Abstract
Distinguishing the epimers iduronic acid (IdoA) versus glucuronic acid (GlcA) has been a longstanding challenge for the mass spectrometry analysis of glycosaminoglycan oligogosaccharides. In this work, electron detachment dissociation (EDD) and Fourier transform ion cyclotron resonance mass spectrometry is shown to provide mass spectral features that can distinguish GlcA from IdoA in heparan sulfate (HS) tetrasaccharides. EDD of HS tetrasaccharide dianions produces a radical species that fragments to produce information-rich glycosidic and cross-ring product ions which can be used to determine the sites of acetylation/sulfation. More significantly, EDD of HS tetrasaccharide epimers produces diagnostic product ions that can be used to distinguish IdoA from GlcA. These diagnostic product ions are not observed in the tandem mass spectra obtained by collisionally activated dissociation (CAD) or infrared multiphoton dissociation (IRMPD) of the tetrasaccharides, suggesting a radical-initiated mechanism for their formation. Differences in the observed product ions obtained by EDD of the tetrasaccharide epimers can be rationalized by simple α-cleavage of an oxy-radical located at C2 or C3 or a radical at C3 or C4. These radicals are proposed to arise from a hydrogen rearrangement in which a hydrogen atom is transferred from the C2 or C3 hydroxyl group or C3 or C4 to a carboxy radical at C5. This hydrogen transfer depends on the proximity of the carboxy radical to the hydroxyl group on C2 or C3 or the hydrogen on C3 or C4, and is thus influenced by C5 stereochemistry. These epimer-sensitive fragmentations should allow this approach to be applied to the structural analysis of a wide variety of GAG oligosaccharides.
INTRODUCTION
Glycosaminoglycans (GAGs) are complex polysaccharides which comprise the carbohydrate portion of proteoglycans, and are found in a variety of organisms ranging from bacteria to humans.1 GAGs participate in a number of significant biological processes, such as cell-cell signaling,2 the regulation of biochemical pathways,3, 4 and the mediation of inflammatory reactions.5 GAGs have also been implicated in the initial step of some pathogenic infections6–8 and have been observed to undergo alteration in some types of cancer.9 There is great interest in correlating the molecular structure of GAGs with their biological function, but the complexity of this class of molecules has slowed progress on this front. Although GAGs are linear biopolymers consisting of alternating acidic sugar and basic sugar residues, they exhibit complexity in a variety of ways. At the proteoglycan level, GAG chains of different length and composition bind core proteins that also exhibit compositional heterogeneity. At the subunit level, GAGs display complexity through their differing degrees of sulfation and N-acetylation. Heparin and heparan sulfate (HS) are the most complex of the GAGs; they are composed of a repeating disaccharide of hexuronic acid and glucosamine. The acidic sugar can be either glucuronic acid (GlcA) or iduronic acid (IdoA) depending on the C5 stereochemistry, and may be sulfated at the C2 hydroxyl group. The glucosamine may be sulfated at the C3 or C6 hydroxyl group, and sulfated, acetylated, or unmodified at the C2 amino group. The structural characterization of GAGs oligosaccharides requires identification of the location of sulfation/acetylation modifications as well as establishing the epimeric nature of the hexuronic acid. Characterizing GAGs is of significant interest since it is believed that the pattern of modification (sulfation, acetylation, GlcA vs. IdoA) affects their biological activity.10–12
Short lengths of heparin/HS oligosaccharides resulting from enzymatic digestion can be analyzed by 1D and 2D NMR.13 These methods can determine the type and location of chain modification and can distinguish IdoA from GlcA. However, analysis by NMR requires milligram quantities of a high purity sample. Unlike proteins and nucleic acids, GAGs are not synthesized by a template-driven mechanism, and so samples must be obtained by isolation of naturally occurring materials that are often available only in low quantity and purity. The development of other techniques for characterizing small amounts of GAGs and GAG mixtures is necessary to advance the goal of identifying structure/function relationships of this class of molecules. Mass spectrometry (MS) meets these requirements, but its development into a general purpose tool for GAG analysis remains incomplete.
Electrospray ionization (ESI) mass spectrometry is useful for obtaining the molecular weight of intact GAG oligosaccharides.14–16 For small oligosaccharides this can be used to determine the degree of sulfation. To establish the specific sites of modification, more advanced methods are required. Tandem mass spectrometry (MS2 and MS3) using collisionally activated dissociation (CAD) of Δ-unsaturated disaccharides resulting from digestion of heparin/HS with heparin lyases has been used to determine the pattern of sulfation/acetylation on short heparin/HS lengths.17 However, such disaccharides have lost chirality at the nonreducing end hexuronic acid, erasing the original epimeric nature of the acid residue at C5.18 Therefore, tetramers or longer oligosaccharides are needed for establishing IdoA versus GlcA composition. Generally speaking, mass spectrometry is not sensitive to chirality in molecules. However, Zaia and coworkers have shown that for chondroiton sulfate (CS) tetrasasaccharides and hexasaccharides, CAD can distinguish between IdoA and GlcA based on the relative abundance of specific X and Y ions.19 For this approach, the relative abundance of key fragment ions as a function of IdoA/GlcA composition was established for a series of tetrasaccharide and hexasaccharide standards, essentially producing a calibration curve for each mixture. This data was then used to determine the fractional abundance of IdoA vs. GlcA in tetrasaccharides and hexasaccharides from an enzymatic digestion of chondroiton/dermatan sulfate proteoglycans.20, 21 The advantage of this approach is that it allows one to establish the epimer composition of an isobaric mixture of GAG oligosaccharides. The drawback is that the fragmentation versus chirality dependence must be established for each different type of GAG, e.g. the fragmentation behavior of CS oligosaccharides could not be applied to HS oligosaccharides. In these CAD experiments, one cannot predict the fragmentation behavior based on the structure of GAG oligosaccharides, and thus this approach is not useful as a general purpose tool for determining the epimeric state of hexuronic acids in other GAG oligosaccharides.
We have recently reported the application of electron detachment dissociation (EDD)22–27 to the analysis of GAG tetrasaccharides.28 EDD produces a radical anion from the closed shell, multiply-charged anionic precursor. The radical anion undergoes distinctly different types of fragmentation than its closed shell counterpart. EDD produces abundant fragmentation, and greatly improves the capability of mass spectrometry to determine the sites of sulfation/acetylation in GAG tetrasaccharides compared to CAD or infrared multiphoton dissociation (IRMPD) of the same precursors. We have proposed a mechanism for the cross-ring fragmentation of hexuronic residues in GAG tetrasaccharides in which electron detachment from the C6 carboxylate anion forms a carboxy radical, which can undergo hydrogen rearrangement from the hydrogens at C3 or C4, or from the hydroxyl groups at C2 or C3.28 The propensity for H atom transfer from C3 or C4 or the C2 versus C3 hydroxyl groups should depend on the proximity of these hydrogen atoms to the carboxy radical, which is a function of the C5 stereochemistry. Moreover, the product ions that result when fragmentation is initiated by an oxy-radical at C2 or a radical at C3 are expected to be different than from an oxyradical at C3 or a radical at C4. Thus, the stereochemistry of the carbon that bears the carboxylic acid group should affect the subsequent fragmentation that is observed. Here we report evidence that GlcA can be distinguished from IdoA in HS tetrasaccharide epimers using EDD. Furthermore, differences in the EDD mass spectra of epimers can be rationalized by simple radical driven fragmentation mechanisms that should allow this approach to be extended to the structure analysis of a wide variety of GAG oligosaccharides.
EXPERIMENTAL
Preparation of Heparan Sulfate Tetrasaccharides
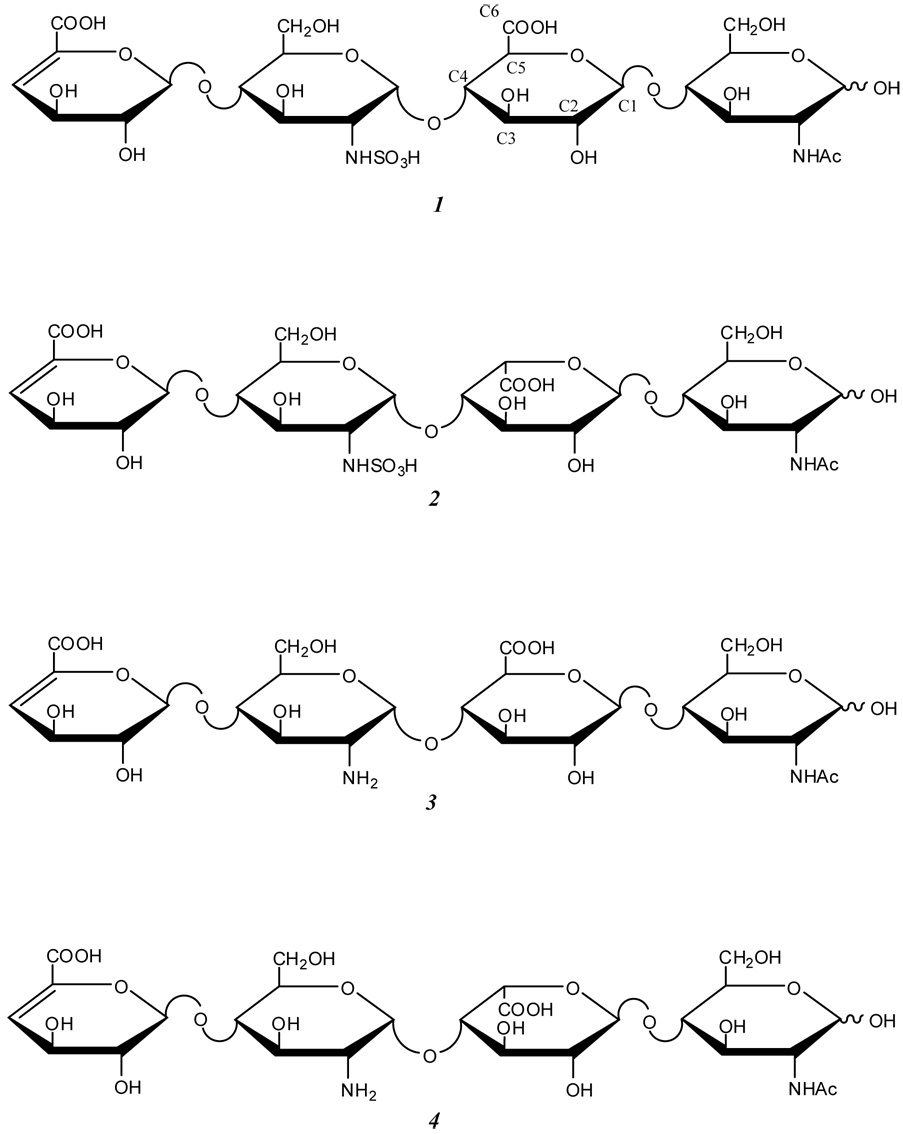
Heparan sulfate sodium salt was obtained from Celsus Laboratories (Cincinnati, OH), and digested with heparinase II (Sigma, St. Louis, MO) and fractionated by gelpermeation chromatography using a P-10 column (Bio-Rad, Hercules, CA) to obtain uniform sized oligosaccharides. The fraction containing tetrasaccharides was desalted on a Bio-Rad Micro Bio-Spin chromatography column packed with 1 mL of Bio-Rad P-2 resin. 50 µL of sample was loaded onto the resin and centrifuged at 1000 × g. After desalting, the fractions containing tetrasaccharides were pooled and concentrated by freeze-drying. Fractions containing individual tetrasaccharides were collected from semi-preparative SAX-HPLC (Shimadzu, Columbia, MD) using a Spherisorb column (Waters Corp, Milford, MA), desalted on a Bio-Rad P-2 column, and freeze-dried.29 For the work reported here, four tetrasaccharides were examined, structures 1 – 4. The structure of tetrasaccharides 2 and 3 were determined by 1D and 2D proton NMR. Tetrasaccharide 1 was prepared from tetrasaccharide 3 by N-sulfonation using the following protocol: 50 µg of tetrasaccharide 3 was dissolved in 12.5 µL of solution containing 10 mg/mL sodium bicarbonate and 10 mg/mL trimethylamine-sulfurtrioxide complex and incubated at 50 °C for 12 h. Equal portion of sodium bicarbonate and trimethylamine-sulfurtrioxide complex was added two more times at 12 h intervals. The solution was then desalted by P-2 spin column and the product was freeze-dried.30 Tetrasaccharide 4 was prepared from tetrasaccharide 2 using the following protocol: tetrasaccharide 2 sodium salt was converted to a pyridinium salt using a Dowex 50W cation exchange column (Sigma, St. Louis, MO). 200 µg of the pyridinium salt of 2 was dissolved in 10 µL of dimethyl sulfoxide (Acros, Geel, Belgium) containing 5% methanol and incubated for 1.5h at 50 °C.31 The desulfated product, 4, was then purified on a Bio-Rad P-2 spin column and freeze-dried. The products of the sulfation and desulfation reactions differ in mass from the starting materials, and they can be easily isolated from each other for the EDD experiments. The sulfation and desulfation reactions do not affect the stereochemistry of the hexuronic residue, and so the chirality of the products has the same purity as that of the reactants, which have been established to be pure by NMR analysis.
STRUCTURES.
Mass Spectrometry Analysis
Experiments were performed with a 7 T Bruker Apex IV QeFTMS fitted with an Apollo II ESI source, a CO2 laser for IRMPD, and an indirectly heated hollow cathode for generating electrons for ECD and EDD. The hollow cathode implementation with the Infinity™ cell has been previously described.27, 32 Solutions of tetrasaccharides 1, 2, and 3 were made at a concentration of 0.1 mg/mL in 50:50 methanol:H2O (Sigma, St. Louis, MO), tetrasaccharide 4 was made at a concentration of 1.0 mg/mL. All tetrasaccharides were ionized by nanospray using a pulled fused silica tip (model# FS360-75-15-D-5, New Objective, Woburn, MA). The sample solutions were infused at a rate of 10 µL/hour. All tetrasaccharides were examined in negative ion mode.
For the EDD experiments, precursor ions were isolated in the external quadrupole and accumulated for 1–2 seconds before injection into the FTMS cell. The isolation/cell fill was repeated up to 6 times. The selection of the precursor ion was further refined by using in-cell isolation with a coherent harmonic excitation frequency (CHEF) event. The precursor ions were then irradiated with electrons for 1 second. For electron irradiation the cathode bias was set to -19 V, the ECD lens was set to -17.5 V±0.5 V, and the cathode heater was set to 5–6 V. The electrons can potentially make multiple passes through the analyzer cell by reflection from the negative potential of the transfer ion optics and the hollow cathode, irradiating the sample at many different energies below the initial value of 19 eV. However, it appears that the efficiency of the EDD process is highly sensitive to the energy of the electrons, as irradiation of GAG tetrasaccharides with electrons at initial energies higher or lower than 19 eV by even a few tenths of an eV results in significantly less fragmentation. 24 acquisitions were averaged per mass spectrum. For each mass spectrum, 512k points were acquired, padded with one zero fill, and apodized using a sinebell window. Background spectra were acquired by leaving all parameters the same but setting the cathode bias to 0 V to ensure that no electrons reached the analyzer cell. All EDD products are reported using the Domon and Costello nomenclature.33
RESULTS AND DISCUSSION
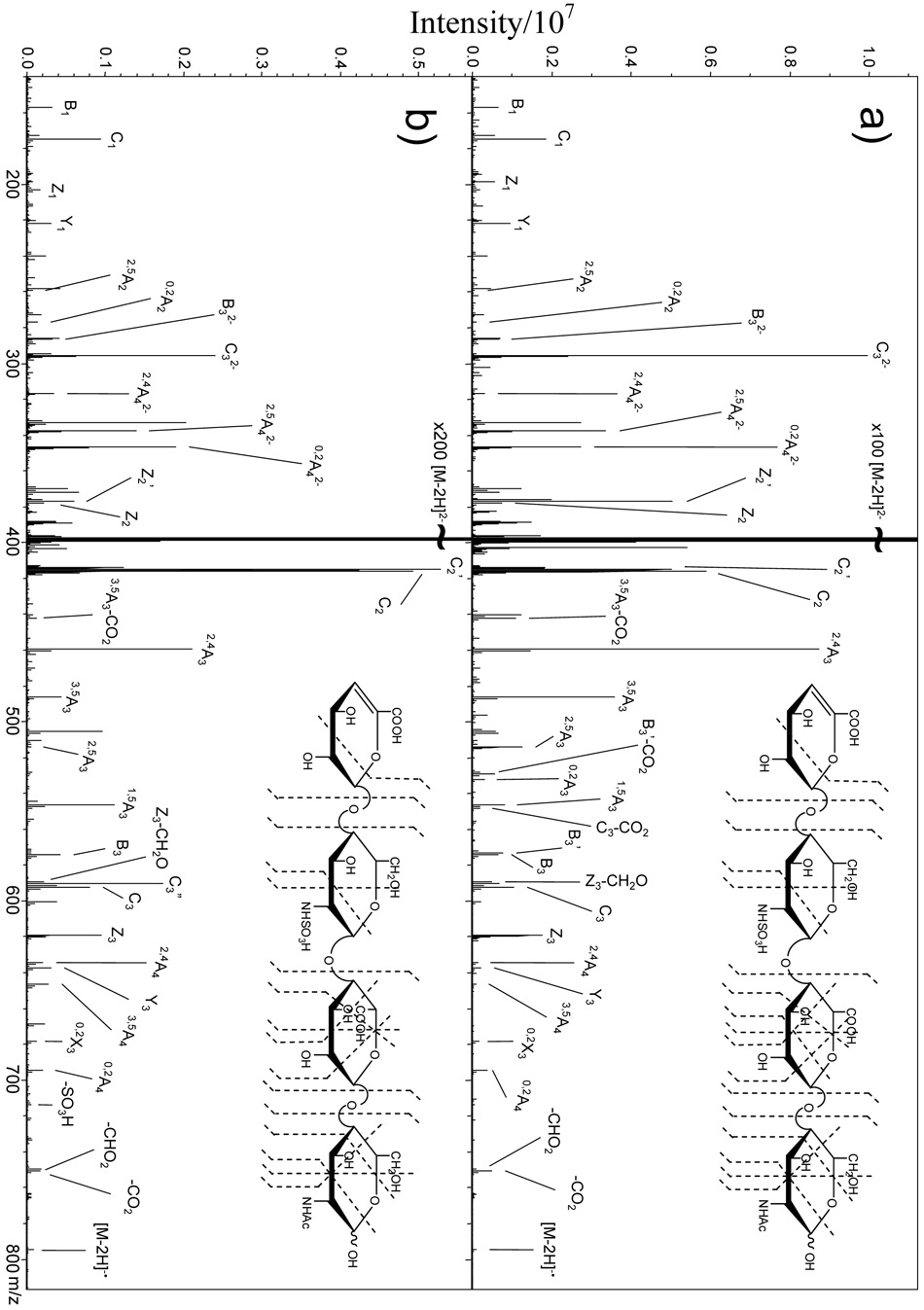
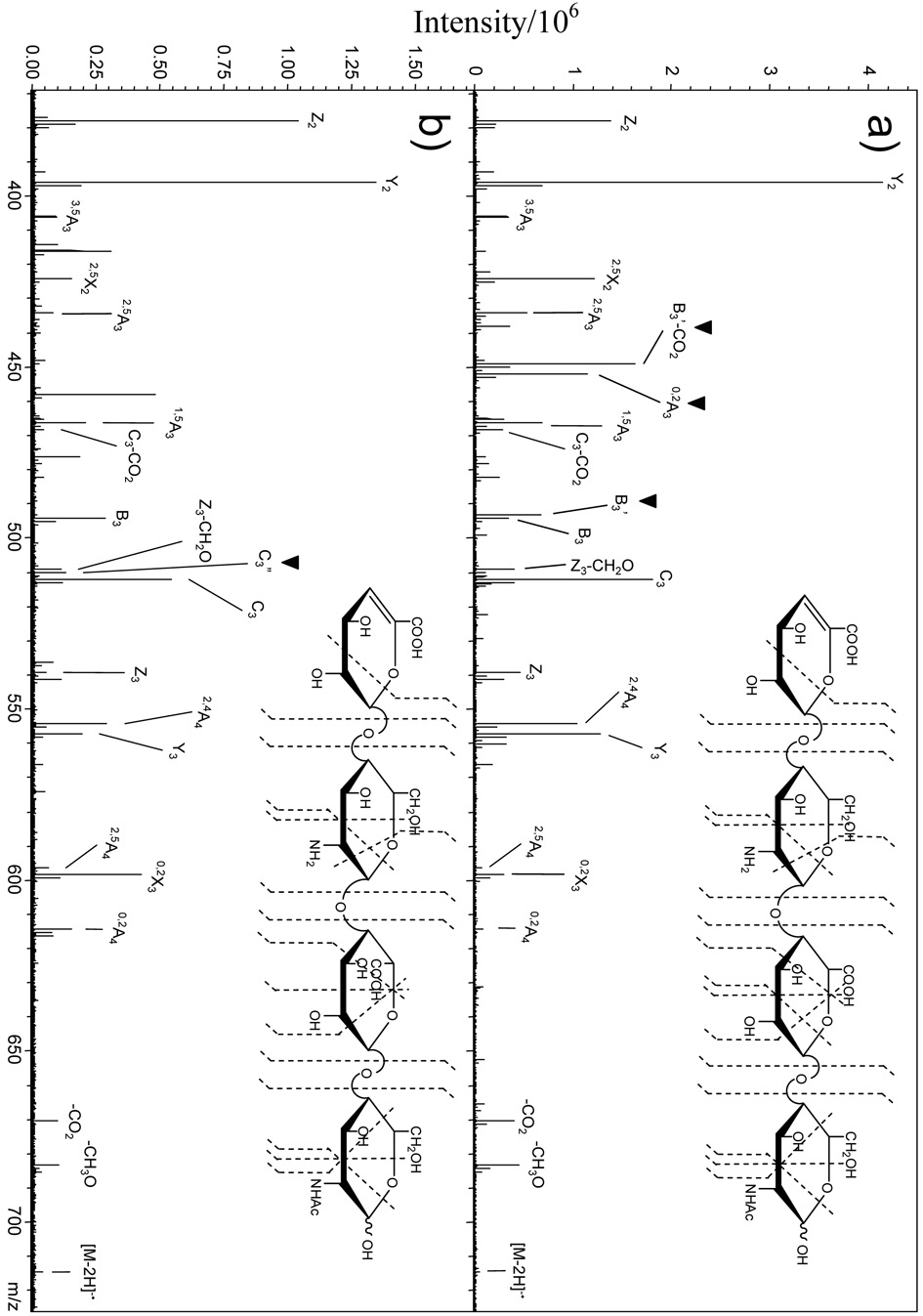
EDD of epimers 1 and 2 produces the mass spectra shown in Figures 1a and 1b, respectively. Glycosidic bond cleavage is the principal type of fragmentation that is observed, but cross-ring cleavage to produce A ions33 is also significant. During EDD, a radical site is produced by detachment of a carboxylate electron in preference to a sulfate electron, as ~1.2 eV less energy is required.28 This is evident from the presence of a [M-2H-CO2]−• product and the absence of a [M-2H-SO3]−• product. The abundance of fragmentation at the hexuronic acid near the reducing end of the tetrasaccharide suggests that the carboxyl group in this residue is a favored site for electron detachment. Many of the product ions observed in EDD of 1 are observed in the EDD of 2 (bond cleavage sites shown in insets, Figure 1), and can be used to establish the sites of modification.28 On the other hand, there are a few peaks that are not common to the spectra of both epimers, for example, the 0,2A3 product ion (m/z 532.059), which is observed only in the EDD of 1, albeit at a relatively low abundance. Closer examination of the EDD mass spectra above the precursor ion (m/z 397.047), Figures 2a and 2b, reveals a number of peaks specific to each epimer. An odd-electron ion at m/z 573.062 is present in EDD of 1 and not in EDD of 2. This product ion differs from the B3 fragment by the exact mass of a hydrogen atom, and is more intense than the B3 product ion in EDD of 1. We have assigned this product as [B3-H]−•, which we denote as B3’. The odd-electron ion at m/z 529.072 is present in EDD of 1 and not in EDD of 2 and differs from B3’ by the exact mass of CO2. We have assigned this product as [B3-H-CO2]−•, which we denote as B 3’-CO2. The ion at m/z 590.065 differs from C3 by the exact mass of two hydrogens, and is assigned as [C3-2H]−, which we denote as C3”. This product ion is observed in both EDD spectra, but is significantly more intense than the C3 product ion in the EDD spectrum of 2, while it is less abundant than the C3 fragment in the EDD spectrum of 1. Likewise, the 3,5A3 product ion is present in both EDD of 1 and 2, but is significantly more intense in the EDD of 1, where it is one of the most abundant product ions in the mass spectrum.
Figure 1.
EDD mass spectra of the [M-2H]2− precursor ion of HS epimers: (a) GlcA-containing tetrasaccharide 1; (b) IdoA containing tetrasaccharide 2. EDD results in cleavage of every glycosidic bond and abundant cross-ring fragmentation without loss of the labile sulfate group, allowing structure determination of the tetrasaccharides. Insets: observed bond cleavages for the HS tetrasaccharides.
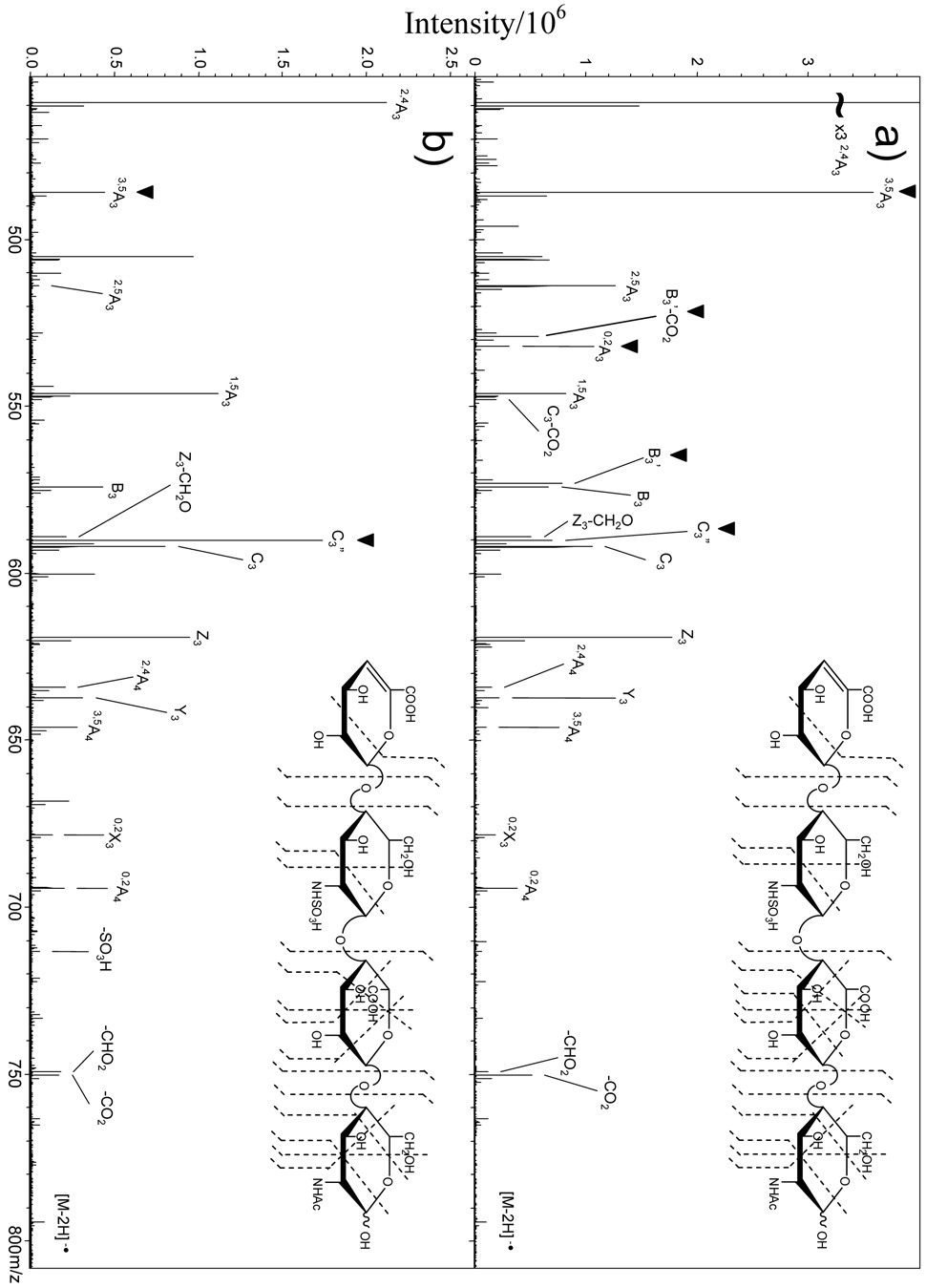
Figure 2.
Expansion of the m/z 450 – 800 region of Figure 1. (a) EDD of 1, GlcA. (b) EDD of 2, IdoA. The ▼ above the peak labels indicates product ions unique to each HS epimer.
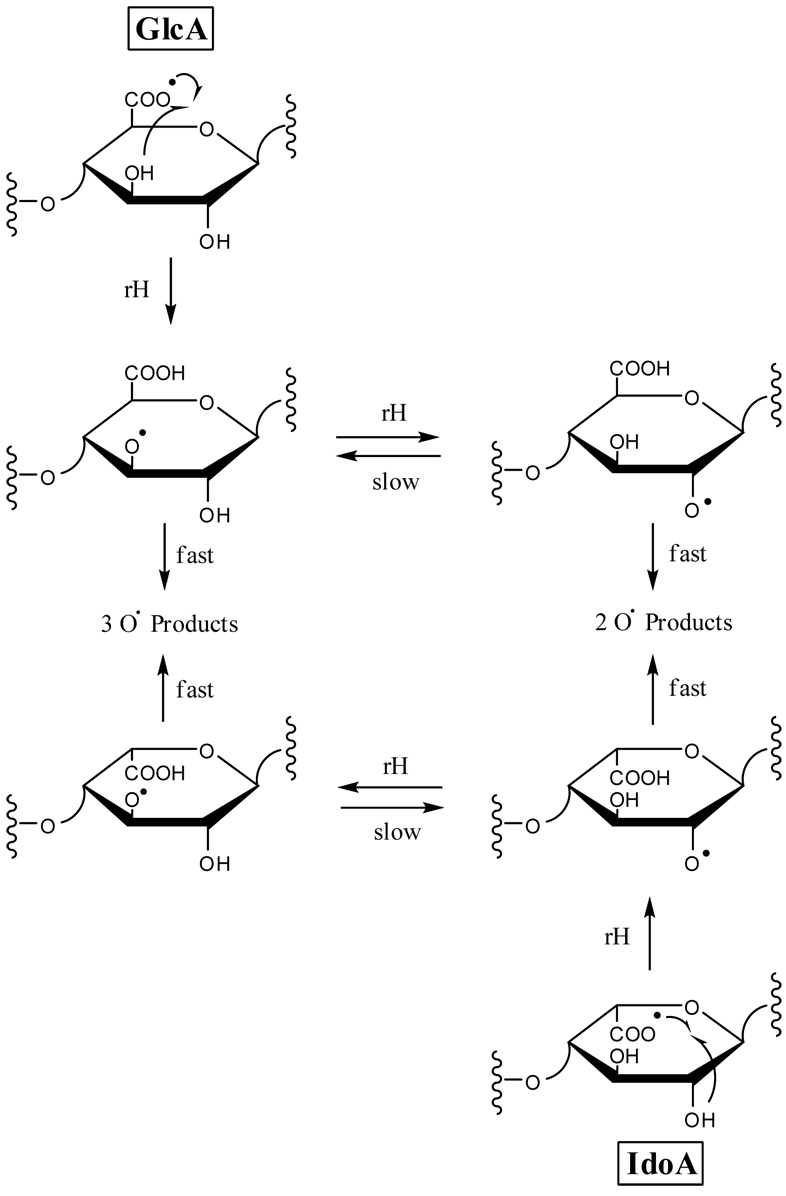
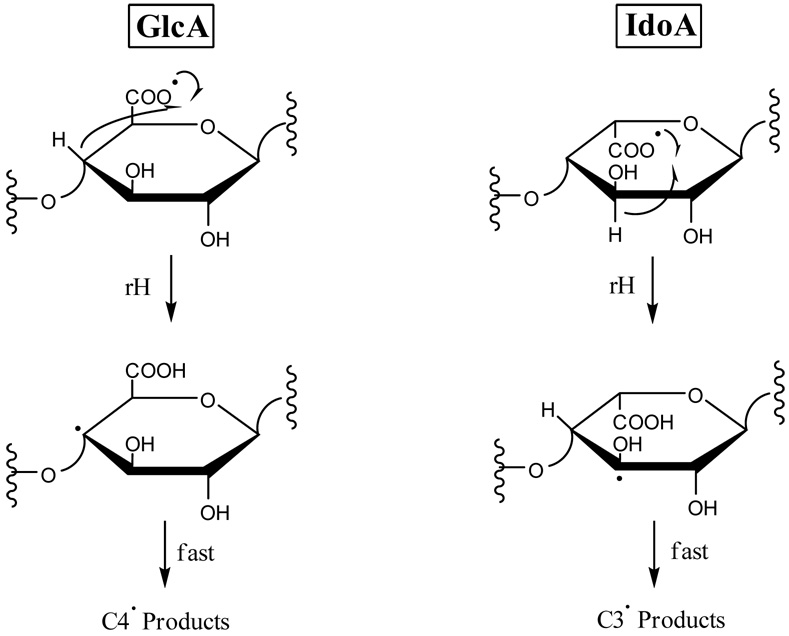
The differences in the observed product ions for the two epimers can be rationalized as arising from hydrogen rearrangement from C4 or the hydroxyl group on C3 of GlcA (tetrasaccharide 1) to the carboxy radical, versus rearrangement of a hydrogen atom from C3 or the C2 hydroxyl group to the carboxy radical for IdoA (tetrasaccharide 2), as proposed in Scheme 1 and Scheme 2 (see structure 1 for carbon atom nomenclature). Hydrogen rearrangement from C3 or the C2 hydroxyl group to the GlcA carboxy radical is not expected to occur for tetrasaccharide 1, as no conformation of the sugar ring brings the carboxy radical into the vicinity of C3 or the C2 hydroxyl group. Likewise, no conformation of tetrasaccharide 2 is expected to bring the IdoA carboxy radical close enough to the C3 hydroxyl group or C4 to permit H atom transfer. Subsequent radical-initiated cleavage of these radicals produces different fragmentations that can distinguish IdoA from GlcA.
Scheme 1.
Scheme 2.
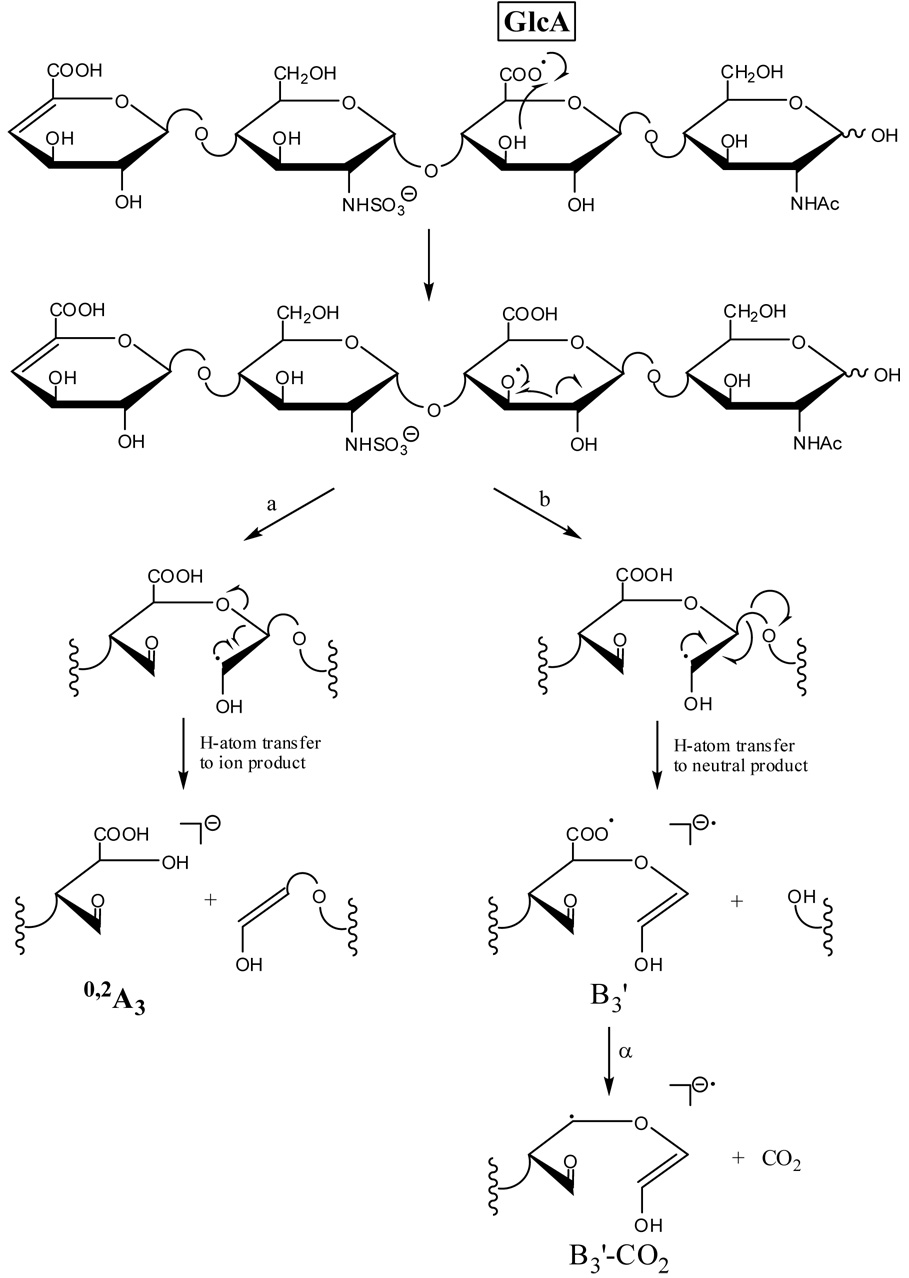
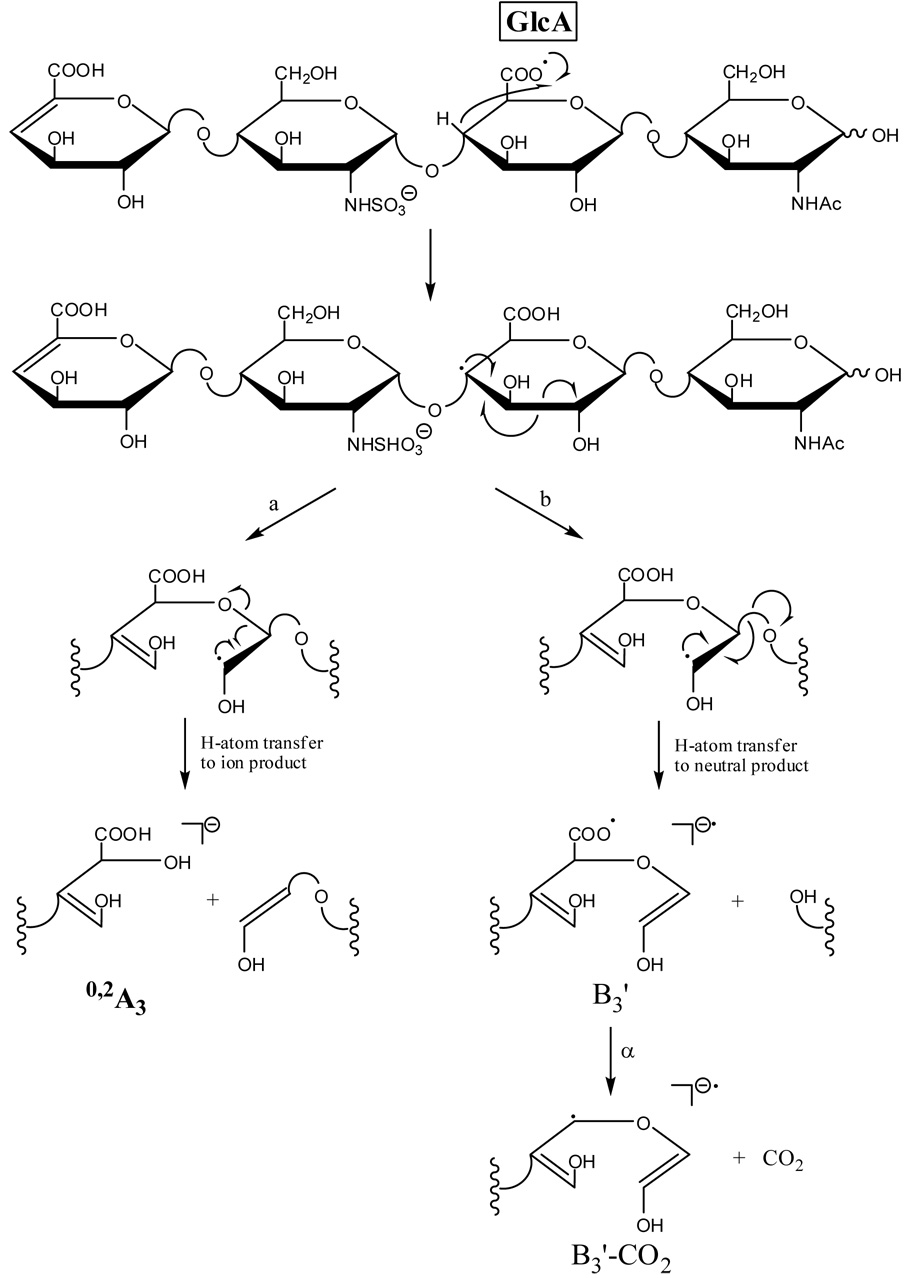
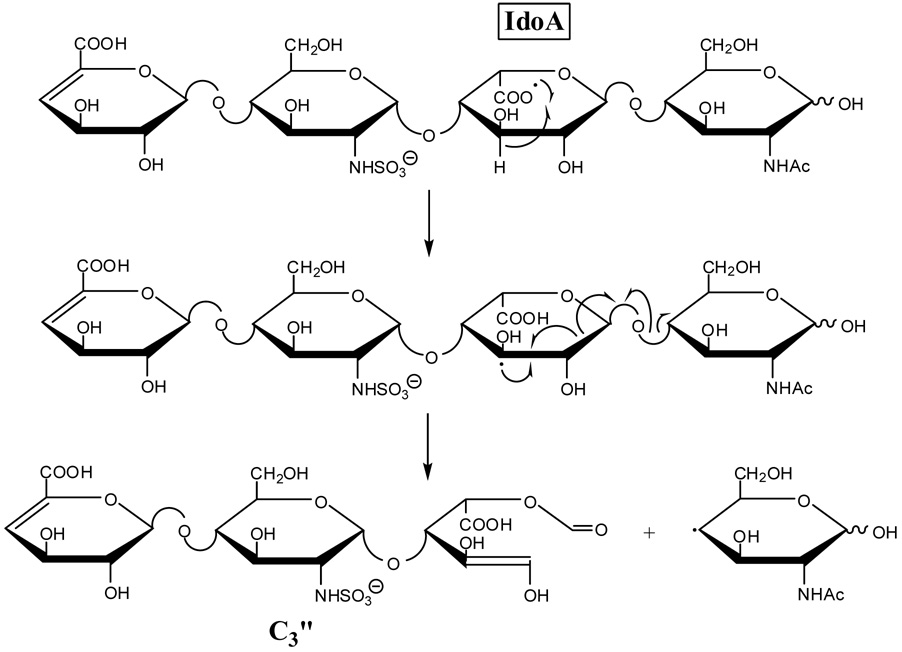
The C3 oxy-radical or the C4 radical that results from the aforementioned hydrogen rearrangement for 1 can undergo α-cleavage to form the 0,2A3 product ion, as proposed in Scheme 3a and Scheme 4a. The observed 0,2A3 fragment ion is even-electron, formed by a mechanism that includes a rearrangement of a hydrogen atom from the departing neutral fragment to the production. H atom transfer to oxygen is often observed in the tandem mass spectra of carbohydrates when a carbon-oxygen bond is broken, for example in the cleavage of glycosidic bonds to form B, C, Y, and Z type ions.33 The 0,2A3 product ion is easily rationalized to result from α-cleavage of an oxy-radical located at C3 or a radical at C4, but not by α-cleavage from an oxy-radical located at C2 or a radical at C3. As H atom transfer is expected to occur between the IdoA carboxy radical and the C2 hydroxyl group or C3, but not with the C3 hydroxyl group or C4, formation of the 0,2A3 product ion is not expected for epimer 2.
Scheme 3.
Scheme 4.
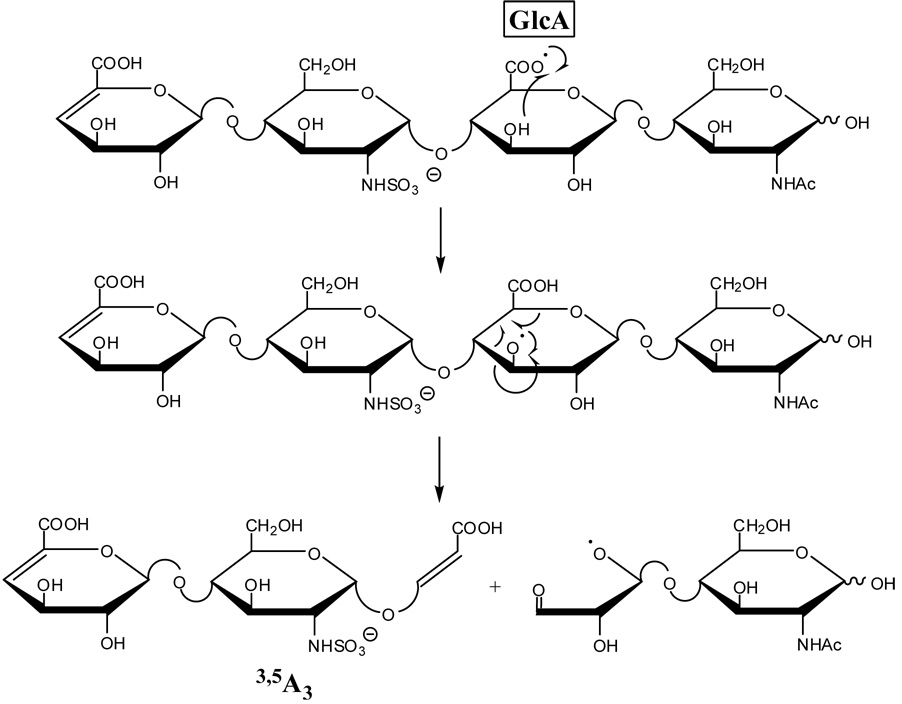
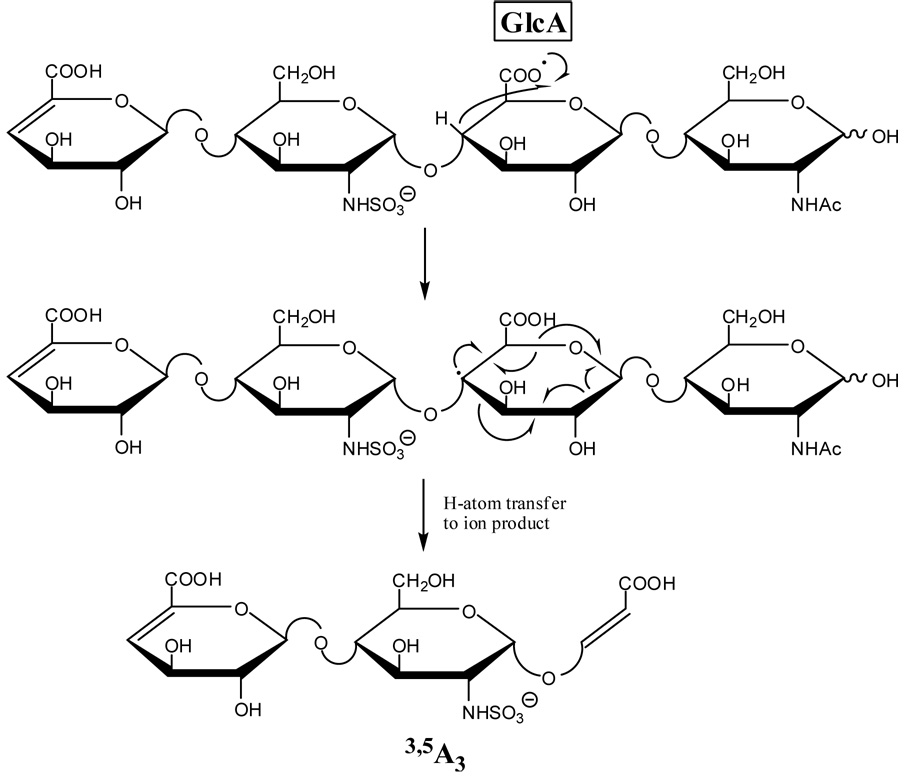
Formation of an oxy-radical on C3 or a C4 radical may initiate a second fragmentation pathway (Scheme 3b and Scheme 4b), resulting in formation of the B3’ product ion. Fragmentation via this pathway occurs with the transfer of a hydrogen atom from the B3’ product ion to the oxygen atom of the neutral fragment. Similar to the formation of the 0,2A3 product ion, formation of the B3’ should occur only when the oxy-radical is on C3, and not when the oxy-radical is located at C2, or when the radical is located at C4 and not C3. We find that B3’ is twice as abundant as B3 in the EDD spectrum of 1, but in the EDD spectrum of 2, the B3’ product abundance is only 12% of the abundance of the B3 product (see table of intensities in supplemental data.) Some of this peak is the isotopic contribution of the adjacent peaks, and very little, if any, is from B3'. Also consistent with this proposed radical mechanism is the observation that B3’ occurs only as a singly-charged ion, while B3 occurs as both a singly- and doubly-charged ion, i.e. only [B3-H]−• is observed, and not [B3-H]2−. Also observed in the EDD spectrum of 1 and not in the EDD spectrum of 2 is the B3’-CO2 ion, which may arise from the B3’ product ion as proposed in Scheme 3b and Scheme 4b. We do not believe that this product arises from the [M-2H-CO2]−• ion, as this species has lost its chirality at C5, and so were the B3’-CO2 to arise from [M-2H-CO2]−• it would be expected to appear in the EDD spectra for both epimers 1 and 2. Similar to B3’, the B3’-CO2 product ion is observed only as a singly-charged ion, consistent with our proposed radical mechanism.
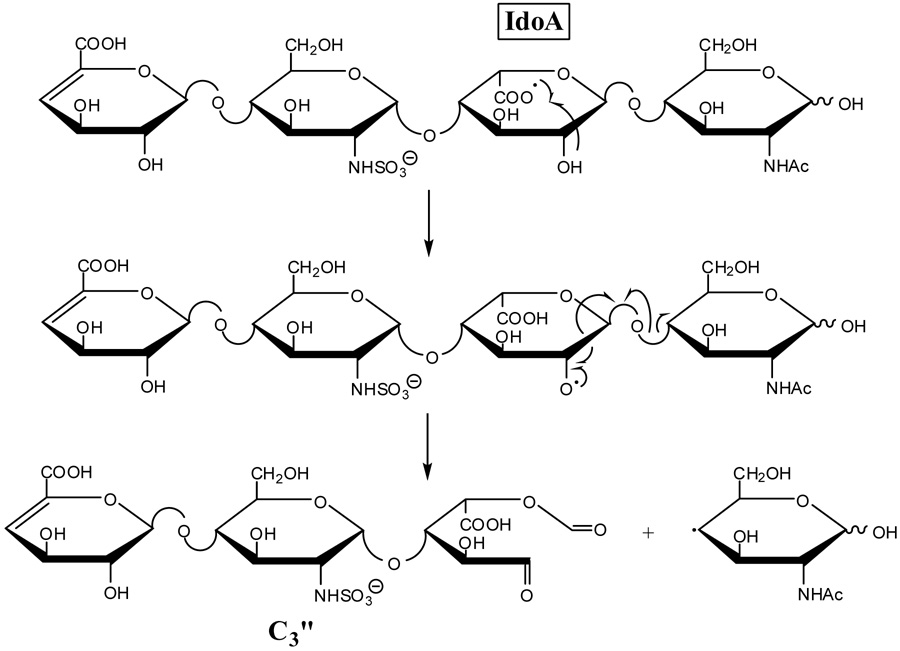
For IdoA (tetrasaccharide 2), hydrogen transfer to form an oxy-radical at C2 or a radical at C3 can lead to the formation of the C3” product ion by α-cleavage as proposed in Scheme 5 and Scheme 6, respectively. If the oxy-radical were located at C3 or the radical at C4, there would be no simple route to form the C3” product. Since this product is observed in the EDD spectrum of 1, albeit at a much lower abundance than in the EDD spectrum of 2, it may also be produced by an additional, secondary fragmentation mechanism. An alternative possibility is that the small amount of C3” product in the EDD of 1 results from hydrogen atom transfer between the oxygen atoms on C2 and C3, as proposed in Scheme 1. This H atom transfer is expected to be slow due to the unfavorable conformation of the sugar ring that would be necessary to align these groups sufficiently to allow this rearrangement to occur. The differences in the abundances of the diagnostic product ions for GlcA versus IdoA suggest that hydrogen transfer between the oxygens of C2 and C3, if it occurs at all, is slow compared to cross-ring and glycosidic fragmentations.
Scheme 5.
Scheme 6.
Similar to C3”, the 3,5A3 product ion is observed in both the EDD of 1 and 2, but is significantly more intense in 1. This product ion may result from α-cleavage of an oxy-radical located at C3 as proposed in Scheme 7 or a radical at C4 as proposed in Scheme 8, but not by α-cleavage from an oxy-radical located at C2 or a radical at C3. The occurrence of the 3,5A3 product at reduced ion intensity in the EDD of 2 can be rationalized as resulting from slow H atom transfer from the C3 hydroxyl group to the oxy-radical on C2 or production of this product via a second fragmentation mechanism.
Scheme 7.
Scheme 8.
A second set of epimers, 3 and 4, were analyzed by EDD (Figure 3a and 3b, respectively) to test these hypothesized fragmentation mechanisms. The epimers 3 and 4 differ from 1 and 2 by absence of N-sulfation on the glucosamine residue adjacent to the non-reducing end. Many of the same product ions observed for 1 and 2 are observed for 3 and 4. Consistent with the observations for 1 and 2, the 0,2A3, B3’, and B3’-CO2 product ions are observed only in EDD of 3, the GlcA epimer, and not in EDD of the IdoA epimer, 4. Also, B3’ is more abundant than the B3 product ion in 3 and the C3” product ion is higher in abundance in the EDD spectrum of 4 versus 3. However, there are few differences between the EDD mass spectra of the two epimer pairs. For example, the C3” product is less abundant than the C3 product in the EDD spectrum of 4, the IdoA epimer, while it was more abundant than C3 for the IdoA epimer, 2. C3” was observed in the EDD spectra of both epimers 1 and 2 (and is more abundant for the IdoA epimer 2 than for the GlcA epimer 1), but for the other epimer pair this product is observed only in the EDD spectrum of the IdoA epimer 4. Since the C3” product ion can distinguish IdoA from GlcA only by being more abundant in the former, it is useful only if one can directly compare EDD mass spectra of both epimers. Differences in the 3,5A3 product ion abundance in 1 and 2 were attributed to the preference of GlcA forming a C3 oxy-radical or a C4 carbon radical. However, the abundance of the 3,5A3 product ion is the same for epimers 3 and 4, indicating that 3,5A3 is not diagnostic of IdoA versus GlcA for these nonsulfated tetrasaccharides. Nevertheless, the EDD spectra of tetrasaccharides 3 and 4 exhibit key diagnostic ions (0,2A3, B3’, and B3’-CO2) that allow GlcA to be distinguished from IdoA. These diagnostic product ions, and their observed relative intensities, are listed in Table 1 for each epimer pair.
Figure 3.
EDD mass spectra of the [M-2H]2− precursor ion of HS epimers expanded to show the region m/z 375 – 725: (a) GlcA containing tetrasaccharide 3; (b) IdoA containing tetrasaccharide 4. The ▼ above the peak labels indicates product ions unique to each HS epimer. Insets: observed bond cleavages for the HS tetrasaccharides.
Table 1.
Diagnostic product ions and their relative ion intensities. The relative ion intensities are reported with respect to the most intense product ion, C2. NO = not observed.
| 0,2A3 | B3' | B3'-CO2 | C2" | 3,5A3 | |
|---|---|---|---|---|---|
| Tetrasaccharide 1 | 0.0524 | 0.1329 | 0.0981 | 0.1181 | 0.6084 |
| Tetrasaccharide 2 | NO | 0.0105 | 0.0087 | 0.3610 | 0.0918 |
| Tetrasaccharide 3 | 1161236 | 681500 | 1630312 | 112749 | 345768 |
| Tetrasaccharide 4 | NO | 21293 | 30715 | 137236 | 98048 |
CONCLUSIONS
EDD produces a radical anion which can undergo radical-driven fragmentation processes that are very different from the fragmentation of even electron anions that are formed by electrospray ionization. We have demonstrated the utility of EDD for distinguishing GlcA versus IdoA in GAG tetrasaccharides. The diagnostic ions for GlcA are the 0,2A3, B3’, and B3’-CO2 product ions. IdoA can be distinguished by the absence of these ions in the EDD mass spectrum. To a lesser extent, the abundance of the C3” product ion can be used to determine C5 stereochemistry. All of the diagnostic product ions can be rationalized as arising from a proposed radical species whose subsequent fragmentation is influenced by C5 stereochemistry. Consistent with the proposed radical mechanisms, these diagnostic ions are not observed by CAD or IRMPD of these GAG tetrasaccharides (data not shown). The EDD mass spectra are highly reproducible, even for acquisitions taken several months apart (see supplemental data), and so the ions that distinguish the epimers from each other have good diagnostic value. Of course, the proposed mechanisms are hypothetical at this point. Validation of these mechanisms will require considerable more effort, for example with isotope labeling experiments. Since it is possible for GAGs to contain a higher degree of sulfation than for the tetrasaccharides studied here, future work will examine the influence of the degree of sulfation on the capability to distinguish GlcA/IdoA epimers by EDD.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge financial support from the National Institutes of Health grant #2R01-GM038060-16.
REFERENCES
- 1.Perrimon N, Bernfield M. Semin. Cell Dev. Biol. 2001;12:65–67. doi: 10.1006/scdb.2000.0237. [DOI] [PubMed] [Google Scholar]
- 2.Linhardt RJ, Toida T. Acc. Chem. Res. 2004;37:431–438. doi: 10.1021/ar030138x. [DOI] [PubMed] [Google Scholar]
- 3.Fannon M, Forsten KE, Nugent MA. Biochemistry. 2000;39:1434–1445. doi: 10.1021/bi991895z. [DOI] [PubMed] [Google Scholar]
- 4.Wu ZL, Zhang L, Yabe T, Kuberan B, Beeler DL, Love A, Rosenberg RD. J. Biol. Chem. 2003;278:17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- 5.Gotte M. The FASEB Journal. 2003;17:575–591. doi: 10.1096/fj.02-0739rev. [DOI] [PubMed] [Google Scholar]
- 6.Williams RK, Straus SE. J. Virol. 1997;71:1375–1380. doi: 10.1128/jvi.71.2.1375-1380.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batinic D, Robey FA. J. Biol. Chem. 1992;267:6664–6671. [PubMed] [Google Scholar]
- 8.Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. Nature Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 9.Liu DF, Shriver Z, Gi YW, Venkataraman G, Sasisekharan R. Seminars in Thrombosis and Hemostasis. 2002;28:67–78. doi: 10.1055/s-2002-20565. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl U, Kusche-Gullberg M, Kjellen L. J. Biol. Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 11.Turnbull J, Powell A, Guimond S. Trends Cell Biol. 2001;11:75–82. doi: 10.1016/s0962-8924(00)01897-3. [DOI] [PubMed] [Google Scholar]
- 12.Salmivirta M, Lidholt K, Lindahl U. The FASEB Journal. 1996;10:1270–1279. doi: 10.1096/fasebj.10.11.8836040. [DOI] [PubMed] [Google Scholar]
- 13.Horne A, Gettins P. Carbohydr. Res. 1992;225:43–57. doi: 10.1016/0008-6215(92)80038-3. [DOI] [PubMed] [Google Scholar]
- 14.Naggar EF, Costello CE, Zaia J. J. Am. Soc. Mass Spectrom. 2004;15:1534–1544. doi: 10.1016/j.jasms.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Takagaki K, Kojima K, Majima M, Nakamura T, Kato I, Endo M. Glycoconjugate J. 1992;9:174–179. doi: 10.1007/BF00731162. [DOI] [PubMed] [Google Scholar]
- 16.Zaia J, Costello CE. Anal. Chem. 2003;75:2445–2455. doi: 10.1021/ac0263418. [DOI] [PubMed] [Google Scholar]
- 17.Saad OM, Leary JA. Anal. Chem. 2005;77:5902–5911. doi: 10.1021/ac050793d. [DOI] [PubMed] [Google Scholar]
- 18.Sweeney MD, Yu Y, Leary JA. J. Am. Soc. Mass Spectrom. 2006;17:1114–1119. doi: 10.1016/j.jasms.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 19.Zaia J, Li X-Q, Chan S-Y, Costello CE. J. Am. Soc. Mass Spectrom. 2003;14:1270–1281. doi: 10.1016/S1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- 20.Miller MJC, Costello CE, Malmstrom A, Zaia J. Glycobiology. 2006;16:502–513. doi: 10.1093/glycob/cwj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hitchcock AM, Costello CE, Zaia J. Biochemistry. 2006;45:2350–2361. doi: 10.1021/bi052100t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anusiewicz I, Jasionowski M, Skurski P, Simons J. J. Phys. Chem. A. 2005;109:11332. doi: 10.1021/jp055018g. [DOI] [PubMed] [Google Scholar]
- 23.Budnik BA, Haselmann KF, Zubarev RA. Chem. Phys. Lett. 2001;342:299–302. [Google Scholar]
- 24.Cooper HJ, Hakansson K, Marshall AG. Mass Spectrom. Rev. 2005;24:201. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 25.Kjeldsen F, Silivra OA, Ivonin IA, Haselmann KF, Gorshkov M, Zubarev RA. Chem. Eur. J. 2005;11:1803–1812. doi: 10.1002/chem.200400806. [DOI] [PubMed] [Google Scholar]
- 26.McFarland MA, Marshall AG, Hendrickson CL, Nilsson CL, Fredman P, Mansson JE. J. Am. Soc. Mass Spectrom. 2005;16:752. doi: 10.1016/j.jasms.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Mo J, Adamson JT, Hakansson K. Anal. Chem. 2005;77:1876–1882. doi: 10.1021/ac048415g. [DOI] [PubMed] [Google Scholar]
- 28.Wolff JJ, Amster IJ, Chi L, Linhardt RJ. J. Am. Soc. Mass Spectrom. 2006 doi: 10.1016/j.jasms.2006.09.020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pervin A, Gallo C, Jandik KA, Han X-J, Linhardt RJ. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 30.Munoz E, Xu D, Avci F, Kemp M, Liu J, Linhardt RJ. Biochem. Biophys. Res. Commun. 2006;339:597–602. doi: 10.1016/j.bbrc.2005.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue Y, Nagasawa K. Carbohydrate Research. 1976;46:87–95. doi: 10.1016/s0008-6215(00)83533-8. [DOI] [PubMed] [Google Scholar]
- 32.Tsybin YO, Witt M, Baykut G, Håkansson P. Rapid Commun. Mass Spectrom. 2004;18:1607–1613. doi: 10.1002/rcm.1525. [DOI] [PubMed] [Google Scholar]
- 33.Domon B, Costello CE. Glycoconjugate J. 1988;5:397–409. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.