca2+ binding (15). Moreover, the second-derivative spectra of both nonoxidized (Fig. 3 C) and oxidized (see Fig. S1 in Data S1) Ca2+-CaM display a minimum at 1554 cm−1 and 1552 cm−1, respectively, further indicating that CaM oxidation does not appreciably disrupt the bidentate coordination of Ca2+ by Glu residues. However, there is an absorbance increase at 1549(+) cm−1 that is consistent with the bidentate coordination of Ca2+ by a carboxylate. Molecular-dynamics simulations report that disruption of the Asp1 interaction with the polypeptide backbone can result in a bidentate ligation of Ca2+ by Asp3 (75). Therefore, it is likely that increased monodentate coordination by Asp1 (discussed above) results in the formation of bidentate coordination by Asp3, as indicated by a band at 1549(+) cm−1.
In apo-CaM, Asy-COO− bands are also observed, indicating that certain Asp residues undergo an oxidation-dependent absorbance shift in the absence of Ca2+ (Fig. 4 D). The band pair at 1600(+)/1590(−) cm−1 suggests a shift in Asp from ionic to bridging or monodentate absorbance frequencies. A potential explanation for this band shift involves the reported interaction between the N-terminal tail and the destabilized central linker of apo-CaM (68). There is a potential for further helix destabilization by oxidation of Met76, which would likely alter interactions between Asp78 and Asp80 (in the central linker of CaM) and the N-terminal residues (68). The broad absorbance increase at 1574(+) may reflect changes in the environment of Glu residues and the possible formation of salt bridges that result in a broadening of absorbance (76). Upon Ca2+ binding, the retention of native fold by Ca2+-CaMox would eliminate N-terminal interactions with the central linker. Therefore, we conclude that the majority of Asy-COO− absorbance changes upon Ca2+-CaM oxidation are related to changes in Ca2+ binding.
An additional 1743(+)/1707(−) cm−1 band pair was detected that is unique to the difference spectrum of Ca2+-CaM (Fig. 4 A). These peaks are too broad to appear in the FSE spectrum. Bands in this region, normally associated with the C=O stretch of protonated carboxyl groups, may occur in the FTIR spectra of ionized carboxylates bound to metal ions in a monodentate mode (36,77–79). We suggest that the 1707(−) cm−1 band represents absorbance by the noncomplexed carbonyl of Asp1 upon Ca2+ binding, reflecting a double bond characteristic that is typical of highly asymmetric monodentate carboxylates (78,79). This scenario can be visualized as the metal cation effectively acting as a proton, pulling electron density from the coordinating oxygen to make the noncomplexed carbonyl more “double bond-like”(78). In CaMox, a further increase in monodentate coordination would again increase the double bond characteristic of the noncoordinating carbonyl, resulting in an absorbance shift to 1743(+) cm−1. Therefore, the two band shifts at 1743(+)/1707(−) cm−1 and 1602(+)/1593(−) cm−1, together with the similarity of the 1602(+) cm−1 band to the Asy-COO− band of Mg2+-CaM (data not shown), lead us to propose that Met oxidation results in the loss of hydrogen-bonding interactions between the monodentate Asp1 of certain EF-loops and the backbone, thereby strengthening the monodentate coordination of Ca2+ by Asp1. Further spectral shifts can be seen in the symmetric COO− stretching band (see Fig. S4 in Data S1). Although we do not assign shifts in this region to specific coordination structures, the oxidation-dependent loss of bands at 1452 cm−1 and 1432 cm−1 is consistent with our assignment of band shifts in two different populations of Asp residues.
In conclusion, band shifts in the Asy carboxylate-stretching band of Ca2+-CaM indicate that methionine oxidation modifies the mode of Ca2+ coordination by carboxylate side chains in the four EF-hand binding motifs of CaM (Fig. 2). We propose that Asp residues are responsible for these results, because the critical bidentate ligation of Ca2+ by the conserved Glu12 of the Ca2+ binding sequence remains intact upon Met oxidation. These findings suggest that altered coordination of Ca2+ by Asp residues in the binding sites may underlie the order-of-magnitude decrease in apparent Ca2+ affinity upon oxidation of CaM methionines (15), and has the potential to explain observed disruptions in Ca2+ signaling under conditions of oxidative stress (26,80,81). Changes in the Asy-COO− band upon oxidation of Ca2+-CaM (Fig. 4) arise from shifts in two classes of carboxylate residues: we suggest that Asp1 shifts to a higher frequency, whereas Asp3 and/or Asp5 shifts to a lower frequency (Fig. 5). We further propose that these band shifts reflect an increase in monodentate coordination by Asp1, which may lead to bidentate Ca2+ ligation by Asp3, and stabilization of a pseudobridging interaction by Asp5.
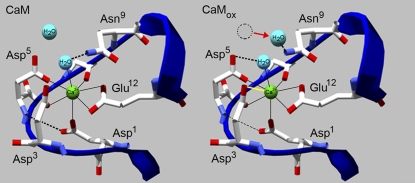
FIGURE 5.
Oxidation-induced destabilization of helices in EF-hand by oxidized methionine (not shown) results in altered conformation of EF-loop that affects Ca2+ liganding. Alterations in Ca2+ liganding involve modifications in hydrogen bonding (dashed lines) and repositioning of a water molecule (red arrow) that displaces hydrogen-bonding interaction between Asn9 and the chelating water ligand. Changes in binding involve diminished hydrogen bonding between Asp1 and the backbone (CaMox, thin dashed line), apparent as an upshift in monodentate Asy COO− absorbance from 1593(−) cm−1 to 1602(+) cm−1 (Fig. 4 B), which may result from destabilization of the EF-loop (not shown). This also permits the formation of a bidentate ligation of Ca2+ by Asp3 (CaMox, yellow line), and a pseudobridging interaction between the available noncoordinating oxygen atom of Asp5 (CaMox, thick dashed line), apparent as a downshift from 1580(−) cm−1 to 1549(+) cm−1 and 1580(−) cm−1 to 1570(+) cm−1, respectively (Fig. 4 B). Model depicts Ca2+ binding site IV (1exr.pdb) (59), with generic numbering of Ca2+ ligands (Fig. 1) before (CaM) and after (CaMox) Met oxidation.
DISCUSSION
Summary
Using FTIR spectroscopy, we resolved spectral shifts in the amide I′ and Asy-COO− stretching bands upon oxidation of methionine in CaM, and assigned these absorbance shifts to 1), an increased solvent exposure of α-helices in CaMox; and 2), an oxidation-induced change in the coordination of Ca2+ by carboxylate side chains in the EF-hands of CaM. We hypothesize that the carboxylate side chains reflect two populations of Ca2+-liganding Asp residues affected by CaM oxidation: one that shifts to a lower frequency (i.e., from 1580 cm−1 to 1570 cm−1 and 1549 cm−1), and one that shifts to a higher frequency (i.e., from 1593 cm−1 to 1602 cm−1; Fig. 4 B). In comparison, methionine oxidation does not affect the band at 1552 cm−1 associated with Ca2+-bound glutamates. This latter result is consistent with retention of the bidentate coordination of Ca2+ by the conserved Glu12 in the Ca2+ binding sequence. The oxidation-induced changes in Asy-COO− absorbance (Fig. 2) are consistent with a change in the mode of Ca2+ coordination by Asp ligands, i.e., Asp1 and Asp3/Asp5 of the EF-loop (Fig. 1). The FTIR spectra also showed increased levels of solvent exposure of α-helical amide groups upon Met oxidation, evident as a spectral shift from 1650 cm−1 to 1642 cm−1 in amide I′ difference spectra after FSE (Fig. 4 B). Thus, whereas methionine oxidation results in significant spectral changes in the absorbance bands associated with carboxylates involved in Ca2+ liganding for Ca2+-CaM, there are also important spectral changes in the amide I′ band, associated with destabilization of α-helical folds. Taken together, these observations suggest that changes in the mode of Ca2+ coordination in CaMox can be explained by an altered conformation of the loop region of the EF-hand, caused by oxidation-induced destabilization of the helices flanking the Ca2+ binding sites.
Conformational changes of CaM upon methionine oxidation
The finding that oxidation of CaM increases the solvent exposure of α-helices corroborates previous results that also found Met oxidation to decrease the dynamic stability of helices and to disrupt certain tertiary-structure contacts in native CaM (16,18,19,22,24). However, these data are at odds with early measurements suggesting that the oxidation of CaM results in global unfolding, with an extensive loss of secondary structure (23). The increase in helix solvation can be interpreted in two compatible ways. In one sense, it may reflect an “opening” of the structure of CaM to expose otherwise solvent-shielded regions of the helix, such as that occurring upon Ca2+ binding (Fig. 3 B). This scenario is possible, but is considered unlikely, given that the α-helices of Ca2+-CaM are already highly solvent-exposed in the wild-type protein before Met oxidation (5,7,51); any additional structural opening upon Met oxidation would likely be insignificant, compared with that caused by Ca2+ binding. Alternately, the increased solvation apparent in the amide I′ band may reflect a partial unwinding or fraying of α-helices by the presence of MetSO, a known helix-destabilizer (16,22,82). In this case, residues in a partially unwound portion of the α-helix would sample conformations similar to those of a classic helix, but with intrahelical hydrogen bonds at distorted angles and/or partially replaced by hydrogen bonds with solvent (61,62), resulting in a downshifted amide I/I′ absorbance frequency. This interpretation of the shift in the amide I′ helix signal agrees with the observed line-broadening in NMR spectra (16) and the observed loss of thermal stability of helices in CaM upon Met oxidation (22,24). Furthermore, a recent study using oxidative surface mapping (70) revealed that in apo-CaM, the reactivity of Met145 toward the hydroxyl radical increased after oxidation of Met144, suggesting that local disordering of the polypeptide backbone occurs when Met is oxidized. Together, these findings strongly suggest that methionine oxidation induces a destabilization, but not an unfolding per se, of CaM helices in the neighborhood of Met residues.
Oxidation modifies the coordination of calcium by carboxylate side chains
The finding that Met oxidation alters the coordination of Ca2+ by EF-hand carboxylate side chains suggests a structural basis for the observed order-of-magnitude decrease in apparent Ca2+ affinity after the oxidation of CaM (15). This effect can be observed after the oxidation of as few as two vicinal C-terminal methionines adjacent to the fourth (highest-affinity) Ca2+ binding site of CaM (15). Our results indicate that the oxidation of methionines in CaM results in a modification of hydrogen-bond interactions of carboxylate ligands that likely involve Asp1, which was previously associated with a loss of Ca2+ affinity. In troponin C, which is structurally homologous to CaM, diminished Ca2+ affinity is evident with the disruption of interactions between Asp1 and the EF-loop backbone (60). Molecular-dynamics simulations of an equivalent perturbation of Asp1 interactions in CaM revealed a rearrangement of Asp3 coordination of Ca2+ from a monodentate to a bidentate mode, resulting in an additional Ca2+ ligand (75). This leads us to suggest that oxidation-induced destabilization of helices results in alterations in Asp coordination of Ca2+, while maintaining a stable Glu bidentate coordination. Although all four Ca2+-binding sites in CaM are geometrically similar (Fig. 1), the sensitivity of Ca2+ binding to the oxidation of CaM may, in part, be attributable to the bracketing of the EF-loop in site IV by three methionine side chains, including the functionally sensitive Met144/Met145 (Fig. 1). Using a recent high-resolution crystal structure of Ca2+-CaM (59), it is possible to construct a model of oxidation-induced perturbations in Ca2+ liganding, suggesting conformational rearrangements consistent with the observed spectral shifts. Although the model reflects the crystal structure of Ca2+ binding site IV (Fig. 5), we use generic numbering (1 through 12) for discussion of the Ca2+ ligands.
Two water molecules appear in the crystal structure: one that acts as a Ca2+ ligand, and is held in place by a hydrogen bond to the side chain −NH2 of Asn9; and a second molecule that is more distal from the cation, but that has the potential to hydrogen-bond to the Asn9 side-chain amide group (Fig. 5, CaM). The Ca2+ liganding water associated with Asn9 is also within hydrogen-bonding distance of the noncoordinating oxygen atoms of the Asp3 and Asp5 carboxylates (Fig. 5). Our data indicate that oxidation results in a destabilization of Met-containing helices proximal to the Ca2+ binding region, altering Ca2+ liganding. We propose that upon Met oxidation, the side-chain interaction between Asp1 and the protein backbone is weakened, which results in the bidentate coordination of Ca2+ by Asp3 (75) and the formation of a stable pseudobridging interaction between Asp5, Ca2+, and the liganding water, permitting Asp9 to shift its interaction to the proximal water molecule (Fig. 5). This change in liganding would explain the shifts in absorbance bands from 1593 cm−1 to 1602 cm−1, and from 1580 cm−1 to 1570 cm−1, and the appearance of a band at 1549 cm−1, which are associated with the monodentate, pseudobridging, and bidentate modes of Ca2+ coordination, respectively (Table 1). Spectral data also indicate that the bidentate coordination of Ca2+ by Glu12 is conserved, because there are no observed shifts in the 1552-cm−1 band associated with Glu coordination (Table 1).
Intriguingly, this change in coordination structure may be limited to the Asp residues of the EF-hands. The conserved Glu residue at position 12 of the EF-hand Ca2+ binding sequence (Fig. 1 B) is known to be essential for Ca2+ selectivity and proper conformational change upon Ca2+ coordination in CaM and other EF-hand proteins (4). The fact that the bidentate coordination of Ca2+ by Glu12 is not disrupted by Met oxidation is consistent with the ability of CaMox to retain its ability to bind (but not fully activate) targets in a Ca2+-sensitive manner (15,19,80). Moreover, the idea that the Asp, but not Glu, coordination of Ca2+ is altered by Met oxidation lends support to the hypothesis that Met oxidation disrupts CaM activation by interfering with the ordered sequence of cation binding and conformational change events necessary for proper target activation (17–19,83,84). In all EF-hands, Ca2+ binding is thought to occur first at the ligands at the N-terminus of the EF-loop (Asp residues in this case), followed by a “wrapping” of the EF-loop around the ion, terminating in the formation of bidentate coordination by Glu12 (72,85). A difference in the preferred coordination structure by Asp residues (i.e., by the N-terminal part of the EF-hand) may slow or otherwise interfere with the early stages of this process, altering the proper sequence of binding and conformational changes needed for proper CaM activation. Consistent with this hypothesis, previous measurements found that the oxidation of methionines in CaM disrupts the cooperativity between Ca2+ binding sites (15).
Our model of selective modulation of Ca2+ binding by Asp residues provides a conceptually interesting means of modulating the Ca2+ activation of sensor proteins such as CaM, because the oxidation of Met is reversible through the action of methionine sulfoxide reductases, and is known to regulate Ca2+ signaling under conditions of oxidative stress (11,26,80). It is known that a loss of Ca2+ coordination by the critical Glu residue effectively abolishes the function of EF-hand proteins (4,73). By allowing this interaction to be retained, Met oxidation of CaM can modulate the function of the protein in a subtle manner, by affecting its affinity for Ca2+ and its relative efficacy at activating targets. The fact that the subtle conformational and cation-binding alterations induced by Met oxidation do not completely abolish the function of CaM, or cause a catastrophic unfolding or aggregation, may explain why this regulatory modification of CaM appears to have persisted through evolution (11) and may be harnessed in some organisms to blunt Ca2+ signaling under conditions of oxidative stress (26). Particularly interesting in this regard may be the role of CaMox in the cellular response to ionizing radiation, which is known both to stimulate Ca2+ signaling and to generate reactive oxygen/nitrogen species (25,27,28); experiments aimed at defining this role are underway in our laboratory. The observation that the oxidative modification of CaM can affect its metal-binding propensity, furthermore, suggests that fine-tuning cation coordination in a metal-binding protein may be a particularly useful way to modulate its function. Sensitivity to such modifications may be a common feature of metal-binding proteins subject to posttranslational regulation in vivo.
CONCLUSIONS AND FUTURE DIRECTIONS
We demonstrated that the oxidation of methionines in CaM selectively alters the coordination of the Ca2+ ion by the carboxylate side chains in the EF-hand binding clefts. We propose that oxidation differentially modifies Ca2+ liganding by two populations of Asp residues, acting to weaken the hydrogen-bond interaction between Asp1 and the EF-loop, and to alter selectively the mode of binding interactions between Asp3 and Asp5 and Ca2+. The overall result is to diminish effectively the Ca2+ binding affinity of CaMox. These findings support the role of destabilized structure and the disruption of tertiary contacts in the inactivation of CaM by oxidation, and further suggest that altered Ca2+ liganding may play a role in the nonproductive association of CaM with a range of target proteins, resulting in large reductions in their function. Future measurements should be directed at resolving residue-specific changes in binding interactions at individual Ca2+ binding sites in CaM, and their effects on both cooperative calcium binding and the mechanism of target-protein association with CaMox.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Drs. D. Bigelow and N. Karin for helpful discussions, Dr. H. Smallwood and Mr. M. Jacoby for the initial purification of calmodulin, and Ms. S. B. Reed for providing Milli-Q deionized water.
This research was supported by a grant from the Low Dose Radiation Research Program, Office of Biological and Environmental Research, United States Department of Energy. Pacific Northwest National Laboratory (PNNL) is operated for the Department of Energy by the Battelle Memorial Institute under contract DE-AC06-76RL0 1830.
Editor: Feng Gai.
References
- 1.Chin, D., and A. R. Means. 2000. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 10:322–328. [DOI] [PubMed] [Google Scholar]
- 2.Clapham, D. E. 2007. Calcium signaling. Cell. 131:1047–1058. [DOI] [PubMed] [Google Scholar]
- 3.Yap, K. L., J. Kim, K. Truong, M. Sherman, T. Yuan, and M. Ikura. 2000. Calmodulin target database. J. Struct. Funct. Genomics. 1:8–14. [DOI] [PubMed] [Google Scholar]
- 4.Gifford, J. L., M. P. Walsh, and H. J. Vogel. 2007. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 405:199–221. [DOI] [PubMed] [Google Scholar]
- 5.Babu, Y. S., C. E. Bugg, and W. J. Cook. 1988. Structure of calmodulin refined at 2.2 Å resolution. J. Mol. Biol. 204:191–204. [DOI] [PubMed] [Google Scholar]
- 6.Babu, Y. S., J. S. Sack, T. J. Greenhough, C. E. Bugg, A. R. Means, and W. J. Cook. 1985. Three-dimensional structure of calmodulin. Nature. 315:37–40. [DOI] [PubMed] [Google Scholar]
- 7.Chattopadhyaya, R., W. E. Meador, A. R. Means, and F. A. Quiocho. 1992. Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol. 228:1177–1192. [DOI] [PubMed] [Google Scholar]
- 8.Chou, J. J., S. Li, C. B. Klee, and A. Bax. 2001. Solution structure of Ca2+-calmodulin reveals flexible hand-like properties of its domains. Nat. Struct. Biol. 8:990–997. [DOI] [PubMed] [Google Scholar]
- 9.Kuboniwa, H., N. Tjandra, S. Grzesiek, H. Ren, C. B. Klee, and A. Bax. 1995. Solution structure of calcium-free calmodulin. Nat. Struct. Biol. 2:768–776. [DOI] [PubMed] [Google Scholar]
- 10.Zhang, M., T. Tanaka, and M. Ikura. 1995. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 2:758–767. [DOI] [PubMed] [Google Scholar]
- 11.Bigelow, D. J. and T. C. Squier. 2005. Redox modulation of cellular signaling and metabolism through reversible oxidation of methionine sensors in calcium regulatory proteins. Biochim. Biophys. Acta Proteins Proteomics. 1703:121–134. [DOI] [PubMed] [Google Scholar]
- 12.Gagnon, C., S. Kelly, V. Manganiello, M. Vaughan, C. Odya, W. Strittmatter, A. Hoffman, and F. Hirata. 1981. Modification of calmodulin function by enzymatic carboxylic methylation. Nature. 291:515–516. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, B. A., J. M. Shirokawa, and D. W. Aswad. 1989. Deamidation of calmodulin at neutral and alkaline pH: quantitative relationships between ammonia loss and the susceptibility of calmodulin to modification by protein carboxyl methyltransferase. Arch. Biochem. Biophys. 268:276–286. [DOI] [PubMed] [Google Scholar]
- 14.Murtaugh, T. J., L. S. Wright, and F. L. Siegel. 1986. Posttranslational modification of calmodulin in rat brain and pituitary. J. Neurochem. 47:164–172. [DOI] [PubMed] [Google Scholar]
- 15.Yao, Y., D. Yin, G. S. Jas, K. Kuczera, T. D. Williams, C. Schoneich, and T. C. Squier. 1996. Oxidative modification of a carboxyl-terminal vicinal methionine in calmodulin by hydrogen peroxide inhibits calmodulin-dependent activation of the plasma membrane Ca-ATPase. Biochemistry. 35:2767–2787. [DOI] [PubMed] [Google Scholar]
- 16.Anbanandam, A., R. J. Bieber Urbauer, R. K. Bartlett, H. S. Smallwood, T. C. Squier, and J. L. Urbauer. 2005. Mediating molecular recognition by methionine oxidation: conformational switching by oxidation of methionine in the carboxyl-terminal domain of calmodulin. Biochemistry. 44:9486–9496. [DOI] [PubMed] [Google Scholar]
- 17.Bartlett, R. K., R. J. Bieber Urbauer, A. Anbanandam, H. S. Smallwood, J. L. Urbauer, and T. C. Squier. 2003. Oxidation of Met144 and Met145 in calmodulin blocks calmodulin dependent activation of the plasma membrane Ca-ATPase. Biochemistry. 42:3231–3238. [DOI] [PubMed] [Google Scholar]
- 18.Chen, B., M. U. Mayer, and T. C. Squier. 2005. Structural uncoupling between opposing domains of oxidized calmodulin underlies the enhanced binding affinity and inhibition of the plasma membrane Ca-ATPase. Biochemistry. 44:4737–4747. [DOI] [PubMed] [Google Scholar]
- 19.Gao, J., Y. Yao, and T. C. Squier. 2001. Oxidatively modified calmodulin binds to the plasma membrane Ca-ATPase in a nonproductive and conformationally disordered complex. Biophys. J. 80:1791–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robison, A. J., D. G. Winder, R. J. Colbran, and R. K. Bartlett. 2007. Oxidation of calmodulin alters activation and regulation of CaMKII. Biochem. Biophys. Res. Commun. 356:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrington, D. A., H. Sun, K. K. Murray, J. Costa, T. D. Williams, D. J. Bigelow, and T. C. Squier. 2001. Selective degradation of oxidized calmodulin by the 20 S proteasome. J. Biol. Chem. 276:937–943. [DOI] [PubMed] [Google Scholar]
- 22.Gao, J., D. H. Yin, Y. Yao, H. Sun, Z. Qin, C. Schoneich, T. D. Williams, and T. C. Squier. 1998. Loss of conformational stability in calmodulin upon methionine oxidation. Biophys. J. 74:1115–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafitte, D., P. O. Tsvetkov, F. Devred, R. Toci, F. Barras, C. Briand, A. A. Makarov, and J. Haiech. 2002. Cation binding mode of fully oxidised calmodulin explained by the unfolding of the apostate. Biochim. Biophys. Acta: Proteins Proteomics. 1600:105–110. [DOI] [PubMed] [Google Scholar]
- 24.Sacksteder, C. A., J. E. Whittier, Y. Xiong, J. Li, N. A. Galeva, M. E. Jacoby, S. O. Purvine, T. D. Williams, M. C. Rechsteiner, D. J. Bigelow, and T. C. Squier. 2006. Tertiary structural rearrangements upon oxidation of methionine145 in calmodulin promotes targeted proteasomal degradation. Biophys. J. 91:1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du, Y. C., S. Gu, J. Zhou, T. Wang, H. Cai, M. A. Macinnes, E. M. Bradbury, and X. Chen. 2006. The dynamic alterations of H2AX complex during DNA repair detected by a proteomic approach reveal the critical roles of Ca2+/calmodulin in the ionizing radiation-induced cell cycle arrest. Mol. Cell. Proteomics. 5:1033–1044. [DOI] [PubMed] [Google Scholar]
- 26.Gao, J., D. Yin, Y. Yao, T. D. Williams, and T. C. Squier. 1998. Progressive decline in the ability of calmodulin isolated from aged brain to activate the plasma membrane Ca-ATPase. Biochemistry. 37:9536–9548. [DOI] [PubMed] [Google Scholar]
- 27.Mikkelsen, R. B., and P. Wardman. 2003. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene. 22:5734–5754. [DOI] [PubMed] [Google Scholar]
- 28.Hallahan, D. E., D. Bleakman, S. Virudachalam, D. Lee, D. Grdina, D. W. Kufe, and R. R. Weichselbaum. 1994. The role of intracellular calcium in the cellular response to ionizing radiation. Radiat. Res. 138:392–400. [PubMed] [Google Scholar]
- 29.Schallreuter, K. U., N. C. Gibbons, C. Zothner, M. M. Abou Elloof, and J. M. Wood. 2007. Hydrogen peroxide-mediated oxidative stress disrupts calcium binding on calmodulin: more evidence for oxidative stress in vitiligo. Biochem. Biophys. Res. Commun. 360:70–75. [DOI] [PubMed] [Google Scholar]
- 30.Barth, A. 2007. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 1767:1073–1101. [DOI] [PubMed] [Google Scholar]
- 31.Byler, D. M., and H. Susi. 1986. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 25:469–487. [DOI] [PubMed] [Google Scholar]
- 32.Krimm, S., and J. Bandekar. 1986. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 38:181–364. [DOI] [PubMed] [Google Scholar]
- 33.Surewicz, W. K., and H. H. Mantsch. 1988. New insight into protein secondary structure from resolution-enhanced infrared spectra. Biochim. Biophys. Acta. 952:115–130. [DOI] [PubMed] [Google Scholar]
- 34.Susi, H., and D. M. Byler. 1986. Resolution-enhanced Fourier transform infrared spectroscopy of enzymes. Methods Enzymol. 130:290–311. [DOI] [PubMed] [Google Scholar]
- 35.Venyaminov, S., and N. N. Kalnin. 1990. Quantitative IR spectrophotometry of peptide compounds in water (H2O) solutions. II. Amide absorption bands of polypeptides and fibrous proteins in alpha-, beta-, and random coil conformations. Biopolymers. 30:1259–1271. [DOI] [PubMed] [Google Scholar]
- 36.Barth, A. 2000. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 74:141–173. [DOI] [PubMed] [Google Scholar]
- 37.Barth, A., and C. Zscherp. 2002. What vibrations tell us about proteins. Q. Rev. Biophys. 35:369–430. [DOI] [PubMed] [Google Scholar]
- 38.Venyaminov, S., and N. N. Kalnin. 1990. Quantitative IR spectrophotometry of peptide compounds in water (H2O) solutions. I. Spectral parameters of amino acid residue absorption bands. Biopolymers. 30:1243–1257. [DOI] [PubMed] [Google Scholar]
- 39.Mizuguchi, M., M. Nara, K. Kawano, and K. Nitta. 1997. FT-IR study of the Ca2+-binding to bovine alpha-lactalbumin. Relationships between the type of coordination and characteristics of the bands due to the Asp COO− groups in the Ca2+-binding site. FEBS Lett. 417:153–156. [DOI] [PubMed] [Google Scholar]
- 40.Mizuguchi, M., M. Nara, Y. Ke, K. Kawano, T. Hiraoki, and K. Nitta. 1997. Fourier-transform infrared spectroscopic studies on the coordination of the side-chain COO− groups to Ca2+ in equine lysozyme. Eur. J. Biochem. 250:72–76. [DOI] [PubMed] [Google Scholar]
- 41.Nara, M., H. Morii, F. Yumoto, H. Kagi, and M. Tanokura. 2006. Fourier transform infrared spectroscopic study on the Ca2+ -bound coordination structures of synthetic peptide analogues of the calcium-binding site III of troponin C. Biopolymers. 82:339–343. [DOI] [PubMed] [Google Scholar]
- 42.Nara, M., M. Tanokura, T. Yamamoto, and M. Tasumi. 1995. A comparative study of the binding effects of Mg2+, Ca2+, Sr2+, and Cd2+ on calmodulin by Fourier-transform infrared spectroscopy. Biospectroscopy. 1:47–54. [Google Scholar]
- 43.Nara, M., M. Tasumi, M. Tanokura, T. Hiraoki, M. Yazawa, and A. Tsutsumi. 1994. Infrared studies of interaction between metal ions and Ca2+-binding proteins. Marker bands for identifying the types of coordination of the side-chain COO− groups to metal ions in pike parvalbumin (pI = 4.10). FEBS Lett. 349:84–88. [DOI] [PubMed] [Google Scholar]
- 44.Nara, M., F. Yumoto, K. Nagata, M. Tanokura, H. Kagi, T. Ojima, and K. Nishita. 2004. Fourier transform infrared spectroscopic study on the binding of Mg2+ to a mutant Akazara scallop troponin C (E142Q). Biopolymers. 74:77–81. [DOI] [PubMed] [Google Scholar]
- 45.Strasburg, G. M., M. Hogan, W. Birmachu, D. D. Thomas, and C. F. Louis. 1988. Site-specific derivatives of wheat germ calmodulin. Interactions with troponin and sarcoplasmic reticulum. J. Biol. Chem. 263:542–548. [PubMed] [Google Scholar]
- 46.Lasch, P., T. Petras, O. Ullrich, J. Backmann, D. Naumann, and T. Grune. 2001. Hydrogen peroxide-induced structural alterations of RNAse A. J. Biol. Chem. 276:9492–9502. [DOI] [PubMed] [Google Scholar]
- 47.van de Weert, M., P. I. Haris, W. E. Hennink, and D. J. A. Crommelin. 2001. Fourier transform infrared spectrometric analysis of protein conformation: effect of sampling method and stress factors. Anal. Biochem. 297:160–169. [DOI] [PubMed] [Google Scholar]
- 48.Sacksteder, C. A., S. L. Bender, and B. A. Barry. 2005. Role for bound water and CH-pi aromatic interactions in photosynthetic electron transfer. J. Am. Chem. Soc. 127:7879–7890. [DOI] [PubMed] [Google Scholar]
- 49.Barth, A. 2000. Fine-structure enhancement—assessment of a simple method to resolve overlapping bands in spectra. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 56:1223–1232. [DOI] [PubMed] [Google Scholar]
- 50.Rainteau, D., C. Wolf, and F. Lavialle. 1989. Effects of calcium and calcium analogs on calmodulin: a Fourier transform infrared and electron spin resonance investigation. Biochim. Biophys. Acta. 1011:81–87. [DOI] [PubMed] [Google Scholar]
- 51.Trewhella, J., W. K. Liddle, D. B. Heidorn, and N. Strynadka. 1989. Calmodulin and troponin C structures studied by Fourier transform infrared spectroscopy: effects of Ca2+ and Mg2+ binding. Biochemistry. 28:1294–1301. [DOI] [PubMed] [Google Scholar]
- 52.Anderle, G., and R. Mendelsohn. 1987. Thermal denaturation of globular proteins. Fourier transform-infrared studies of the amide III spectral region. Biophys. J. 52:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai, S., and B. R. Singh. 2004. A distinct utility of the amide III infrared band for secondary structure estimation of aqueous protein solutions using partial least squares methods. Biochemistry. 43:2541–2549. [DOI] [PubMed] [Google Scholar]
- 54.Fu, F. N., D. B. DeOliveira, W. R. Trumble, H. K. Sarkar, and B. R. Singh. 1994. Secondary structure estimation of proteins using the amide III region of Fourier transform infrared spectroscopy: application to analyze calcium-binding-induced structural changes in calsequestrin. Appl. Spectrosc. 48:1432–1441. [Google Scholar]
- 55.Seaton, B. A., J. F. Head, R. C. Lord, and G. A. Petsko. 1983. Studies of calmodulin structure—laser Raman-spectroscopy of biomolecules. Biochemistry. 22:973–978. [DOI] [PubMed] [Google Scholar]
- 56.Susi, H., and D. M. Byler. 1983. Protein structure by Fourier transform infrared spectroscopy: second derivative spectra. Biochem. Biophys. Res. Commun. 115:391–397. [DOI] [PubMed] [Google Scholar]
- 57.Ozawa, T., M. Fukuda, M. Nara, A. Nakamura, Y. Komine, K. Kohama, and Y. Umezawa. 2000. How can Ca2+ selectively activate recoverin in the presence of Mg2+? Surface plasmon resonance and FT-IR spectroscopic studies. Biochemistry. 39:14495–14503. [DOI] [PubMed] [Google Scholar]
- 58.Jackson, M., P. I. Haris, and D. Chapman. 1991. Fourier transform infrared spectroscopic studies of Ca2+-binding proteins. Biochemistry. 30:9681–9686. [DOI] [PubMed] [Google Scholar]
- 59.Wilson, M. A., and A. T. Brunger. 2000. The 1.0 Å crystal structure of Ca2+-bound calmodulin: an analysis of disorder and implications for functionally relevant plasticity. J. Mol. Biol. 301:1237–1256. [DOI] [PubMed] [Google Scholar]
- 60.Babu, A., H. Su, Y. J. Ryu, and J. Gulati. 1992. Determination of residue specificity in the EF-hand of troponin-C for Ca2+ coordination, by genetic-engineering. J. Biol. Chem. 267:15469–15474. [PubMed] [Google Scholar]
- 61.Manas, E. S., Z. Getahun, W. W. Wright, W. F. DeGrado, and J. M. Vanderkooi. 2000. Infrared spectra of amide groups in alpha-helical proteins: evidence for hydrogen bonding between helices and water. J. Am. Chem. Soc. 122:9883–9890. [Google Scholar]
- 62.Reisdorf, W. C., Jr., and S. Krimm. 1996. Infrared amide I′ band of the coiled coil. Biochemistry. 35:1383–1386. [DOI] [PubMed] [Google Scholar]
- 63.Walsh, S. T., R. P. Cheng, W. W. Wright, D. O. Alonso, V. Daggett, J. M. Vanderkooi, and W. F. DeGrado. 2003. The hydration of amides in helices: a comprehensive picture from molecular dynamics, IR, and NMR. Protein Sci. 12:520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marchand, S., and B. Roux. 1998. Molecular dynamics study of calbindin D9k in the apo and singly and doubly calcium-loaded states. Proteins. 33:265–284. [PubMed] [Google Scholar]
- 65.Georget, D. M., and P. S. Belton. 2006. Effects of temperature and water content on the secondary structure of wheat gluten studied by FTIR spectroscopy. Biomacromolecules. 7:469–475. [DOI] [PubMed] [Google Scholar]
- 66.Chehin, R., I. Iloro, M. J. Marcos, E. Villar, V. L. Shnyrov, and J. L. Arrondo. 1999. Thermal and pH-induced conformational changes of a beta-sheet protein monitored by infrared spectroscopy. Biochemistry. 38:1525–1530. [DOI] [PubMed] [Google Scholar]
- 67.Arrondo, J. L., and F. M. Goni. 1999. Structure and dynamics of membrane proteins as studied by infrared spectroscopy. Prog. Biophys. Mol. Biol. 72:367–405. [DOI] [PubMed] [Google Scholar]
- 68.Faga, L. A., B. R. Sorensen, W. S. VanScyoc, and M. A. Shea. 2003. Basic interdomain boundary residues in calmodulin decrease calcium affinity of sites I and II by stabilizing helix-helix interactions. Proteins. 50:381–391. [DOI] [PubMed] [Google Scholar]
- 69.Takahashi, R., and T. Noguchi. 2007. Criteria for determining the hydrogen-bond structures of a tyrosine side chain by Fourier transform infrared spectroscopy: density functional theory analyses of model hydrogen-bonded complexes of p-cresol. J. Phys. Chem. B. 111:13833–13844. [DOI] [PubMed] [Google Scholar]
- 70.Sharp, J. S., and K. B. Tomer. 2007. Analysis of the oxidative damage-induced conformational changes of apo- and holocalmodulin by dose-dependent protein oxidative surface mapping. Biophys. J. 92:1682–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohki, S., M. Ikura, and M. Zhang. 1997. Identification of Mg2+-binding sites and the role of Mg2+ on target recognition by calmodulin. Biochemistry. 36:4309–4316. [DOI] [PubMed] [Google Scholar]
- 72.Evenas, J., A. Malmendal, E. Thulin, G. Carlstrom, and S. Forsen. 1998. Ca2+ binding and conformational changes in a calmodulin domain. Biochemistry. 37:13744–13754. [DOI] [PubMed] [Google Scholar]
- 73.Evenas, J., E. Thulin, A. Malmendal, S. Forsen, and G. Carlstrom. 1997. NMR studies of the E140Q mutant of the carboxy-terminal domain of calmodulin reveal global conformational exchange in the Ca2+-saturated state. Biochemistry. 36:3448–3457. [DOI] [PubMed] [Google Scholar]
- 74.Evenas, J., S. Forsen, A. Malmendal, and M. Akke. 1999. Backbone dynamics and energetics of a calmodulin domain mutant exchanging between closed and open conformations. J. Mol. Biol. 289:603–617. [DOI] [PubMed] [Google Scholar]
- 75.Likić, V. A., E. E. Strehler, and P. R. Gooley. 2003. Dynamics of Ca2+-saturated calmodulin D129N mutant studied by multiple molecular dynamics simulations. Protein Sci. 12:2215–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muga, A., W. K. Surewicz, P. T. Wong, H. H. Mantsch, V. K. Singh, and T. Shinohara. 1990. Structural studies with the uveopathogenic peptide M derived from retinal S-antigen. Biochemistry. 29:2925–2930. [DOI] [PubMed] [Google Scholar]
- 77.Deacon, G. B., and R. J. Phillips. 1980. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord. Chem. Rev. 33:227–250. [Google Scholar]
- 78.Nara, M., H. Torii, and M. Tasumi. 1996. Correlation between the vibrational frequencies of the carboxylate group and the types of its coordination to a metal ion: an ab initio molecular orbital study. J. Phys. Chem. 100:19812–19817. [Google Scholar]
- 79.Tackett, J. E. 1989. FT-IR characterization of metal acetates in aqueous-solution. Appl. Spectrosc. 43:483–489. [Google Scholar]
- 80.Sun, H., J. Gao, D. A. Ferrington, H. Biesiada, T. D. Williams, and T. C. Squier. 1999. Repair of oxidized calmodulin by methionine sulfoxide reductase restores ability to activate the plasma membrane Ca-ATPase. Biochemistry. 38:105–112. [DOI] [PubMed] [Google Scholar]
- 81.Yin, D., K. Kuczera, and T. C. Squier. 2000. The sensitivity of carboxyl-terminal methionines in calmodulin isoforms to oxidation by H2O2 modulates the ability to activate the plasma membrane Ca-ATPase. Chem. Res. Toxicol. 13:103–110. [DOI] [PubMed] [Google Scholar]
- 82.Liu, D., D. Ren, H. Huang, J. Dankberg, R. Rosenfeld, M. J. Cocco, L. Li, D. N. Brems, and R. L. Remmele, Jr. 2008. Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry. 47:5088–5100. [DOI] [PubMed] [Google Scholar]
- 83.Boschek, C. B., T. E. Jones, H. S. Smallwood, T. C. Squier, and D. J. Bigelow. 2008. Loss of the calmodulin-dependent inhibition of the RyR1 calcium release channel upon oxidation of methionines in calmodulin. Biochemistry. 47:131–142. [DOI] [PubMed] [Google Scholar]
- 84.Sun, H., and T. C. Squier. 2000. Ordered and cooperative binding of opposing globular domains of calmodulin to the plasma membrane Ca-ATPase. J. Biol. Chem. 275:1731–1738. [DOI] [PubMed] [Google Scholar]
- 85.Kobayashi, C., and S. Takada. 2006. Protein grabs a ligand by extending anchor residues: molecular simulation for Ca2+ binding to calmodulin loop. Biophys. J. 90:3043–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kretsinger, R. H., and C. D. Barry. 1975. The predicted structure of the calcium-binding component of troponin. Biochim. Biophys. Acta. 405:40–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.