Abstract
Thymidylate deprivation brings about “thymineless death” in prokaryotes and eukaryotes. Although the precise mechanism for thymineless death has remained elusive, inhibition of the enzyme thymidylate synthase (TS), which catalyzes the de novo synthesis of TMP, has served for many years as a basis for chemotherapeutic strategies. Numerous studies have identified a variety of cellular responses to thymidylate deprivation, including disruption of DNA replication and induction of DNA breaks. Since stalled or collapsed replication forks and strand breaks are generally viewed as being recombinogenic, it is not surprising that a link has been demonstrated between recombination induction and thymidylate deprivation in bacteria and lower eukaryotes. A similar connection between recombination and TS inhibition has been suggested by studies done in mammalian cells, but the relationship between recombination and TS inhibition in mammalian cells had not been demonstrated rigorously. To gain insight into the mechanism of thymineless death in mammalian cells, in this work we undertook a direct investigation of recombination in human cells treated with raltitrexed (RTX), a folate analog that is a specific inhibitor of TS. Using a model system to study intrachromosomal homologous recombination in cultured fibroblasts, we provide definitive evidence that treatment with RTX can stimulate accurate recombination events in human cells. Gene conversions not associated with crossovers were specifically enhanced several-fold by RTX. Additional experiments demonstrated that recombination events provoked by a double-strand break (DSB) were not impacted by treatment with RTX, nor was error-prone DSB repair via nonhomologous end-joining. Our work provides evidence that thymineless death in human cells is not mediated by corruption of DSB repair processes and suggests that an increase in chromosomal recombination may be an important element of cellular responses leading to thymineless death.
1. Introduction
The enzyme thymidylate synthase (TS) catalyzes the reductive methylation of dUMP by 5,10-methylenetetrahydrofolate to produce TMP, which is required for DNA replication in proliferating cells. The reaction catalyzed by TS represents the sole pathway for de novo TMP synthesis, and targeted inhibition of this pathway has served for many years as a common strategy in chemotherapeutic approaches. For example, the antimetabolite 5-fluorouracil (5-FU) has been put to use for many years in the treatment of a variety of cancers [1,2,3]. 5-FU can be metabolized to FdUMP which inhibits TS by forming a ternary complex with TS and 5,10-methylenetetrahydrofolate. 5-FU is often administered in combination with leucovorin which is metabolized to 5,10-methylenetetrahydrofolate. In turn, the formation of the ternary complex between FdUMP, TS, and 5,10-methylenetetrahydrofolate is enhanced and the anti-cancer action of 5-FU is potentiated. Although inhibition of TS is undoubtedly a major means by which 5-FU exerts it anti-cancer effects, some metabolites of 5-FU (namely, FUTP and FdUTP) do not target TS but, rather, readily incorporate into RNA and DNA. Incorporation of fluoropyrimidines into nucleic acids engenders additional cellular effects that make important contributions to the cytotoxicity of 5-FU [1,2]. In addition to fluoropyrimidine-based TS inhibitors, folate analogs have also been put to use as TS inhibitors in chemotherapeutic approaches. Raltitrexed (RTX) is an example of a folate analog that was rationally designed as a specific inhibitor of TS [3–6]. Unlike fluoropyrimidines, RTX apparently acts exclusively through TS inhibition. The use of RTX in the treatment of colorectal cancer has been approved in Europe, Canada, Australia and Japan [7,8].
Interestingly, and perhaps surprisingly, despite the fact that inhibition of TS has served for many years in chemotherapeutic strategies, the mechanism by which “thymineless stress” ultimately leads to “thymineless death” has yet to be fully elucidated. Several documented effects of TS inhibition, alone or in combination, likely contribute to cellular demise. One notable consequence of TS inhibition is the generation of nucleotide pool imbalance, which may disrupt DNA synthesis and DNA repair and lead to lethal DNA lesions [1,9–12]. Another consequence of TS inhibition that has received much attention involves the intracellular accumulation of dUMP due to the blockage of its conversion to TMP. Accumulated dUMP can be converted to dUTP which, in combination with the depletion of TTP pools, may result consequently in the misincorporation of uracil into DNA [1,3,9,12,13]. Subsequent action of base excision repair to remove uracil from DNA has been hypothesized to result in futile cycles of repair since, under conditions of thymidylate deprivation, uracil will continue to be misincorporated into DNA during repair. The hypothesized futile cycling in turn leads to DNA strand breakage which may lead directly or indirectly to cell death [1,3,9,12,13]. It must be noted that the role that misincorporation and/or excision of uracil plays in the induction of cell death through TS inhibition remains controversial as there have been a number of reports suggesting that uracil incorporation and excision is not required for thymineless death [14–18]. It is, however, well-established that thymineless stress induces single and double-strand breaks (DSBs) in chromosomal DNA [9,12,14,18–24]. Whether such DNA strand breakage is an early event that plays a causal role in thymineless death, or a late event occurring after apoptosis has already been initiated, is an issue that has proven problematic to resolve.
One interesting potential impact of TS inhibition that has received some attention in the literature is the possibility that thymidylate starvation induces homologous recombination (HR) among chromosomal sequences. Since TS inhibition leads to DNA breakage and interferes with DNA synthesis, it seems plausible to suspect that HR may concomitantly be induced because DNA breaks and replication blockage are generally viewed as being recombinogenic [25]. Potentially high levels of HR could, in turn, engender an instability that may directly or indirectly contribute to cell death or lead to genetic abnormalities following recovery from treatment with TS inhibitors. A link between HR and thymidylate starvation has been confirmed in prokaryotes and lower eukaryotes [9,26–33]. A similar link has been suggested, but not demonstrated definitively, in higher eukaryotes [14,19,34–36].
In order to better understand the relationship between HR and TS inhibition in human cells, and to gain further insight into the mechanism of thymineless death, we initiated a study of the effect of TS inhibition on intrachromosomal HR. In this paper, we describe the development of a model system using cultured human fibroblasts to attack this issue directly. Our system involves the use of novel integrated DNA substrates that enable the study of both spontaneous HR as well as HR provoked by induction of a genomic DSB. We report that treatment of human cells with the TS inhibitor RTX can increase the rate of spontaneous intrachromosomal gene conversion several-fold. In contrast, RTX treatment had no apparent effect on either DSB-induced HR or repair of genomic DSBs by error-prone nonhomologous end-joining (NHEJ). Our results suggest that a global increase in recombination activity may be an integral component of cellular responses leading to thymineless death.
2. Materials and methods
2.1. Cell culture
Normal human fibroblast cell line GM00637 (immortalized by SV40) was obtained from the NIGMS. Cells were cultured in alpha-modified minimum essential medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum. All cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
2.2. Recombination and repair substrates
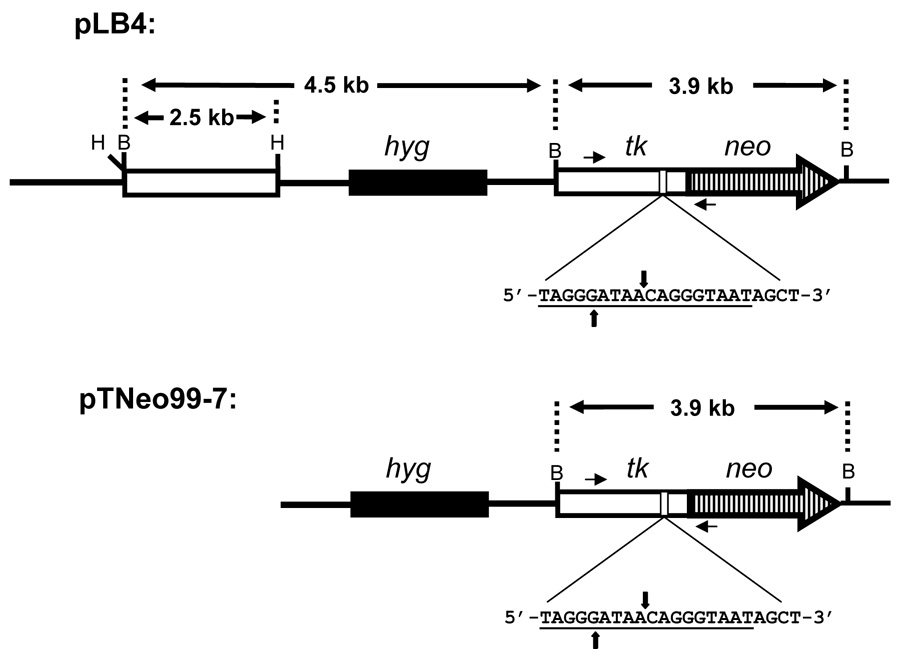
Plasmids pLB4 and pTNeo99-7 (Fig. 1) were described previously [37,38]. Substrate pLB4 was derived from pTNeo99-7 by inserting a 2.5 kb HindIII fragment containing a complete functional herpes simplex virus type 1 (HSV-1) thymidine kinase (tk) gene into the unique HindIII site on pTNeo99-7. This inserted “donor” fragment on pLB4 shares about 1.7 kb of homology with the tk portion of the fusion gene. Due to several scattered mismatches, the donor on pLB4 displays about 1% sequence divergence with the tk portion of the tk-neo fusion gene (Fig. 2).
Fig. 1.
Recombination and NHEJ substrates pLB4 and pTNeo99-7. Each substrate contains a functional hygromycin resistance gene (hyg), used to select for stably transfected cells. The 18 bp recognition site for endonuclease I-SceI is underlined, and the sites of staggered cleavage by I-SceI are indicated. The orientation, with respect to transcription, of both the tk gene and/or tk-neo fusion gene in each substrate is from left to right as drawn. PCR primers AW85 and AW91 are indicated by short horizontal arrows and are located 1.4 kb apart in the tk-neo fusion gene. BamHI (B) and HindIII (H) sites are shown. See text for details.
Fig. 2.
Alignment of tk-neo fusion gene sequence with donor tk sequence from pLB4. Nucleotides 308–1622, numbering according to [39], of the tk portion of the tk-neo fusion gene are aligned with the corresponding donor sequence from pLB4. The span of tk sequence shown comprises the tk portion of PCR products generated by primers AW85 and AW91. Mismatches between donor and tk-neo fusion gene sequences are highlighted. The 22 bp oligonucleotide containing the 18 bp I-SceI recognition sequence inserted in the fusion gene (absent from donor) is depicted in bold, with the actual I-SceI recognition sequence in uppercase. AluI recognition sites (agct) are underlined and are indicated by downward arrows.
2.3. Insertion of recombination and repair substrates into cell lines
5 × 106 GM00637 cells were electroporated with 3 µg of plasmid substrate using a Bio-Rad Gene Pulser set at 1000V, 25 µF. Cells were plated into 150 cm2 flasks and allowed to grow for two days under no selection. Cells were then plated at a density of 1 × 106 cells per 75 cm2 flask into medium supplemented with hygromycin at 100 µg/ml. After 4 days of growth, hygromycin resistant colonies were picked. These clones were propagated and screened by Southern blot analysis to identify cell lines containing a single stably integrated copy of the transfected plasmid substrate.
2.4. Determination of effect of RTX on cell viability and/or growth rate
RTX was generously provided by AstraZeneca, UK. To assess the effect of RTX on cell viability and/or growth, GM00637 cells were plated into 25 cm2 flasks at a density of 3.3 × 105 cells per flask. Twenty four hours later, the medium was replaced with medium supplemented with various doses of RTX over a broad range of concentrations ranging from less than 1 nM to greater than 1 µM. Three flasks of cells were used for each dose tested. Cells were exposed to RTX for 24 hours, at which time the cells were refed with medium containing no RTX. Forty-eight hours after feeding with drug-free medium, cells were harvested and counted. The cell counts for the cells exposed to the various RTX doses were compared with the cell count for control cells not exposed to RTX as a measure of the impact of RTX on cell viability and/or growth rate.
2.5. Determination of spontaneous intrachromosomal HR frequencies
Spontaneous recombination frequencies were determined for cells treated or not treated with RTX. To perform such an analysis on a cell line, 10 subclones were first generated from the parental cell line. Subclones were propagated separately to several million cells at which point cells from each subclone were plated into several 150 cm2 flasks at a density of 2 × 106 cells per flask. Twenty-four hours after plating the cells, the flasks for each subclone were divided into two groups; one group of flasks was refed with medium supplemented with 1.5 nM RTX while the other group was fed with medium containing no RTX. Twenty-four hours later, cells from each group (RTX-treated and untreated) were harvested by trypsinization and plated at a density of 5 × 105 cells per 75 cm2 flask into medium containing 1000 µg/ml G418 in order to select for G418R segregants. At the same time, aliquots of cells from each subclone were plated into medium under no selection to assess cell viability. Specifically, for each subclone and condition (RTX-treated and untreated), 200 cells were plated into each of two 25 cm2 flasks. After 14 days growth, colonies recovered under G418 selection or under no selection were counted. Based on the number of colonies recovered under no selection, the number of viable cells that had been plated into G418 selection was calculated. Recombination frequency for each subclone and condition was then calculated by dividing the number of G418R colonies recovered by the number of viable cells plated into G418. G418R colonies were harvested for further propagation and analysis.
2.6. Recovery of DSB-induced HR and nonhomologous end-joining (NHEJ) events
Plasmid CMV3xnls-I-SceI (“pSce”) expresses endonuclease I-SceI in mammalian cells and was generously provided by M. Jasin (Sloan Kettering). This plasmid was transiently introduced into cells to induce a DSB at the I-SceI site within an integrated DNA construct. To accomplish this, cells from an appropriate cell line were plated into 150 cm2 flasks at a density of 2 × 106 cells per flask. Twenty-four hours after plating cells, the growth medium was changed to medium supplemented with 1.5nM RTX or medium containing no RTX supplement for controls. Twenty-four hours later, cells were harvested; 5 × 106 cells and 20 µg of pSce were resuspended in 800 µl of phosphate buffered saline and placed in a 0.4 cm gap electroporation cuvette. A Bio-Rad Gene Pulser was used to deliver a pulse of 700 V and 25 µF. Following electroporation with pSce, cells were plated into a 150 cm2 flask under no selection. Cells were harvested 48 hours later and plated into medium containing 1000 µg/ml G418. As above, aliquots of cells were also plated into medium under no selection to assess cell viability.
2.7. PCR amplification and DNA sequence analysis
A segment of the tk-neo fusion gene spanning the I-SceI site was amplified from 500 ng of genomic DNA isolated from G418R clones using primers AW85 (5’-TAATACGACTCACTATAGGGCCAGCGTCTTGTCATTGGCG- 3’) and AW91 (5’-GATTTAGGTGACACTATAGCCAAGCG GCCGGAGAACCTG-3’). AW85 is composed of nucleotides 308–327 of the coding sequence of the HSV-1 tk gene, numbering according to [39], with a T7 forward universal primer appended to the 5’ end of the primer. AW91 is composed of 20 nucleotides from the noncoding strand of neomycin gene mapping 25 through 44 bp downstream from the neomycin start codon, with an Sp6 primer appended to the 5’ end of the primer. PCR was carried out using Ready-To-Go PCR beads (GE Healthcare) and a “touchdown” PCR protocol. The annealing temperature was initially set to 72°C and was progressively decreased in steps of 2° down to 62° with two cycles at each temperature. An additional 20 cycles were run at an annealing temperature of 60°C. For each experiment, the PCR products were expected to be ~1.4 kb in length unless detectable deletions or insertions had occurred. Prior to sequencing, PCR products were treated with shrimp alkaline phosphatase and exonuclease I (USB). PCR products were then sequenced from a T7 or Sp6 primer on a LICOR 4000L at the DNA Sequencing and Synthesis Core Facility in the Department of Biological Sciences at the University of South Carolina. At least 800 bp of DNA sequence were determined for each PCR product sequenced.
2.8. Cell cycle analysis
Cell cycle analysis was done essentially as previously described [40]. Briefly, cells were plated at a density of 3.3 × 105 cells per 25 cm2 flask. One day later, cells were fed with medium containing 1.5 nM RTX or containing no RTX in the case of controls. Twenty-four hours later, cells were collected in growth medium and fixed with 70% ethanol. Fixed cells were stained at 4°C with a solution containing propidium iodide (50 µg/ml), RNase A (0.1 mg/ml), and 1% BSA in phosphate-buffered saline. DNA content was assessed using an EPICs XL-MCL flow cytometer (Beckman Coulter, Fullerton, CA) at the South Carolina Cancer Center, University of South Carolina.
3. Results
3.1. An experimental system for monitoring intrachromosomal HR in human cells
It was our aim to assess the effect that treatment with RTX has on intrachromosomal HR in human cells. To do so, we used an experimental system we developed that incorporates a genetic selection for HR events in cultured human fibroblasts. We made use of human fibroblast cell line GM00637, an immortalized cell line derived from an apparently healthy individual. Recombination substrate pLB4 (Fig. 1) was inserted into the genome of GM00637 cells by stable transfection, and a cell line (designated pLB4/11) containing a single integrated copy of pLB4 was isolated. pLB4 contains a tk-neo fusion gene disrupted by a 22 bp oligonucleotide inserted into the tk portion of the fusion gene. The oligonucleotide contains the 18 bp recognition site for yeast endonuclease I-SceI. Also contained in pLB4 is a complete, functional tk gene. HR between the tk gene and the tk-neo fusion gene can restore function to the fusion gene, and such an event can be recovered by selection for G418R segregants. Because of the presence of the ISceI site in the tk-neo fusion gene in pLB4, a DSB can be introduced into the fusion gene by transiently transfecting a cell line containing a copy of pLB4 with the I-SceI expression plasmid pSce. A cell line containing pLB4 is therefore useful for studying both spontaneous as well as DSB-induced HR. Restoration of function of the neo portion of the tk-neo fusion gene on pLB4 does not require accurate removal of the 22 bp oligonucleotide inserted into the upstream tk portion since neo expression requires only that an appropriate reading frame be restored to the fusion gene. This feature is of relevance to studies involving DSB-induction since it allows recovery of error-prone NHEJ events in addition to accurate HR. Only one third of NHEJ events would be expected to restore the correct reading frame, so the true number of NHEJ events is likely three times greater than the number recoverable with pLB4.
3.2. Sensitivity of cells to RTX
Before we could assess the effect that RTX treatment has on HR in human cells, we first had to determine an appropriate dose of RTX to use in our studies. Our experimental protocol incorporated a 24 hour exposure to RTX. We wanted to use a dose of RTX that was sufficiently high to have a measurable impact on cells (i.e., induce thymineless stress), but not high enough to kill a majority of the cell population. Sensitivity of GM00637 cells to RTX increases as a function of cell density, with higher sensitivities recorded at higher cell densities. (Although the reason for this phenomenon is not known, it may be due to a more rapid depletion of thymidine from the medium at higher cell densities. It is also conceivable that there is a “bystander effect” in which dying cells release one or more toxic substances into the medium.) It was important that we measured RTX dose response at the same cell density that was to be used in subsequent experiments. The relevant cell density was too high to allow the recovery of individual colonies, so we could not assess cell survival using a colony counting assay. We therefore adopted the strategy described in Materials and Methods to asses the impact of RTX treatment on cell viability and/or growth. A 24 hour treatment with 1.5 nM RTX resulted in a total cell number that was reduced by about 30% compared to untreated cells when cells were counted two days after removal of RTX. We selected 1.5 nM RTX as a suitable dose for subsequent experiments. Cell cycle analysis performed on cells immediately following a 24 hour treatment with 1.5 nM RTX indicated that cells were arrested in early S-phase (data not shown), consistent with previously reported effects of thymidylate deprivation [15]. Although recovery from cell cycle arrest upon removal of RTX was not explicitly investigated, recovery from arrest was evident based on cell counts determined 48 hours after removal of RTX from the growth medium.
3.3. Induction of HR in human cells by RTX treatment
Cell line pLB4/11 was derived from GM00637 cells and contains a single integrated copy of recombination substrate pLB4 (Fig. 1). To assess the effect of RTX treatment on intrachromosomal HR, three independent experiments, each involving 10 independent subclones of pLB4/11, were carried out. A portion of each subclone was treated with 1.5 nM RTX for 24 hours prior to selection for G418R recombinants. As presented in Table 1, in each of the three experiments the cells treated with RTX produced a greater frequency of G418R colonies than did untreated cells. Overall, the frequency of G418R colonies was found to be about five-fold greater among cells treated with RTX compared with untreated cells. The difference in colony frequency for RTX-treated versus untreated cells was highly statistically significant for the pooled data from all experiments (p = 0.0003 by a paired t test performed on data from all subclones).
Table 1.
Recovery of spontaneous recombination events
| Experiment numbera |
RTXb | Total no. coloniesc |
Colony frequencyd (10−7) |
Recombination frequencye (10−7) |
|---|---|---|---|---|
| 1 | − | 10 | 1.6 | 1.5 |
| + | 32 | 5.2 | 4.7 | |
| 2 | − | 12 | 3.3 | 2.7 |
| + | 34 | 17.4 | 13.6 | |
| 3 | − | 7 | 1.7 | 1.2 |
| + | 24 | 10.9 | 10.1 | |
Three independent experiments, each involving10 independent subclones of cell line pLB4/11, were conducted in order to measure recombination frequency.
Portions of cells from each subclone in each experiment were untreated (−) or treated (+) for 24 hours with 1.5 nM RTX prior to plating into G418 selection as described in Materials and Methods.
Total number of G418R colonies recovered from all subclones for a given experiment and condition.
Calculated by dividing the total number of G418R colonies by the total number of viable cells plated into G418 selection.
Calculated by multiplying colony frequency by the percentage of colonies that arose from bona fide recombination events as assessed by PCR and AluI restriction analysis as described in text.
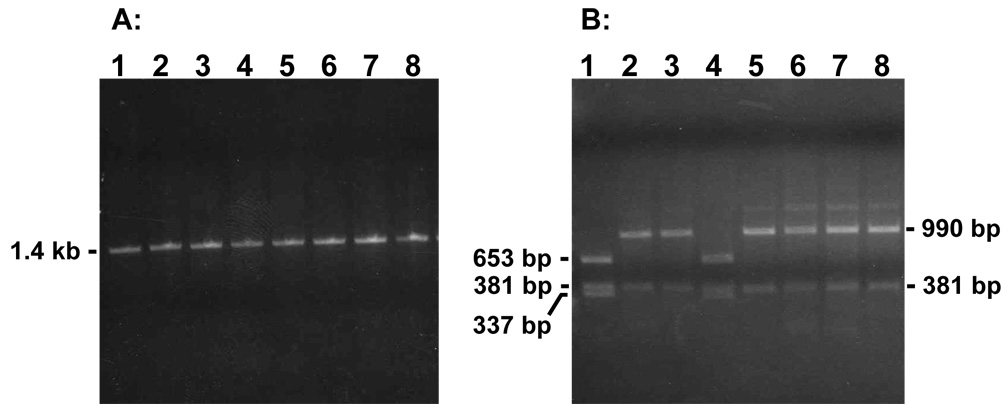
To ascertain that G418R segregants arose from HR events, samples of genomic DNA isolated from G418R clones were analyzed by PCR and restriction enzyme analysis (Fig. 3). A portion of the tk-neo fusion gene encompassing the original position of the disrupting 22 bp oligonucleotide was PCR-amplified, and a 1.4 kb PCR product was obtained from each clone as expected (see Fig. 1). PCR products were digested with AluI (Fig. 3B). In the parental cell line, two AluI sites flank the I-SceI site while a third AluI site is about 300 bp downstream from the ISceI site (Fig. 2). If HR had taken place in a given clone, the 22 bp insert containing the I-SceI site and the two flanking AluI sites would have been eliminated and the PCR product would produce a 990 bp fragment and a 381 bp fragment upon digestion with AluI (for example, Fig. 3B, lanes 2,3,5–8). In contrast, retention of the parental AluI restriction sites in a PCR product would produce 653 bp, 381 bp, 337 bp, and 22 bp fragments upon AluI digestion (Fig. 3B, lanes 1 and 4). The latter outcome was expected for the parental cell line as well as clones that arose via a mechanism other than accurate HR. In total, 38 G418R colonies that arose from RTX-treated cells and 25 colonies that arose from untreated cells were analyzed. We found that 87% (33 out of 38) of the clones from RTX-treated cells and 88% (22 out of 25) of the clones from untreated cells appeared to have arisen from HR. Nonrecombinant clones presumably arose from mutations that restored function to the tk-neo fusion gene, and these clones were not analyzed further.
Fig. 3.
Representative analysis of PCR products from G418R segregants. (A) Using primers AW85 and AW91, PCR products were generated from genomic DNA samples isolated from parental cell line pLB4/11 (lane 1) or representative G418R segregants arising from pLB4/11 (lanes 2–8) following treatment with RTX. (B) Lanes 1–8 display the PCR products from the corresponding lanes in (A) following digestion with AluI. The digested PCR product from parental cell line pLB4/11 in lane1 illustrates the nonrecombinant pattern of restriction fragments. The clone displayed in lane 4 produced a nonrecombinant pattern, while the remaining clones each produced the expected recombinant AluI digest pattern. Although not shown, for the gels in (A) and (B) a HindIII digest of lambda DNA and a HaeIII digest of phi X DNA were also run as molecular weight markers to confirm PCR fragment sizes.
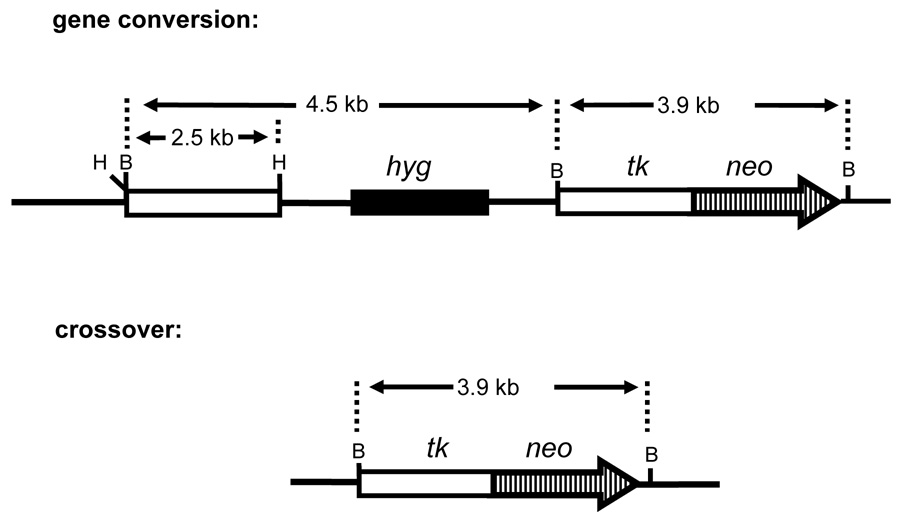
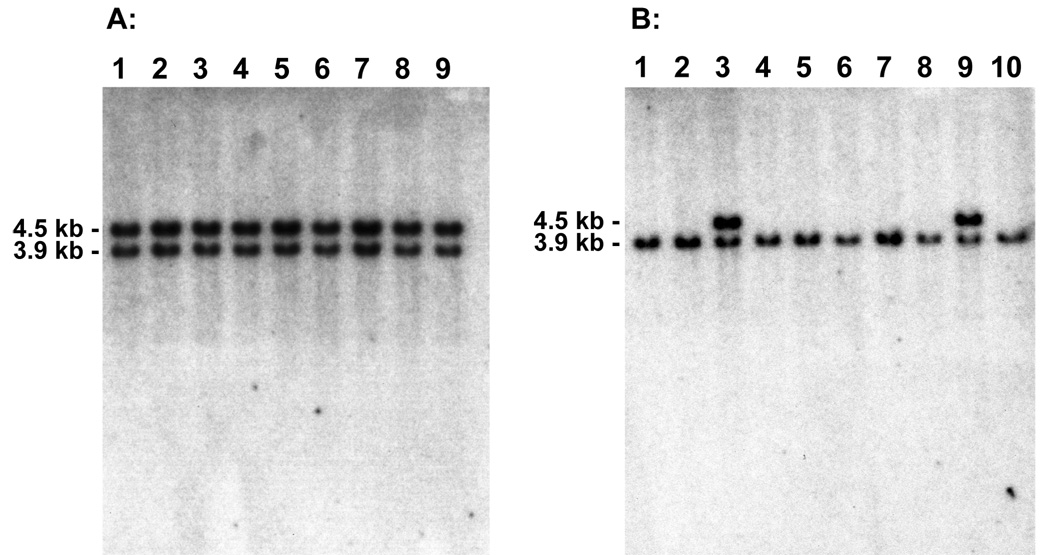
3.4. RTX selectively induces gene conversions (noncrossovers)
Both gene conversions and crossovers were theoretically recoverable in our experimental system. Gene conversions and crossovers could readily be distinguished on a Southern blot using a tk-specific probe since a BamHI digest of DNA from a gene conversion clone should produce both a 4.5 kb and a 3.9 kb band while a similar digest from a crossover clone should produce only the 3.9 kb band (Fig. 4). A representative Southern blot analysis is presented in Fig. 5, and the results from such analyses are summarized in Table 2. The data presented in Table 2 revealed a striking shift toward gene conversion following treatment of cells with RTX. For untreated cells, more than 75% of the G418R colonies analyzed arose from crossovers, with the balance arising from gene conversions. For RTX-treated cells, every G418R colony analyzed arose from a gene conversion. When the data from all three experiments were pooled, the difference in the relative occurrence of gene conversion and crossover clones for RTX-treated versus untreated cells was highly statistically significant (p = 9 × 10−10 by a Fisher exact test). If we considered only the minimal number of HR events that were unambiguously independent and necessarily were not siblings (that is, we counted only the number of distinct clones recovered from each subclone), the difference in the recovery of gene conversions and crossovers for treated versus untreated cells was again highly significant (p = 3.3 × 10−5 by a Fisher exact test). Indeed, we did not recover a single crossover event among 33 analyzed events that were recovered from RTX-treated cells.
Fig. 4.
Schematics of products of gene conversion and crossover events that reconstruct a functional tk-neo fusion gene. Diagnostic expected BamHI fragments are shown.
Fig. 5.
Representative Southern blot analysis of recombination events. Genomic DNA samples (8 µg) from G418R colonies recovered from pLB4/11 cells either treated with RTX (panel A) or untreated (panel B) were digested with BamHI and hybridized with a probe specific for HSV-1 tk to distinguish gene conversions from crossovers. Samples displaying both the 4.5 kb and 3.9 kb band were categorized as gene conversions while the samples displaying only the 3.9 kb band were categorized as crossovers (see Fig. 4). Cells treated with RTX displayed a significant shift toward gene conversion events.
Table 2.
Characterization of spontaneous recombination events
| Experiment number |
RTXa | Colonies Analyzed |
Gene Conversionsb | Crossoversb |
|---|---|---|---|---|
| 1 | − | 7 | 2 (2)c | 5 (4) |
| + | 12 | 12 (5) | 0 | |
| 2 | − | 10 | 2 (1) | 8 (4) |
| + | 9 | 9 (6) | 0 | |
| 3 | − | 4 | 1 (1) | 3 (2) |
| + | 12 | 12 (5) | 0 | |
In each expeiment, portions of cells from each subclone were untreated (−) or treated (+) for 24 hours with 1.5 nM RTX prior to plating into G418 selection as described in Materials and Methods.
Characterization of events as gene conversions (noncrossovers) versus crossovers was based on Southern blotting analysis of BamHI digests as described in the text.
Numbers in parentheses indicate the minimal number of independent recombination events.
The nucleotide sequence was determined for PCR products amplified from 12 apparent gene conversions from RTX-treated cells and from all five apparent gene conversions from untreated cells. We used the scattered nucleotide mismatches between the donor tk sequence and the fusion gene (Fig 2) to aid in sequence analysis. Sequence analysis confirmed unequivocally that in each clone the 22 bp insertion had been removed from the tk-neo fusion gene and that an accurate exchange of nucleotides had occurred between the tk donor and the tk-neo fusion gene in pLB4. The minimum lengths of gene conversion tracts were estimated based on the number of mismatched nucleotides that were transferred from the donor to the fusion gene. Conversion tracts appeared to be continuous and ranged from a minimum of 21 bp to over 924 bp in length, with the minimum length of most tracts estimated at 21 bp. There were no notable differences in conversion tracts recovered from RTX-treated versus untreated cells.
3.5. DSB-induced HR is unaffected by RTX treatment
The results described above indicated that RTX treatment of human cell line pLB4/11 stimulated HR and produced a profound shift away from crossovers and toward gene conversions. Substrate pLB4 contains a recognition site for endonuclease I-SceI within the tk portion of the tk-neo fusion gene, allowing the study of DSB-induced HR following transient expression of I-SceI. To assess the impact of RTX treatment on DSB-induced HR, cells from the pLB4 /11 cell line were treated with 1.5 nM RTX for 24 hours and then electroporated with pSce. After 48 hours of recovery under no selection, cells were plated into G418 selection to recover recombinants. As reported in Table 3, RTX-treated cells produced a similar number of DSB-induced G418R colonies as did untreated cells. Comparison of DSB-induced colony frequencies in Table 3 with spontaneous colony frequencies shown in Table 1 revealed that the I-SceI DSB increased colony frequency substantially, by as much as 10,000- fold.
Table 3.
Recovery of DSB-induced HR events from cell line pLB4/11
| Experiment No. |
RTXa | Cells platedb (105) |
Colonies recoveredc |
Colony frequencyd (10−3) |
|---|---|---|---|---|
| 1 | − | 2.66 | 566 | 2.13 |
| + | 2.39 | 457 | 1.91 | |
| 2 | − | 1.70 | 589 | 3.46 |
| + | 1.05 | 438 | 4.17 | |
Cells were untreated (−) or treated (+) with 1.5 nM RTX prior to electroporation with pSce, as described in Materials and Methods.
Total number of viable cells plated into G418 selection two days after electroporation with pSce.
Total number of G418R colonies recovered.
Number of G418R colonies divided by number of cells plated into G418.
To ascertain that DSB-induced G418R colonies arose from HR, genomic DNA isolated from representative clones was subject to PCR and AluI digestion as described above. Nine out of ten G418R clones from RTX-treated and ten out of 12 G418R clones from untreated cells were found to have arisen from HR. The balance of clones was presumed to have arisen via NHEJ.
To classify recovered HR events as crossovers or gene conversions, Southern blot analysis of BamHI digests of genomic DNA samples were carried out as above for representative HR clones. The distribution of recovered crossovers and gene conversions was similar for the RTX-treated and untreated cells. Specifically, four out of nine HR clones from RTX-treated cells and four out of ten HR clones from untreated cells were found to be gene conversions, with the balance being classified as crossovers. Sequence analysis of the eight gene conversion events recovered from untreated and RTX-treated cells revealed unequivocally that an accurate exchange of genetic information occurred in all clones. Based on the nucleotide mismatches between the tk donor and the tk-neo fusion gene, conversion tracts appeared to be continuous and the minimal length of conversion tracts ranged from 21 bp to 795 bp (data not shown). To summarize, our experiments provided no evidence that RTX impacted DSB-induced HR.
3.6 NHEJ is unaffected by RTX treatment
Although the above experiments indicated that RTX had no discernible impact on DSB-induced HR, it remained a possibility that DSB repair via NHEJ was affected by RTX. This issue was of interest to investigate because DSBs are induced under conditions of thymidylate starvation and NHEJ is a major DSB repair pathway in mammalian cells. To explore the possibility that NHEJ is compromised by TS inhibition, we made use of cell line 9-2, which was derived by stably transfecting GM00637 cells with pTNeo99-7 (Fig. 1). Substrate TNeo99-7 lacks a donor sequence and allowed us to monitor NHEJ events in the absence of any confounding influence of HR. The 9-2 cells were treated with 1.5 nM RTX for 24 hours, or left untreated, and then electroporated with pSce. After 48 hours of recovery under no selection, as above, cells were plated into G418 selection to recover clones that had undergone NHEJ. As recorded in Table 4, there was no appreciable difference in the recovery of G418R colonies from untreated versus RTX-treated cells.
Table 4.
Recovery of NHEJ events from cell line 9-2
| Experiment | RTXa | Cells platedb (105) |
Colonies recoveredc |
Colony frequencyd (10−3) |
|---|---|---|---|---|
| 1 | − | 17.62 | 3313 | 1.88 |
| + | 12.89 | 3559 | 2.76 | |
| 2 | − | 2.20 | 712 | 3.23 |
| + | 1.60 | 544 | 3.40 | |
Cells were untreated (−) or treated (+) with 1.5 nM RTX prior to electroporation with pSce, as described in Materials and Methods.
Total number of cells plated into G418 selection two days after electroporation with pSce, corrected for viability.
Total number of G418R colonies recovered.
Number of G418R colonies divided by number of viable cells plated into G418. In control experiments in which cell line 9-2 was mocked electroporated in phosphate buffered saline alone, colony frequency was less than 10−7.
To study the recovered NHEJ events further, genomic DNA samples isolated from representative clones were PCR amplified as above, and nucleotide sequences across NHEJ repair junctions were determined. In total, 17 NHEJ events recovered from RTX-treated cells and 20 NHEJ events recovered from untreated cells were analyzed at the nucleotide level and the results of such analyses are summarized in Table 5 and Table 6. We found no consistent difference in the size of deletions, frequencies of insertions, or use of microhomologies among the NHEJ events from RTX-treated versus untreated cells.
Table 5.
NHEJ events recovered from cells treated with RTX
| Clone name | Deletion size (bp) | Microhomologya |
|---|---|---|
| 4ar | 4 | none |
| 2ar | 4b | none |
| 5ar | 4 | none |
| 6ar | 7 | a |
| 8cr | 7 | none |
| 7ar | 10 | a |
| 6cr | 22c | none |
| 10ar | 28 | ggg |
| 8ar | 28 | ggg |
| 2br | 31 | c |
| 5cr | 49 | none |
| 3br | 55 | ctt |
| 10br | 100d | none |
| 1ar | 244 | c |
| 1br | 244 | gc |
| 5br | 403 | c |
| 9ar | 1141 | accc |
For NHEJ junctions displaying microhomology (segments of sequence identity) at the joined DNA termini, the actual sequence of microhomology shared between the joined termini is shown.
Clone 2ar displayed a 4 bp deletion plus a run of 12 consecutive T residues inserted at the DSB site.
Clone 6cr displayed a net deletion of 22 bp, including an insertion of a three bp sequence (tta) at the DSB site.
Clone 10br displayed a net deletion of 100 bp, including an insertion of a 16 bp sequence (taataactgaaaacag) at the DSB site.
Table 6.
NHEJ events recovered from cells without RTX treatment
| Clone name | Deletion size (bp) | Microhomologya |
|---|---|---|
| 5c | 4 | none |
| 8b | 4 | none |
| 4b | 7 | a |
| 7a | 10 | gg |
| 8c | 22 | agct |
| 6b | 22 | agg |
| 5b | 33b | ctcab |
| 3a | 37 | g |
| 2b | 37 | none |
| 10b | 49 | ggc |
| 6a | 115 | gg |
| 9b | 169 | none |
| 3b | 186 | g |
| 1b | 199 | none |
| 3c | 214 | ac |
| 2a | 244 | gg |
| 4a | 244 | gggt |
| 9a | 256 | ggcc |
| 8a | 328 | none |
| 10a | 1087 | none |
For NHEJ junctions displaying microhomology (segments of sequence identity) at the joined DNA termini, the actual sequence of microhomology shared between the joined termini is shown.
Clone 5b displayed a 33 bp deletion plus an insertion of a 149 bp sequence originating from the hygromycin resistance gene (present in pTNeo99-7). The 4 bp microhomology (ctca) was located at the junction between the insert and the right-most DNA terminus from the DSB.
4. Discussion
In this paper, we present what we believe is the first direct assessment of the influence of a TS inhibitor on HR in human chromosomes. Our main finding was that treatment of human fibroblasts with the folate analog RTX increased the rate of intrachromosomal HR about five-fold overall (Table 1). More specifically, RTX appeared to selectively increase the frequency of spontaneous recombination events that resolved as gene conversions (Table 2). Our work substantiates studies that have suggested a link between HR and cellular response to thymidylate deprivation in higher eukaryotes, a link that previously had been established in prokaryotes and lower eukaryotes.
It was reported that E. coli. strains deficient in the recF recombination pathway were resistant to thymineless death while strains carrying mutations in genes in the RecBC pathway retained sensitivity to thymidylate starvation [9,26–28]. Evidence has been presented for the formation of recombinational structures between sister DNA duplexes following replication under conditions of thymidylate starvation in bacteria [29]. From the work in bacteria, it can be inferred that thymidylate starvation induces lesions (possibly single strand gaps or nicks) following stalling of DNA replication, and that these lesions can serve as substrates for the RecBC or RecF recombinational repair pathways. Repair via the RecBC pathway may frequently be successful and allow survival whereas repair via the RecF pathway may lead to the formation of irreparable, lethal structures. Nakayama et. al [29] presented evidence that such structures are branched and contain single-stranded regions and tails. These structures may lead to formation of DSBs which may also contribute to lethality. It is also of interest to note that RecQ, a DNA helicase from E. coli that participates in the RecF pathway, was identified in a search for mutant strains resistant to thymineless death [27]. BLM helicase, which is a human homolog of RecQ, suppresses sister chromatid exchanges (SCEs) in human cells.
In the yeast Saccharomyces cerevisiae, Kunz et al. [30] had shown that starvation for thymidylates, either by genetic lesion or by inhibition of TS activity by FdUMP or antifolates, brought about killing and strongly induced mitotic recombination between homologous chromosomes. Both the recombinogenic and lethal effects of FdUMP and antifolates were reversed by dTMP. Other studies by Kunz et al. [33], involving a DNA substrate conceptually similar to the substrate used in our current work, demonstrated that antifolate treatment of yeast induced intrachromosomal HR between closely linked sequences by about 20-fold overall. TS inhibition in yeast brought about a particularly large increase in crossover events relative to gene conversions. Indeed, the frequency of crossover events specifically was induced 110-fold by antifolate treatment [33]. Such findings were consistent with earlier reports [31] demonstrating induction of unequal crossing-over in the yeast ribosomal DNA gene cluster upon inhibition of dTMP synthesis. The findings for yeast contrast with our finding that RTX treatment selectively induced gene conversions in human cell line pLB4/11 (Table 2). The precise basis for the differential stimulation of crossovers seen in yeast versus the stimulation of gene conversions reported here for a human fibroblast cell line is not presently clear. Several potential explanations exist, and these include the possibility of general differences between nucleic acid metabolism in lower versus higher eukaryotes and/or differences between the specific genetic loci studied. It is possible that the use of additional human cell lines and/or the integration of recombination reporters at other loci might reveal stimulation of crossovers by RTX treatment. Importantly, however, the observations for yeast and human cells lead to the common inference that TS inhibition can stimulate HR among chromosomal sequences and impact the nature of how recombination events are resolved.
In mammalian cells, a connection between thymineless stress and HR had been made previously but the earlier work was more suggestive than definitive. For example, it was shown that SCEs are induced in mammalian cells under conditions of thymidylate starvation [14,19,24,34]. However, the precise relationship between cytologically observed SCEs and bona fide HR events has not been established rigorously. Recent work provided evidence that differential sensitivity of different mouse cell lines to thymidylate deprivation correlated better with SCE induction rather than with the extent of uracil incorporation into DNA [14]. In the latter study, it was suggested that recombination may play a role in the mechanism of thymineless death. However, recombination was measured cytologically as SCEs and so the question of whether chromosomal HR was truly induced was not fully explored.
Some evidence for HR as a response to thymidylate starvation in mammalian cells can be gleaned from published observations that are not based on cytologically monitored SCEs. In particular, thymineless stress induced in mouse cells by methotrexate treatment was reported to result in a high frequency of deletion of an integrated transfected marker gene [36]. The mechanism of deletion involved a specific chromosomal rearrangement that appeared consistent with intrachromosomal HR and it was speculated that recombination may have occurred between two AluI sequences contained within the marker gene sequence [36]. These putative recombination events were not studied at the nucleotide level, however, so evidence for thymineless stress-induced intrachromosomal HR remained only suggestive. Additional evidence for thymineless stress-induced HR may perhaps be inferred from work by Murphy et al. [8]. This latter work reports the curious observation that RTX treatment actually increased tumorigenesis in the ApcMin/+ mouse which the investigators were using as an animal model for colon and intestinal cancer. Significantly, the vast majority of the RTX-induced tumors displayed loss of heterozygosity (LOH) at the Apc locus. HR would seemingly provide a plausible explanation for the observed LOH. Unfortunately, the molecular basis for the LOH was not examined in detail. The outcome reported by Murphy et al. does, nonetheless, raises the possibility that TS inhibition might induce HR and/or other genomic rearrangements with potentially deleterious consequences (e.g., tumor induction or genesis of genetic abnormalities). Our current data lends credence to such a notion.
A question remains regarding the mechanistic connection between cytologically visualized SCEs and the intrachromosomal HR events recovered in our current study. We found that RTX stimulated noncrossover events whereas, on the face of it, induced SCEs [14,19,24,34] seemingly signify crossovers. However, these observations can perhaps be reconciled by the realization that recombination events that resolve as noncrossover events at collapsed replication forks can actually manifest themselves as cytological SCEs since leading and lagging strands are swapped and template DNA becomes fused to newly synthesized DNA [see Fig. 3 in reference 25]. It is also possible that the HR events we recovered are unrelated to cytological SCEs,
Our experiments involving DSB induction by I-SceI revealed that a DSB can bring about as much as a 10,000-fold increase in HR and that the DSB-induced HR events were not affected quantitatively (Table 3) or qualitatively by RTX treatment. Regardless of RTX treatment, about one half of the HR events recovered following DSB induction were crossover events, while all spontaneous HR events recovered following RTX treatment were gene conversions (Table 2 and data not shown). Indeed, a comparison of HR events recovered from cells treated with RTX (without I-SceI treatment) versus HR events recovered from cells treated with I-SceI (irrespective of RTX treatment) revealed a highly significant difference (p < 1.3 × 10−6 by a Fisher exact test) in the relative numbers of crossovers versus gene conversions. This difference may be interpreted as evidence that RTX-induced events are mechanistically distinct from DSB-induced events, suggesting that the RTX-induced events were not provoked directly by DSBs occurring in or near the integrated pLB4 recombination substrate. We suggest that RTX enhanced HR “in trans” due to a global elevation of HR activity. Consistent with the notion of global HR induction was the report of elevated in vitro HR activity measured using extracts prepared from mouse cells under thymidylate stress [35]. We have also observed a modest induction in levels of rad51 protein in human cell lines treated with RTX (unpublished observations). Global HR elevation may constitute part of a cellular response to an interruption in replication caused by thymidylate deprivation. The apparent selective increase in gene conversions that we have seen is consistent with expectations for the type of recombination events most likely to be induced at stalled or collapsed replication forks [25]. We note that collapse of a replication fork is predicted to produce a “one ended” DSB rather than a classical “two-ended DSB” as is produced by action of I-SceI [25]. It therefore is plausible to propose that RTX-stimulated recombination are mediated through the production of one-ended DSBs and that this type of DSB is processed in a manner to specifically produce gene conversions. Along these lines, it is imaginable that recombination is induced specifically in the vicinity of stalled or collapsed replication forks. It is also conceivable that RTX-induced arrest in S phase per se is responsible for elevated HR activity, independent of actual fork blockage or collapse.
Our experiments involving (two-ended) DSB induction in cell line pLB4/11 suggest that the balance of DSB repair accomplished via HR versus NHEJ is not significantly altered by RTX treatment. Additional experiments with cell line 9-2 (Table 4–Table 6) also indicate that DSB repair via NHEJ is not altered quantitatively or qualitatively by RTX treatment. Collectively our data imply that although DNA breakage has commonly been reported as one consequence of TS inhibition [9,12,14,18–24], the manner in which cells metabolize DSBs is not notably impacted by TS inhibition. Our work therefore suggests that “thymineless death” is not mediated by corruption of DSB processing. This conclusion is consistent with the earlier finding that the sensitivity of cells to TS inhibitors does not appear to correlate with the level of detectable DSBs but, instead, correlates with levels of SCEs [14]. Along these lines, it is at least conceivable that some DNA breaks induced by thymidylate starvation are not primary lesions generated by disrupted replication or by futile cycles of base excision repair but, rather, are produced through the processing of recombination intermediates.
While our data directly demonstrate that treatment with the TS inhibitor RTX can lead to an increased rate of intrachromosomal HR, it presently is not clear how such changes in genome dynamics contribute to cell death. It is possible that an increased level of intrachromosomal HR, like an increased level of cytologically detected SCE, is a “marker” of a more general genomic calamity that ends in the demise of the cell. Of relevance to this possibility, Hori et al. [24] reported an induction of chromosomal aberrations and gross rearrangements in mouse cells deprived of thymidylate. Interestingly, the breakpoints for these chromosomal rearrangements seemed to be focused at or near replication forks [24], consistent with the notion that lesions at stalled forks might provoke a plethora of DNA transactions including the types of accurate HR events that we monitored in the current work. Similar to what was observed in bacteria, it is possible that certain pathways for processing recombination intermediates may lead to the formation of lethal DNA structures. Such a scenario suggests that certain mutations in mammalian proteins implicated in recombination may confer resistance to thymineless death while other mutations may confer increased sensitivity. Studies of the impact of TS inhibitors on mammalian cells mutated in recombination functions may thus provide additional insight into the mechanism of thymineless death. By establishing that treatment with a TS inhibitor induces HR in human cells, our current work can thus serve as a springboard to bring us to a better understanding of thymineless death, an understanding that has remained elusive for decades.
Acknowledgements
This work was supported by Public Health Service grants CA100450 and GM47110 from the National Cancer Institute and from the National Institute for General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Longley DB, Harkin DP, Johnston, P.G. PG. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 2.Peters GJ, Kohne, C.H. CH. Fluoropyrimidines as antifolate drugs. In: Jackman AL, editor. Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press; 1999. pp. 101–145. [Google Scholar]
- 3.Assaraf YG. Molecular basis of antifolate resistance. Cancer Metastasis Rev. 2007;26:153–181. doi: 10.1007/s10555-007-9049-z. [DOI] [PubMed] [Google Scholar]
- 4.Jackman AL, Taylor GA, Gibson W, Kimbell R, Brown M, Calvert AH, Judson IR, Hughes LR. ICI D1694, a quinazoline antifolate thymidylate synthase inhibitor that is a potent inhibitor of L1210 tumor cell growth in vitro and in vivo: a new agent for clinical study. Cancer Res. 1991;51:5579–5586. [PubMed] [Google Scholar]
- 5.Hughes LR, Stephens TC, Boyle FT, Jackman AL. Raltitrexed (Tomudex™), a highly polyglutamatable antifolate thymidylate synthase inhibitor: design and preclinical activity. In: Jackman AL, editor. Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press; 1999. pp. 147–165. [Google Scholar]
- 6.Beale P, Clarke S. Tomudex: clinical development. In: Jackman AL, editor. Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press; 1999. pp. 167–181. [Google Scholar]
- 7.McGuire JJ. Anticancer antifolates: current status and future directions. Curr. Pharm. Des. 2003;9:2593–2613. doi: 10.2174/1381612033453712. [DOI] [PubMed] [Google Scholar]
- 8.Murphy JT, Tucker JM, Davis C, Berger FG. Raltitrexed increases tumorigenesis as a single agent yet exhibits anti-tumor synergy with 5-fluorouracil in ApcMin/+ mice. Cancer Biol. Ther. 2004;3:1169–1176. doi: 10.4161/cbt.3.11.1220. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 10.Houghton JA, Tillman DM, Harwood FG. Ratio of 2'-deoxyadenosine-5'-triphosphate/thymidine-5'-triphosphate influences the commitment of human colon carcinoma cells to thymineless death. Clin. Cancer Res. 1995;1:723–730. [PubMed] [Google Scholar]
- 11.Yoshioka A, Tanaka S, Hiraoka O, Koyama Y, Hirota Y, Ayusawa D, Seno T, Garrett C, Wataya Y. Deoxyribonucleoside triphosphate imbalance. 5-Fluorodeoxyuridine-induced DNA double strand breaks in mouse FM3A cells and the mechanism of cell death. J. Biol. Chem. 1987;262:8235–8241. [PubMed] [Google Scholar]
- 12.Aherne GW, Brown S. The role of uracil misincorporation in thymineless death. In: Jackman AL, editor. Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press; 1999. pp. 409–421. [Google Scholar]
- 13.Goulian M, Bleile B, Tseng BY. Methotrexate-induced misincorporation of uracil into DNA. Proc. Natl. Acad. Sci. U S A. 1980;77:1956–1960. doi: 10.1073/pnas.77.4.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Connor EE, Berger SH, Wyatt MD. Determination of apoptosis, uracil incorporation, DNA strand breaks, and sister chromatid exchanges under conditions of thymidylate deprivation in a model of BER deficiency. Biochem. Pharmacol. 2005;70:1458–1468. doi: 10.1016/j.bcp.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Luo Y, Walla M, Wyatt MD. Uracil incorporation into genomic DNA does not predict toxicity caused by chemotherapeutic inhibition of thymidylate synthase. DNA Repair (Amst) 2008;7:162–169. doi: 10.1016/j.dnarep.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welsh SJ, Hobbs S, Aherne GW. Expression of uracil DNA glycosylase (UDG) does not affect cellular sensitivity to thymidylate synthase (TS) inhibition. Eur. J. Cancer. 2003;39:378–387. doi: 10.1016/s0959-8049(02)00610-x. [DOI] [PubMed] [Google Scholar]
- 17.Andersen S, Heine T, Sneve R, König I, Krokan HE, Epe B, Nilsen H. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26:547–555. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- 18.Ayusawa D, Shimizu K, Koyama H, Takeishi K, Seno T. Accumulation of DNA strand breaks during thymineless death in thymidylate synthase-negative mutants of mouse FM3A cells. J. Biol. Chem. 1983;258:12448–12454. [PubMed] [Google Scholar]
- 19.Seno T, Ayusawa D, Shimizu K, Koyama H, Takeishi K, Hori T. Thymineless death and genetic events in mammalian cells. Basic Life Sci. 1985;31:241–263. doi: 10.1007/978-1-4613-2449-2_15. [DOI] [PubMed] [Google Scholar]
- 20.Canman CE, Radany EH, Parsels LA, Davis MA, Lawrence TS, Maybaum J. Induction of resistance to fluorodeoxyuridine cytotoxicity and DNA damage in human tumor cells by expression of Escherichia coli deoxyuridinetriphosphatase. Cancer Res. 1994;54:2296–2298. [PubMed] [Google Scholar]
- 21.Matsui SI, Arredondo MA, Wrzosek C, Rustum YM. DNA damage and p53 induction do not cause ZD1694-induced cell cycle arrest in human colon carcinoma cells. Cancer Res. 1996;56:4715–4723. [PubMed] [Google Scholar]
- 22.Schöber C, Gibbs JF, Yin MB, Slocum HK, Rustum YM. Cellular heterogeneity in DNA damage and growth inhibition induced by ICI D1694, thymidylate synthase inhibitor, using single cell assays. Biochem. Pharmacol. 1994;48:997–1002. doi: 10.1016/0006-2952(94)90370-0. [DOI] [PubMed] [Google Scholar]
- 23.Tonkinson JL, Marder P, Andis SL, Schultz RM, Gossett LS, Shih C, Mendelsohn LG. Cell cycle effects of antifolate antimetabolites: implications for cytotoxicity and cytostasis. Cancer Chemother. Pharmacol. 1997;39:521–531. doi: 10.1007/s002800050608. [DOI] [PubMed] [Google Scholar]
- 24.Hori T, Ayusawa D, Shimizu K, Koyama H, Seno T. Chromosome breakage induced by thymidylate stress in thymidylate synthase-negative mutants of mouse FM3A cells. Cancer Res. 1984;44:703–709. [PubMed] [Google Scholar]
- 25.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama K, Shiota S, Nakayama H. Thymineless death in Escherichia coli mutants deficient in the RecF recombination pathway. Can. J. Microbiol. 1988;34:905–907. doi: 10.1139/m88-157. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt PC. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Gen. Genet. 1984;3:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama H, Nakayama K, Nakayama R, Nakayama Y. Recombination-deficient mutations and thymineless death in Escherichia coli K12: reciprocal effects of recBC and recF and indifference of recA mutations. Can. J. Microbiol. 1982;28:425–430. doi: 10.1139/m82-064. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama K, Kusano K, Irino N, Nakayama H. Thymine starvation-induced structural changes in Escherichia coli DNA. Detection by pulsed field gel electrophoresis and evidence for involvement of homologous recombination. J. Mol. Biol. 1994;243:611–620. doi: 10.1016/0022-2836(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 30.Kunz BA, Barclay BJ, Game JC, Little JG, Haynes RH. Induction of mitotic recombination in yeast by starvation for thymine nucleotides. Proc. Natl. Acad. Sci. USA. 1980;77:6057–6061. doi: 10.1073/pnas.77.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunz BA, Taylor GR, Konforti B, Glickman BW, Haynes RH. Inhibition of thymidylate biosynthesis induces mitotic unequal sister chromatid recombination in Saccharomyces cerevisiae. Curr.Genet. 1984;8:211–217. doi: 10.1007/BF00417818. [DOI] [PubMed] [Google Scholar]
- 32.Kunz BA, Taylor GR, Haynes RH. Mating-type switching in yeast is induced by thymine nucleotide depletion. Mol. Gen. Genet. 1985;199:540–542. doi: 10.1007/BF00330772. [DOI] [PubMed] [Google Scholar]
- 33.Kunz BA, Taylor GR, Haynes RH. Induction of intrachromosomal recombination in yeast by inhibition of thymidylate biosynthesis. Genetics. 1986;114:375–392. doi: 10.1093/genetics/114.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hori T, Ayusawa D, Seno T. Thymidylate stress and sister chromatid exchanges. Basic Life Sci. 1984;29:149–159. doi: 10.1007/978-1-4684-4889-4_12. [DOI] [PubMed] [Google Scholar]
- 35.Mishina Y, Ayusawa D, Seno T, Koyama H. Thymidylate stress induces homologous recombination activity in mammalian cells. Mutat. Res. 1991;246:215–220. doi: 10.1016/0027-5107(91)90124-7. [DOI] [PubMed] [Google Scholar]
- 36.Ayusawa D, Koyama H, Shimizu K, Kaneda S, Takeishi K, Seno T. Induction, by thymidylate stress, of genetic recombination as evidenced by deletion of a transferred genetic marker in mouse FM3A cells. Mol. Cell. Biol. 1986;6:3463–3469. doi: 10.1128/mcb.6.10.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith JA, Bannister LA, Bhattacharjee V, Wang Y, Waldman BC, Waldman AS. Accurate homologous recombination is a prominent double-strand break repair pathway in mammalian chromosomes and is modulated by mismatch repair protein Msh2. Mol. Cell. Biol. 2007;27:7816–7827. doi: 10.1128/MCB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bannister LA, Waldman BC, Waldman AS. Modulation of error-prone double-strand break repair in mammalian chromosomes by DNA mismatch repair protein Mlh1. DNA Repair. 2004;3:465–474. doi: 10.1016/j.dnarep.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Wagner MJ, Sharp JA, Summers WC. Nucleotide sequence of the thymidine kinase of herpes simplex virus type 1. Proc. Natl. Acad. Sci. USA. 1981;78:1441–1445. doi: 10.1073/pnas.78.3.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Berger SH, Wyatt MD. Involvement of base excision repair in response to therapy targeted at thymidylate synthase. Mol. Cancer. Ther. 2004;3:747–753. [PubMed] [Google Scholar]