Abstract
We engineered a mammary-specific knockout model for Brca1 deficiency that also lacks the majority of one chromosome 11 to determine whether tumor susceptibility loci reside on this chromosome that cooperate with the loss of Brca1 during mammary cancer formation. Brca1-deficient females that are haploinsufficient in 60 cM of chromosome 11 exhibited accelerated mammary tumorigenesis in comparison to Brca1 conditional knockout mice. On the histopathologic level, these tumors were either adenocarcinomas or benign, inflammatory lesions. Like human BRCA1-associated breast cancers, mammary carcinomas in this new mouse model were ERα-negative and of basal epithelial origin. Brca1 deficiency and haploinsufficiency in 60 cM of chromosome 11 caused widespread genome instability as determined by spectral karyotyping analysis. In addition to the verification of the long-range deletion event, the spectral karyotyping analysis revealed that the duplication of the genome and higher degree of aneuploidy occur rather late in tumor progression. Despite chromosomal rearrangements near the Trp53 locus as determined by fluorescence in situ hybridization, the Trp53 gene was transcriptionally active. The analysis of the coding sequence and expression pattern of p53 and p21 suggests that loss-of-heterozygosity of Trp53 caused by somatic mutations contributes to accelerated mammary tumorigenesis in this model.
Introduction
Germ line mutations of the breast cancer-associated gene-1 (BRCA1) are responsible for a 55%to 85%cumulative lifetime risk of breast cancer by age 70 [1–3]. BRCA1 is a multidomain protein that is suggested to play a role in a variety of cellular functions including maintenance of genomic stability, DNA double-strand break repair, transcriptional regulation, and cell cycle and spindle checkpoint control [4,5]. Although the precise mechanisms for BRCA1's ability to prevent breast cancer initiation are not clearly defined, targeted gene deletion models of Brca1 provided important insights into the biological functions of this gene for embryonic development, tissue homeostasis, and cancer initiation [6]. Our previous studies on a conditional knockout model of Brca1 demonstrated that the ablation of this tumor-suppressor gene from the mammary epithelium is sufficient to induce neoplastic transformation and mammary cancer after a long latency. Haploinsufficiency in p53 greatly accelerates mammary carcinogenesis in this model for hereditary human breast cancer [7].
Epidemiological evidence in humans as well as genetic studies in Brca1-deficient murine cancer models suggest that subsequent somatic mutations, including p53 and possibly additional modifier loci, are responsible for variations in the latency of Brca1-associated mammary tumorigenesis. Although BRCA1 gene mutations are rare in sporadic breast cancers, the expression of full-length BRCA1 transcripts and the protein is reduced in a subset of sporadic malignancies [8,9], suggesting that genetic or epigenetic alterations in noncoding, regulatory regions near BRCA1 play a role in sporadic breast cancer. On the basis of a meta-analysis for commonly deleted regions in sporadic breast cancers that also identified a modified chromosomal segment on 17q21 near BRCA1 [10], Biggs et al. [11] suggested that there might be tumor-suppressor genes near Brca1 that may play a role in cancer initiation. The latter research team used Cre/loxP-based chromosome engineering as a method to delete large portions (up to 5Mb) of the mouse chromosome 11, which contains the Brca1 and p53 genes, to identify additional putative tumor susceptibility genes that cooperate with these more prominent tumor-suppressor loci. These large deletions, however, tend to cause lethality in homozygous mutants, and segmental haploidy in the germ line can cause abnormalities in other organs that may interfere with the analysis of tumor susceptibility loci in specific adult tissues such as the mammary gland.
To address whether putative tumor susceptibility loci on chromosome 11 are able cooperate with the loss of Brca1 during mammary carcinogenesis, we generated a mammary-specific knockout of Brca1 that is also haploid-deficient in nearly the entire chromosome 11. Females deficient in Brca1 that also lack approximately 60 cM of one chromosome 11 develop mammary cancer after a significantly shorter latency compared to females that carry only a conditional mutation of Brca1. Like familiar BRCA1-associated breast cancers in humans, the adenocarcinomas that appeared in Brca1-deficient mice that are haploinsufficient in chromosome 11 are ERα-negative and of basal epithelial origin. Unlike previous reports, these lesions do not exhibit an up-regulation of ErbB2. Using spectral karyotyping (SKY) analysis, we demonstrate that these mammary cancers have a highly unstable genome that includes chromosomal rearrangements in the central region of the homologous chromosome 11, which did not undergo the long-range deletion event. Using fluorescence in situ hybridization (FISH), we determined that the chromosomal breaks occurred near the Trp53 locus. However, the Trp53 gene itself was not translocated to other chromosomes and was transcriptionally active. The sequence analysis of the coding region of p53 suggests that, in a significant subset of cases, loss-of-heterozygosity of Trp53 caused by somatic mutations is a contributing factor to accelerated tumorigenesis in this breast cancer model.
Materials and Methods
Mice
The generation of WAP-Cre and MMTV-Cre transgenic lines as well as the conventional Wap knockout mice and the Brca1 conditional knockout model was described previously [7,12,13]. All animals used in this study were treated humanely and in accordance with institutional guidelines and federal regulations.
Whole Mount Staining of Mammary Glands and Histologic Analysis of Mammary Tumors
Protocols for the preparation of mammary gland whole mounts and hematoxylin and eosin-stained sections of formalin-fixed tissues were described previously [14]. The entire hematoxylin and eosin-stained sections of representative mammary gland lesions were digitized at high resolution using a whole-slide scanning microscope (Zeiss, Jena, Germany) with image capture software from MicroBrightField, Inc. Composite images were analyzed and annotated by Dr. Bob Cardiff using an Internet-based image database with integrated virtual microscopy software (Zoomify) at the Center for Comparative Medicine, University of California, Davis.
Immunostaining
A basic protocol for immunohistochemistry and immunofluorescent staining of paraffin-embedded mammary gland specimens was described previously [15]. We used the following primary antibodies: α-CK5 (1:500 dilution), α-CK6 (1:500 dilution), and >-CK14 (1:1000 dilution) from Covance (Berkeley, CA); α-CK8 (TROMA-I; 1:250 dilution) from the Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA; α-SMA (NCL-SMA; 1:50 dilution) from Novocastra Laboratories, Ltd (Newcastle upon Tyne, United Kingdom); α-pStat3 (58E12, 1:50 dilution) from Cell Signaling Technology (Danvers, MA); and α-ERα (MC-20, 1:500 dilution) and α-p21 (C-19, 1:200 dilution) from Santa Cruz Biotechnology (Santa Cruz, CA). The cytokeratins were visualized with mouse-specific or rabbit-specific Alexa Fluor 594 and 488-conjugated secondary antibodies (1:1000 dilution) from Invitrogen (Carlsbad, CA). The slides were mounted with Vectashield containing 1.5 µg of 4′,6-diamidino-2-phenylindole (DAPI, Vector Laboratories, Inc., Burlingame, CA). For immunohistochemistry of ERα, pStat3, and p21, we used the corresponding biotinylated secondary antibodies (1:200 to 1:1000 dilution) and Vectastain Elite ABC kit (Vector). 3,3-Diaminobenzidine (DAB) was used as a chromogen, and slides were counterstained with Mayer's hematoxylin. Bright field and fluorescence images of histologic slides were taken on a Zeiss Axio Imager microscope equipped with a SPOT FLEX camera (Diagnostic Instruments, Inc., Sterling Heights, MI).
Spectral Karyotyping and FISH
The SKY and FISH analyses were performed on short-term cultured mammary cancer cells that were derived from primary tumors similar to a published protocol by Medina and Kittrell [16]. The annotation of the karyotypes was performed at the Cytogenetic Core facility of the VanAndel Institute, Grand Rapids, MI. The composite karyotypes for each tumor were assigned based on the individual analysis of more than 20 metaphases. The FISH analyses were carried out at the Genome Imaging Facility at Albert Einstein College of Medicine. The genomic probe for the Trp53 locus was kindly provided by Thomas Ried (National Institutes of Health, Bethesda, MD). The methodology for the chromosomal labeling of Trp53 and Septin 9 (Sept9) were described previously [7,17].
Reverse Transcription-Polymerase Chain Reaction and Sequencing of the Trp53 Coding Region
To determine the transcriptional activation of the Trp53 gene, we isolated total RNA from tumor cell pellets using standard guanidinium thiocyanate-phenol-chloroform extraction. A SuperScript II kit from Invitrogen with oligo dT primers was used to perform the first strand synthesis. The primer sequences and PCR conditions for the amplification of a 1166-bp region of the Trp53 coding sequence were described previously [18]. Polymerase chain reaction products were gel-purified and cloned into the TOPOTA vector (Invitrogen). A minimum of three clones per tumor sample were sequenced from both directions using standard M13 forward and M13 reverse primers. The contigs were assembled and analyzed using the Sequencher 4.7 software (Gene Codes Corporation, Ann Arbor, MI).
Western Blot Analysis
The preparation of whole-cell extracts of clarified cell lysates and the experimental procedures for Western blot analysis were described previously [19]. The following antibodies were used: α-ActB (I-19; 1:2000 dilution) and α-p53 (FL-393; 1:1000 dilution) from Santa Cruz Biotechnology; α-p21 (sx118; 1:1000 dilution) from BD Biosciences (San Jose, CA); α-ErbB2 (1:200 dilution) from Abcam (Cambridge, MA); and α-p19/Arf (Ab-1; 1:1000 dilution) and α-Mdm2 (Ab-2; 1:1000 dilution) from EMD Biosciences (San Diego, CA).
Results
Accelerated Mammary Tumorigenesis in Brca1-Deficient females That Lack 60 cM of Chromosome 11
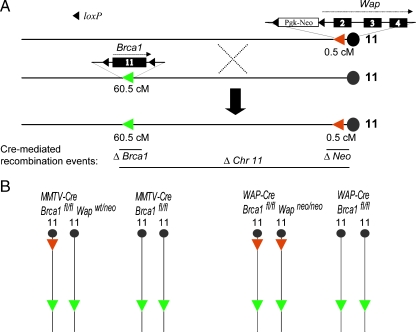
To determine whether tumor susceptibility loci reside on chromosome 11 that cooperate with the loss of Brca1 during mammary cancer formation, we have engineered a mammary-specific knockout model for Brca1 deficiency that also lacks nearly the entire chromosome 11. Figure 1A illustrates the generation of a mouse chromosome 11 that contains loxP sites that are approximately 60 cM apart. The targeted replacement of exon 1 of the whey acidic protein (Wap) gene with a floxed neomycin selection marker was described previously [13]. The Wap gene is located in close proximity (approximately 0.5 cM) to the centromere of chromosome 11. Similar to the Wap locus, the Brca1 gene resides on the antisense strand but at a distance of approximately 60.5 cM away from the centromere. We previously described the generation of a Brca1 conditional knockout allele that contains loxP sites adjacent to exon 11 [7]. Homozygous Brca1 floxed (Brca1fl/fl) male mice were bred with females containing the Wap knockout locus to generate female mice that carry both targeted chromosomes 11. These females were subsequently mated with homozygous Wap knockout (Wapneo/neo) males to generate Wapneo/neo offspring that also carry a Brca1 floxed allele as a result of crossover events between both genetically modified chromosomes 11 during meiosis in the oocytes.The resulting Brca1fl/wt Wapneo/neo mice were bred to obtain the floxed chromosomes 11 in homozygosity (Brca1fl/fl Wapneo/neo). The presence of the floxed loci on both homologous chromosomes prevents the segregation of the targeted loci through subsequent crossover events that occur quite frequently within the 60-cM region between Wap and Brca1. Because pups thrive poorly on Wap-deficient dams [13], the maintenance of this new strain also requires a prolonged lactation period of at least 4 weeks.
Figure 1.
Generation of conditional knockout mice that are Brca1-deficient and that lack 60 cM of chromosome 11. (A) A crossover event between the two homologous chromosomes 11 that carry a targeted Wap locus and a conditional knockout allele of Brca1, respectively, produces a single chromosome with the two targeted loci. Because Wap and Brca1 are directly oriented on the antisense strand of chromosome 11, the expression of Cre recombinase is able to catalyze two short-range excision events (i.e., exon 11 of Brca1 and the PGK-neo selection marker of Wap) and a long-range deletion of approximately 60 cM between the two targeted loci. (B) A mammaryspecific deletion of Brca1 and a very large portion of chromosome 11 is achieved in experimental animals that carry the MMTV-Cre or WAP-Cre transgenes in a homozygous Brca1 floxed background where one or both homologous chromosomes 11 contain a targeted Wap locus (Wapwt/neo and Wapneo/neo, respectively). MMTV-Cre and WAP-Cre-based Brca1 conditional knockout females that carry two wild type Wap alleles served as controls.
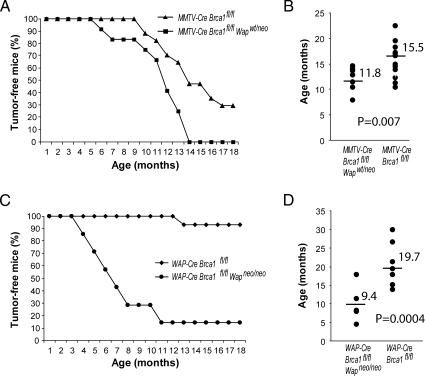
To generate a mammary-specific knockout of Brca1 and a long-range deletion on chromosome 11, Brca1fl/fl Wapneo/neo females were crossed with MMTV-Cre (line A) or WAP-Cre transgenic males that also possess two Brca1 floxed alleles. This breeding strategy avoids the segregation of the mutant Brca1 and Wap loci during the introduction of the Cre transgenes. MMTV-Cre and WAP-Cre Brca1fl/fl Wapwt/neo males were subsequently mated with Brca1fl/fl Wapneo/neo females in an attempt to create mice that carry the respective Cre transgenes in a homozygous chromosome 11 floxed background. This strategy worked for the WAP-Cre transgene in the expected mendelian fashion, but we only obtained seven viable MMTV-Cre Brca1fl/fl Wapneo/neo mice after genotyping more than 40 litters. Three of the seven MMTV-Cre Brca1fl/fl Wapneo/neo mice succumbed to T-cell proliferative disorders and exhibited a wasting syndrome between the ages of 4 and 7 months. One mouse developed a mammary cancer, one mouse died during a surgical procedure, and two mice are still alive to date. Because the MMTV-Cre transgene is not located on chromosome 11 and the MMTV-Cre-mediated deletion of Brca1 does not cause embryonic lethality, this observation might suggest that the long-range deletion event might occur quite efficiently during embryogenesis in various cell types that efficiently express Cre under the MMTV-LTR [20]. Because of embryonic lethality, we maintained MMTV-Cre Brca1fl/fl Wapwt/neo along with WAP-Cre Brca1fl/fl Wapneo/neo females as the two experimental groups (Figure 1B) and compared their tumor latency to the two control cohorts of regular Brca1 conditional knockouts (i.e., MMTV-Cre and WAP-Cre Brca1fl/fl mice). All females were multiparous and had delivered two to three litters. As illustrated in Figure 2, the MMTV-Cre-mediated long-range deletion event on just one of the two homologous chromosomes 11 was sufficient to accelerate Brca1-associated mammary tumorigenesis. All experimental animals had succumbed to mammary cancer by the age of 14 months (Figure 2A). The mean tumor latency was shortened by approximately 4 months (Figure 2B). The most noticeable difference was observed in the WAP-Cre-based experimental group. We previously reported that, in comparison to MMTV-Cre Brca1fl/fl females, WAP-Cre-based conditional Brca1 knockout mice have a reduced incidence in mammary tumor formation and a prolonged latency [7]. This fact was confirmed in our current control cohort where only very few WAP-Cre Brca1fl/f\l females developed mammary cancer by the age of 18 months (Figure 2C). The likelihood that a long-range deletion event occurs on one of the two homologous chromosomes 11 had a significant impact on the tumor-free survival of the experimental females, and their average life expectancy was shortened by approximately 10 months (Figure 2, C and D).
Figure 2.
Tumor-free survival of Brca1-deficient females that lack 60 cM of chromosome 11 (MMTV-Cre Brca1fl/fl Wapwt/neo, n = 12, and WAP-Cre Brca1fl/fl Wapneo/neo, n = 7) in comparison to Brca1 conditional knockout mice (MMTV-Cre Brca1fl/fl, n = 17, and WAP-Cre Brca1fl/fl, n = 15). Mice were monitored twice weekly for a period of 18 months and killed when a tumor became palpable. (A, C) Kaplan-Meier curves. (B, D) Mean age of onset of palpable mammary tumors (horizontal bars). Each marker represents the age of onset of the first palpable tumor per mouse.
Mammary Tumors That Arise in MMTV-Cre and WAP-Cre-Mediated Chromosome 11 Deletion Models Exhibit Differences in Their Histopathologic Features
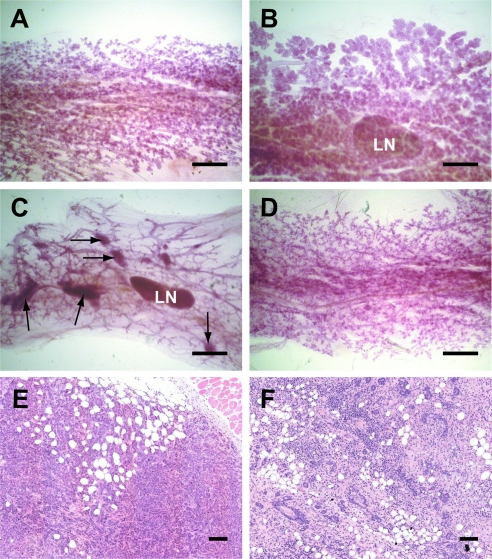
The mammary-specific deletion of Brca1 causes a selective growth inhibition of Brca1-deficient epithelial cells and impaired mammogenesis [7]. The severity of the phenotypic abnormalities during ductal elongation and alveologenesis can vary between animals and depends greatly on the expression pattern of the Cre recombinase driven by the MMTV-LTR or the Wap gene promoter. None of the MMTV-Cre Brca1fl/fl Wapwt/neo females was able to lactate during the first and second gestation periods owing to impaired alveologenesis (Figure 3A), and some animals also exhibited a reduction in ductal branching (not shown). The developmental abnormalities in these experimental animals were probably a direct consequence of Brca1 deficiency in basal and luminal epithelial cells throughout the developing ductal system [20]. In contrast, the expression of the WAP-Cre transgene occurs mainly during alveolar development. This transgene is also known to exhibit a mosaic expression pattern, which, depending on the deleted gene, can result in a negative selection of knockout cells. These knockout cells can be replaced in the developing alveolus by proliferating alveolar progenitors that do not express Cre [15]. Therefore, most WAP-Cre Brca1fl/fl Wapneo/neo females were able to lactate despite a reduction in the overall alveolar density (Figure 3B). The subsequent analysis of mammary glands from parous, tumor-bearing animals showed that many MMTV-Cre Brca1fl/fl Wapwt/neo females exhibited multifocal lesions throughout the ductal tree (Figure 3C), and a subset of these animals had several palpable mammary cancers. Because multifocal tumorigenesis was a rare event in the control cohort, this observation suggests that the MMTV-Cre-mediated long-range deletion event occurs in a significant subset of epithelial cells. Similar to the controls, WAP-Cre Brca1fl/fl Wapneo/neo females generally did not exhibit multiple macroscopic and microscopic lesions (Figure 3D). This was likely the result of a more restricted expression of the WAP-Cre transgene in luminal epithelial cells of developing alveoli during pregnancy and lactation. Also, the histopathologic analysis of large mammary cancers revealed important differences in both models. The mammary cancers that arose in the MMTV-Cre Brca1fl/fl Wapwt/neo females were predominantly adenocarcinomas that invaded into the surrounding stroma (Figure 3E). In contrast, a significant number of WAP-Cre-mediated mammary tumors were classified as benign lesions (Figure 3F). These palpable tumors were soft and contained large inflammatory regions and reactive hyperplasia. On the histopathologic level, these lesions exhibited features of severe acute and chronic mastitis with fibrosis. These large mammary tumors, which tend to appear in WAP-Cre Brca1fl/fl Wapneo/neo females within a month after weaning of their last litter, had a significant negative impact on the general health of the animals that consequently needed to be euthanized. None of the WAP-Cre Brca1fl/fl control mice exhibited such distinct mammary lesions. We are currently unable to fully explain why only WAP-Cre Brca1fl/fl Wapneo/neo females with accelerated mammary tumorigenesis predominantly developed benign, inflammatory lesions. The Stat3 gene, which is located in very close proximity to Brca1, is required for the initiation of apoptosis of differentiated alveolar cells after weaning of the young. Females that lack Stat3 in secretory epithelial cells exhibit a delay in mammary gland remodeling and have a higher incidence in mastitis [21]. We therefore considered that a long-range deletion event on both homologous chromosomes 11 might result in Stat3-deficiency in differentiated epithelial cells. In support of this notion, active Stat3 was not present in most epithelial cells within hyperplastic lesions from WAP-Cre Brca1fl/fl Wapneo/neo females. In contrast, nuclear Stat3 was expressed in nearly all fibrous stromal cells adjacent to these benign lesions (Figure W1, A and B). However, we never observed impaired remodeling and prolonged survival of a significant subset of secretory epithelial cells throughout the mammary glands of WAP-Cre Brca1fl/fl Wapneo/neo females immediately after weaning of the offspring. This suggests that such long-range recombination events occur rarely and do not lead to widespread milk stasis and mastitis.
Figure 3.
Differences in mammogenesis and the histopathology of mammary tumors in MMTV-Cre-based (A, C, E) and WAP-Cre-based (B, D, F) Brca1-deficient females that also carry a 60 cM floxed region of chromosome 11. (A–D) Carmine red-stained mammary gland whole mounts in postpartum dams several hours after delivering the offspring (A, B) and in 1-year-old multiparous females (C, D). LN indicates lymph node. Bar, 1 mm. Multifocal lesions were only present in the MMTV-Cre-based deletion models (C, arrows). (E, F) Hematoxylin and eosin-stained sections of mammary tumors. Bar, 100 µm.
Mammary Cancers That Are Deficient in Brca1 and Haploinsufficient in Chromosome 11 Express Basal Epithelial Cell Markers
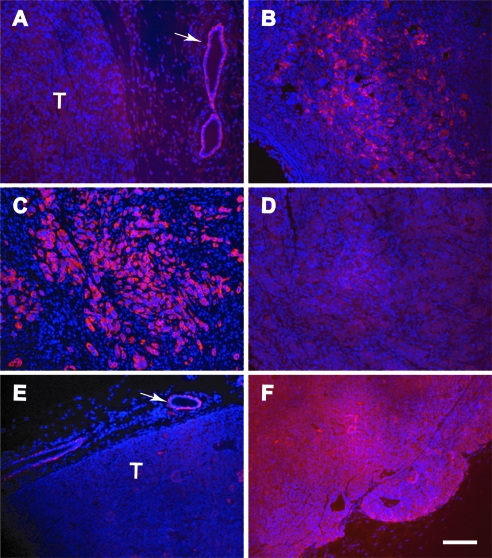
The histopathologic examination of large mammary tumors that arose in the WAP-Cre Brca1fl/fl Wapneo/neo females revealed that these tumors were mostly benign, inflammatory lesions. A subsequent analysis of expression patterns of cytokeratins (CKs) 5, 6, 8, and 14 as well as smooth muscle actin (SMA) showed that all different epithelial subtypes were present in these hyperplastic and fibrocystic lesions (not shown). We therefore focused our attention on the expression of these markers in the invasive mammary cancers that predominantly arose in the MMTV-Cre Brca1fl/fl Wapwt/neo mice. These cancer cells expressed low levels of CK5 compared to adjacent normal epithelial cells (Figure 4A), and they expressed moderate levels of CK6 (Figure 4B). The bulk of these tumors contained many cancer cells that expressed high levels of CK14 but not CK8 (Figure 4, C and D). In addition, we used mammary cancers from MMTV-neu mice as positive and negative controls for the examination of these cellular markers. In contrast, MMTV-neu tumors arise in the luminal epithelial compartment, and consequently, they express higher levels of CK8 and little CK14 compared to adjacent normal mammary ductal cells (Figure 4, E and F). Analogous to CK14, MMTV-neu-overexpressing mammary cancer cells exhibited very little expression of SMA (Figure W2B). The expression of this myoepithelial marker was significantly higher in Brca1-deficient mammary cancers, in particular, in cells that invaded into the stroma. Within the bulk of the tumor, the SMA staining was somewhat reduced and seemed to be weaker than the immunoreactivity in well-differentiated ductal cells that were engulfed by the invading cancer cells (Figure W2A). Collectively, the analysis of the cytokeratin markers and SMA expression pattern shows that mammary cancers that are deficient in Brca1 and haploinsufficient in chromosome 11 are of basal epithelial origin, and they are distinctly different from Her2/neu-overexpressing mammary cancers and benign lesions that arose in the WAP-Cre Brca1fl/fl Wapneo/neo females. Like Her2/neu-overexpressing tumors,mammary cancers from MMTV-Cre Brca1fl/fl Wapwt/neo females lack the expression of the estrogen receptor (Figure W2, C and D). However, ERα-positive mammary epithelial cells were present in hyperplastic lesions in WAP-Cre Brca1fl/fl Wapneo/neo mice (Figure W2E).
Figure 4.
Mammary cancers that are deficient in Brca1 and haploinsufficient in chromosome 11 are of basal epithelial origin. Immunostaining of cytokeratins 5 (A), 6 (B), 14 (C, E), and 8 (D, F) using Alexa Fluor 594-conjugated secondary antibodies (red) in mammary cancers from MMTV-Cre Brca1fl/fl Wapwt/neo females (A–D) and Her2/neu-overexpressing controls (E, F). The slides were counterstained with DAPI (blue nuclei). Arrows in panels A and E indicate the positive staining of cytokeratins 5 and 14 in normal mammary epithelial cells adjacent to the primary neoplasm. Bar, 100 µm.
Brca1 Deficiency and Haploinsufficiency in 60 cM of Chromosome 11 Cause Widespread Genome Instability
Previous studies demonstrated that Brca1-deficient mammary cancer cells were aneuploid, and chromosomal translocations as well as loss and gain of chromosomal regions were observed [22,23]. To assess the extent of genome instability in mammary cancers that arose in MMTV-Cre Brca1fl/fl Wapwt/neo females, we performed a SKY analysis on short-term cultured mammary cancer cells that were derived from two tumor-bearing animals (Figure 5). The SKY analysis confirmed that these cancers originated from mammary epithelial cells that were haploinsufficient in a very large portion of one chromosome 11, suggesting that such large segmental deletion events of more than 60 cM can be achieved through Cre-mediated recombination. According to information obtained from the Mouse Genome Informatics database, more than 1500 annotated genes and expressed sequences reside between Wap and Brca1 on chromosome 11. The fact that neoplastic mammary epithelial cells are able to survive with a single copy of such a large number of genes is remarkable, and this finding clearly underlines the extraordinary plasticity that cancer cells possess. It might also suggest that mammary epithelial cells are capable of developing mechanisms that will allow them to compensate for the lack of important loci that are expressed in these cells. Normal cells in other tissues and organs might not have that ability, and they express different genes located at chromosome 11 that are essential for their survival. For example, segmental deletions and the resulting haploidy of just 3 to 4 cM within the same region of chromosome 11 cause early embryonic lethality [24]. In addition, mutagenesis screens have identified 59 lethal mutations between the Trp53 gene and the Wnt3 locus, which resides approximately 2.5 cM distal of Brca1 [25].
Figure 5.
The SKY analysis of adenocarcinomas from two MMTV-Cre Brca1fl/fl Wapwt/neo females. Arrows indicate chromosomes 11 in both karyotypes that underwent Cre-mediated long-range deletion events. In addition, the SKY analysis revealed that both homologous chromosomes 11 carried translocations with chromosomes 8 and 14, respectively.
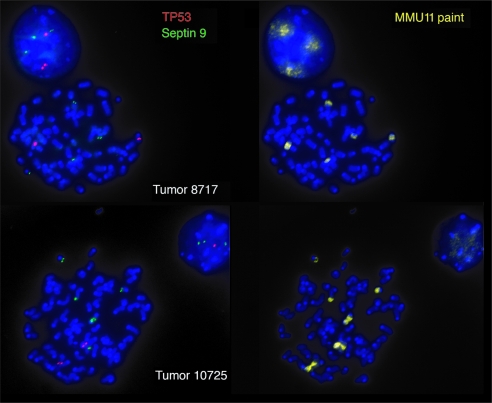
Besides confirming the long-range deletion event, the SKY analysis and examination of more than 20 metaphase spreads per tumor also demonstrates that Brca1 deficiency and chromosome 11 haploinsufficiency cause widespread genome instability with numerous chromosomal aberrations that are also summarized in Table 1. A comparison of the specific aberrations between 2n and 4n cells [for example, the translocation event T(8C1;11C)] shows that the duplication of the genome and a higher degree of aneuploidy occur rather late in tumor progression. In addition to the long-range deletion event, both mammary cancers exhibited chromosomal breaks and translocations within the central region of the homologous chromosome 11, which is known to contain the Trp53 tumor-suppressor gene.We used FISH analysis to determine whether the Trp53 locus was affected or lost as a consequence of the chromosomal rearrangements (Figure 6). In addition to the Trp53 probe and the chromosome 11 paint, we used a genomic probe encompassing the Septin 9 (Spet9) locus to label the distal region of chromosome 11. This particular chromosomal segment, which is orthologous to human 17q25.3, has been reported to be amplified and overexpressed in a subset of murine adenocarcinomas and human breast cancer cell lines [17]. The results of this study show that the chromosomal breaks and translocations seemed to have occurred in both tumors in close proximity of the Trp53 locus. In addition, the dissociation of Trp53 from the Sept9 locus indicates that, unlike the distal region of chromosome 11, the p53 gene itself was not translocated. Because Sept9 is located outside the long-range deletion event, it was still present on the two short chromosomes 11 in 4n metaphase spreads.
Table 1.
Composite Karyotypes of Two Mammary Cancers After Examination of More Than 20 Metaphases Per Tumor Using SKY Analysis.
| Composite karyotype of tumor 8717 |
| 2n cells: 40,XX[cp8]/38,X,Der(4)T(1E;4D3)[6],Del(4D)[2],+Del(6B)[8],-7[8], Del(8C3)[8],T (8C1;11C)[8],Dic(9A.13A)[8],-10[3],Del(10A)[3],Del(10C)[2], Del(11A2-D)[8],+Del(12C) [3],Der(12;12)T(12A2;12A1)Del(12A2)[8],Del(13C)[8], Der(14)T(4D3;14B)[8],-16[8][cp8] |
| 4n cells: 67∼77<4n>,X or XX,-1[3],-2[4],-3[4],Der(4)T(1E;4D3)X1[1], Der(4)T(1E;4D3)X2 [5],+Del(6B)X2[7],-7X1[1],-7X2[5],Del(8C3)X2[7], T(8C1;11C)X1[1],T(8C1;11C)X2[6], Dic(9A.13A)X1[1],Dic(9A.13A)X2[6],-10[2],Del(10A)X1[1], Del(10A)X2[2],Del(10C)[4], Del(11A-D)X2[7],Der(12;12) T(12A2;12A1)Del(12A2)X1[2], Der(12;12)T(12A2;12A1)Del (12A2)X2[5],-13[3],Del(13C)X1[4],Del(13C)X2[3], Der(14)T(4D3;14B)X1[2],Der(14) T (4D3;14B)X2[4],-15[5],-16X1[2],-16X2[5],-17[3][cp7] |
| Composite karyotype of tumor 10725 |
| 2n Cells: 40,XX[cp4] |
| 4n Cells: 67∼83<4n>,XX,Del(XB)X1∼2[17],-1[5],+1[2],Del(2F)X1∼2[17],-3[5], Der(3)T (3D;9E)X1∼2[16],Der(3)T(3D;9EF;8C)[6],-4X2[16],Der(4)T(4D;15E)[4], T(4D;15E)X1∼2 [16],Der(5)T(5E;3;11)X2[17],Del(6B)[3],+Der(6)T(5E;6B)X2[17],-7X2[17], Del(8B)[3],Der (8)T(4;8C)X2∼3[17],-9X2[17],10X2[17],T(10C;12C)X2[2], Del(11A2-D)X2[17], Der(11)T (11C;14B)[2],T(11C;14B)X1∼2[17],-12X2[17],-13X1∼2[10],Der(14)T(11C;14B)[2],-15[4],-16 [7],-18[4],+19[2][cp17] |
The long-range deletion event in chromosome 11 is highlighted in bold font. Please note that in contrast to tumor 8717, tumorigenic cells with a 2n karyotype were not present in tumor 10725. It is likely that the normal karyotype of 2n cells in this tumor represents that of dividing fibroblasts that were present in the short-term culture.
Figure 6.
Fluorescence in situ hybridization to determine the localization of the Trp53 gene (red) and the Sept9 locus (green) in adenocarcinomas from two MMTV-Cre Brca1fl/fl Wapwt/neo females. A specific paint (MMU11) was used to counterstain individual fragments of chromosome 11.
Somatic Mutations in p53 Contributed to Brca1-Associated Mammary Tumorigenesis
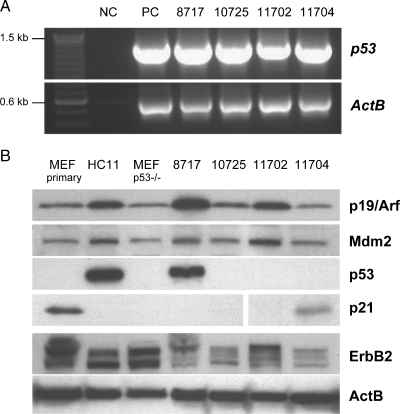
The previous work on the mammary-specific Brca1 deletion model shows that rearrangements in chromosome 11 and aberrant p53 expression contribute to neoplastic transformation [7]. In accordance with this finding, Weaver et al. [23] reported that Brca1-deficient mammary cancers showed a loss of the proximal portion of chromosome 11, which may have included the Trp53 gene. Because both tumors that were examined by SKY and FISH exhibited chromosomal breaks and translocations within the central region of chromosome 11, we assumed that these translocations might have affected the expression of p53. Surprisingly, the results of a reverse transcription-polymerase chain reaction assay show that this gene is transcriptionally active in these two and two additional cancers (Figure 7A). One of four tumors examined expresses a p53 protein, which seems to be transcriptionally inactive based on the lack of p21 expression (Figure 7B). The notion that this tumor contains a somatic mutation in the coding region of Trp53 is supported by the fact that the p53 protein is highly upregulated. HC11 cells that possess a transcriptionally inactive mutant of p53 and that lack p21 expression were used as appropriate controls. After examining the expression of p53, we sequenced the entire coding region of the p53 transcripts in the four different cancer cases (Table 2). Somatic mutations were detected in three mammary cancers, and two of these mutations lead to a premature termination of the coding region. Tumor 8717, which expresses the nonfunctional p53 protein, exhibited a deletion of three nucleotides that caused the absence of a single amino acid (R246) from the DNA binding domain of p53. We were unable to identify a somatic mutation of p53 in tumor 10725, and the underlying alteration of the function of the encoded p53 protein (as assessed by the lack of p21 expression) remains elusive. Neither the down-regulated expression of p19/Arf (Cdkn2a) nor the overexpression of Mdm2 contributed to the functional inhibition of p53 in all four cancers (Figure 7B). It should also be noted that the examination of p53 and p21 expression can be skewed by the presence of stromal cells within the primary cancer cell cultures. In one instance, this was evident in explanted tumor cells of from animal 11704, which exhibited a weak expression of p21 due to the presence of fibroblasts. P21, however, could not be detected by immunostaining in the nuclei of neoplastic epithelial cells of the corresponding primary lesion from this animal (Figure W3). In summary, our study shows that, despite chromosomal rearrangements in the central portion of the chromosome 11, which did not undergo Cre-mediated excision, the Trp53 locus was still transcriptionally active. Somatic mutations in p53 were responsible for the absence or functional inhibition of this tumor suppressor in a subset of Brca1-associated mammary cancers in our animal model.
Figure 7.
Somatic mutations caused p53 deficiency in mammary cancers that are deficient in Brca1 and haploinsufficient in chromosome 11. (A) Reverse transcription-polymerase chain reaction analysis of the transcriptional activation of the Trp53 gene in four mammary cancers from MMTV-Cre Brca1fl/fl Wapneo/neo mice; NC indicates negative control [total RNA from p53-deficient mouse embryonic fibroblasts (MEFs)]; PC, positive control (total RNA from primary wild type MEFs). (B) Western blot analysis to determine the expression of the p53 protein and its downstream target p21Cip1 as well as the expression of upstream regulators of p53 (p19/Arf and Mdm2) and the ErbB2 receptor tyrosine kinase. Beta-actin (ActB) served as a loading control. Cell lysates from primary wild type and p53-deficient MEFs were used as positive and negative controls for p21 and p53 expression. Because wild type p53 is barely detectable in primary MEFs, we used HC11 cell extracts as a positive control for the expression of mutant p53, which is known to be expressed at much higher levels. HC11 cells also served as a control for the baseline expression of ErbB2 in untransformed mammary epithelial cells.
Table 2.
Mutations in the Coding Region of the Trp53 Gene in Four Independent Tumor Samples of Brca1-Deficient Mammary Cancers That Are Haploinsufficient in Chromosome 11.
| Tumor No. | Mutation | Location | |
| 8717 | Del(735–737) | Del(R246) | DNA binding domain |
| 10725 | wild type | — | — |
| 11702 | T927G transversion | Y324Stop | Homo-oligomerization domain |
| 11704 | C421T transition | Q141Stop | DNA binding domain |
Human Brca1-associated breast cancers more frequently exhibit a basal phenotype, and they lack expression of ERα, PR, and ErbB2 [26–28]. In contrast, Brodie et al. [22] reported that most Brca1-deficient mammary tumors in mice overexpressed ErbB2. In addition to this apparent inconsistency between humans and mice, we also asked whether the expression of ErbB2 could have been altered by the long-range Cre-mediated deletion event or the observed translocations that involve the homologous chromosome 11. According to the current Mouse Genome Informatics sequence information, the ErbB2 gene resides proximal of Brca1 on chromosome 11 (i.e., 57 cM from the centromere) and therefore within the long-range deletion segment. The Western blot analysis, however, shows that the ErbB2 protein expression was not significantly altered in Brca1-deficient cells that were haploinsufficient in a large portion of chromosome 11 (Figure 7B). The levels of ErbB2 expression in the mammary cancer cells were equal to or lower than the expression level observed in nontumorigenic, immortalized mammary epithelial cells (Figure 7B, HC11 cells in lane 2). Therefore, the previously reported up-regulation of ErbB2 may not be a common feature or necessity in Brca1-deficient murine mammary cancers.
Discussion
In the Brca1 conditional knockout model generated by Xu et al. [7], the Cre-mediated deletion of exon 11 of Brca1 results in a shift from a predominantly full-length transcript to a shorter mRNA splice variant that encodes a Brca1 protein of approximately 90 kDa [29]. This short Brca1 isoform occurs, albeit at much lower levels, in the normal mammary gland. The sole expression of the Brca1 Δexon 11 isoform in mammary epithelial cells is sufficient to initiate neoplastic transformation after a long latency [7]. It has been recently suggested that the estrogen receptor (ERα) plays a role in premalignant transformation in this model [30]. Elevated levels of estrogen resulted in accelerated mammary cancer formation in Brca1-deficient mice that lacked one functional Trp53 allele. Similar to this report, we also observed that ERα was predominantly expressed in the benign, inflammatory lesions that originated in WAP-Cre Brca1fl/fl Wapneo/neo females. Although these mammary tumors were palpable and relatively large, they were distinctly different from adenocarcinomas of similar size that originated in the MMTV-Cre-mediated Brca1-deficient mammary cancer model that was haploinsufficient in chromosome 11. The latter model seems to more closely resemble key features of human Brca1-associated breast cancers. They exhibit a basal phenotype as assessed by the expression profile of keratins, they are ERα-negative, and they do not exhibit an up-regulation of ErbB2. The differences between these two models, therefore, do not support the notion that Brca1-deficient mammary cancers originate from ERα-positive precursor lesions. Recently, published findings by Jones et al. [31] demonstrate that, although the overexpression of ERα accelerates Brca1-associated mammary tumorigenesis, exogenous levels of this steroid hormone receptor increases the incidence in preneoplastic lesions and mammary cancers that are ERα-negative. Because it has been shown that ERα can stimulate the proliferation of human and murine mammary epithelial cells in a juxtracrine or paracrine manner [32–34], it is possible that ERα promotes the neoplastic growth of adjacent Brca1-deficient mammary epithelial cells that lack ERα.
ERα-negative adenocarcinomas that are Brca1-deficient and haploinsufficient in chromosome 11 exhibited normal or reduced levels of ErbB2, and we therefore concluded that the previously reported upregulation of ErbB2 is not a requirement for Brca1-associated mammary cancer in mice. Similarly, the conditional deletion of the 3-prime region of Brca1 (i.e., exons 22–24) resulted in ErbB2-negative lesions as determined by immunostaining [35]. Whereas the long-range, Cre-mediated deletion event in our model will result in a complete ablation of exons 11 through 24 of Brca1 (Figure 1A), the homologous chromosome 11, however, will retain the expression of the Δexon 11 isoform of Brca1. Hence, the sole expression of this splice form may not be responsible for any differences in ErbB2 expression in Brca1-deficient mammary cancers as suggested by McCarthy and colleagues [35].
Females deficient in Brca1 that also lack approximately 60 cM of chromosome 11 develop mammary cancer after a significantly shorter latency compared to females that carry only a conditional mutation of Brca1. Nonetheless, the increase in tumorigenicity and penetrance of the phenotype is quite similar to Brca1 conditional knockout mice that lack one copy of the Trp53 gene [7]. This observation suggests that the loss of heterozygosity of Trp53 is the predominant genetic event responsible for an accelerated onset of the disease. In agreement with this conclusion, somatic mutations in Trp53 in the remaining homologous chromosome 11 were tightly linked to the occurrence of mammary cancers. Conversely, this finding might also suggest that this large portion of chromosome 11 does not contain additional tumor-suppressor genes that play a role in the initiation of Brca1-deficient mammary cancer as previously hypothesized. Consequently, the search for tumor-suppressor loci that modify the disease onset has to include other regions of the genome.
Supplementary Material
Acknowledgments
The authors thank Gordon Todd (University of Nebraska Medical Center) for digitizing whole slides of selected tumor specimens. The authors also thank Robert Cardiff (UC Davis) for providing his expertise in pathology and for the histopathologic annotation of high-resolution images. The authors would also like to thank the Genome Imaging Facility at Albert Einstein College of Medicine for performing the fluorescence in situ hybridization analyses.
Footnotes
Financial support was provided to K.U.W. by the Nebraska Research Initiative.
This article refers to supplementary materials, which are designated by Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Easton DF, Narod SA, Ford D, Steel M. The genetic epidemiology of BRCA1. Breast Cancer Linkage Consortium. Lancet. 1994;344:761. doi: 10.1016/s0140-6736(94)92256-x. [DOI] [PubMed] [Google Scholar]
- 2.Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, Timmerman MM, Brody LC, Tucker MA. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 3.Blackwood MA, Weber BL. BRCA1 and BRCA2: from molecular genetics to clinical medicine. J Clin Oncol. 1998;16:1969–1977. doi: 10.1200/JCO.1998.16.5.1969. [DOI] [PubMed] [Google Scholar]
- 4.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 5.Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 7.Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 8.Magdinier F, Ribieras S, Lenoir GM, Frappart L, Dante R. Down-regulation of BRCA1 in human sporadic breast cancer; analysis of DNA methylation patterns of the putative promoter region. Oncogene. 1998;17:3169–3176. doi: 10.1038/sj.onc.1202248. [DOI] [PubMed] [Google Scholar]
- 9.Staff S, Isola J, Tanner M. Haplo-insufficiency of BRCA1 in sporadic breast cancer. Cancer Res. 2003;63:4978–4983. [PubMed] [Google Scholar]
- 10.Osborne RJ, Hamshere MG. A genome-wide map showing common regions of loss of heterozygosity/allelic imbalance in breast cancer. Cancer Res. 2000;60:3706–3712. [PubMed] [Google Scholar]
- 11.Biggs PJ, Vogel H, Sage M, Martin LA, Donehower LA, Bradley A. Allelic phasing of a mouse chromosome 11 deficiency influences p53 tumorigenicity. Oncogene. 2003;22:3288–3296. doi: 10.1038/sj.onc.1206384. [DOI] [PubMed] [Google Scholar]
- 12.Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Triplett AA, Sakamoto K, Matulka LA, Shen L, Smith GH, Wagner KU. Expression of the whey acidic protein (Wap) is necessary for adequate nourishment of the offspring but not functional differentiation of mammary epithelial cells. Genesis. 2005;43:1–11. doi: 10.1002/gene.20149. [DOI] [PubMed] [Google Scholar]
- 14.Wagner KU, Young WS, Liu X, Ginns EI, Li M, Furth PA, Hennighausen L. Oxytocin and milk removal are required for post-partum mammary-gland development. Genes Funct. 1997;1:233–244. doi: 10.1046/j.1365-4624.1997.00024.x. [DOI] [PubMed] [Google Scholar]
- 15.Wagner KU, Krempler A, Triplett AA, Qi Y, George NM, Zhu J, Rui H. Impaired alveologenesis and maintenance of secretory mammary epithelial cells in Jak2 conditional knockout mice. Mol Cell Biol. 2004;24:5510–5520. doi: 10.1128/MCB.24.12.5510-5520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medina D, Kittrell FS. Establishment of mouse mammary cell lines. In: Ip MM, Ash BB, editors. Methods in Mammary Gland Biology and Breast Cancer. Chap 13. New York, NY: Kluwer Academic/Plenum; 2000. pp. 137–145. [Google Scholar]
- 17.Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T, Hissong B, Weaver Z, Ried T. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63:179–2187. [PubMed] [Google Scholar]
- 18.Krempler A, Henry MD, Triplett AA, Wagner KU. Targeted deletion of the Tsg101 gene results in cell cycle arrest at G1/S and p53-independent cell death. J Biol Chem. 2002;277:43216–43223. doi: 10.1074/jbc.M207662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto K, Creamer BA, Triplett AA, Wagner KU. The Janus kinase 2 is required for expression and nuclear accumulation of cyclin D1 in proliferating mammary epithelial cells. Mol Endocrinol. 2007;21:1877–1892. doi: 10.1210/me.2006-0316. [DOI] [PubMed] [Google Scholar]
- 20.Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- 21.Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, Akira S, Clarke AR, Watson CJ. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604–2616. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brodie SG, Xu X, Qiao W, Li WM, Cao L, Deng CX. Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene. 2001;20:7514–7523. doi: 10.1038/sj.onc.1204929. [DOI] [PubMed] [Google Scholar]
- 23.Weaver Z, Montagna C, Xu X, Howard T, Gadina M, Brodie SG, Deng CX, Ried T. Mammary tumors in mice conditionally mutant for Brca1 exhibit gross genomic instability and centrosome amplification yet display a recurring distribution of genomic imbalances that is similar to human breast cancer. Oncogene. 2002;21:5097–5107. doi: 10.1038/sj.onc.1205636. [DOI] [PubMed] [Google Scholar]
- 24.Liu P, Zhang H, McLellan A, Vogel H, Bradley A. Embryonic lethality and tumorigenesis caused by segmental aneuploidy on mouse chromosome 11. Genetics. 1998;150:1155–1168. doi: 10.1093/genetics/150.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hentges KE, Nakamura H, Furuta Y, Yu Y, Thompson DM, O'Brien W, Bradley A, Justice MJ. Novel lethal mouse mutants produced in balancer chromosome screens. Gene Expr Patterns. 2006;6:653–665. doi: 10.1016/j.modgep.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Noguchi S, Kasugai T, Miki Y, Fukutomi T, Emi M, Nomizu T. Clinicopathologic analysis of BRCA1- or BRCA2-associated hereditary breast carcinoma in Japanese women. Cancer. 1999;85:2200–2205. [PubMed] [Google Scholar]
- 27.Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, Trudel M, Akslen LA. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 28.Lakhani SR, van de Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Sgagias MK, Wagner KU, Hamik B, Stoeger S, Spieker R, Huber LJ, Chodosh LA, Cowan KH. Brca1-deficient murine mammary epithelial cells have increased sensitivity to CDDP and MMS. Cell Cycle. 2004;3:1451–1456. doi: 10.4161/cc.3.11.1211. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Xiao C, Vonderhaar BK, Deng CX. A role of estrogen/ERalpha signaling in BRCA1-associated tissue-specific tumor formation. Oncogene. 2007;26:7204–7212. doi: 10.1038/sj.onc.1210527. [DOI] [PubMed] [Google Scholar]
- 31.Jones LP, Tilli MT, Assefnia S, Torre K, Halama ED, Parrish A, Rosen EM, Furth PA. Activation of estrogen signaling pathways collaborates with loss of Brca1 to promote development of ERalpha-negative and ERalpha-positive mammary preneoplasia and cancer. Oncogene. 2008;27:794–802. doi: 10.1038/sj.onc.1210674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clarke RB, Howell A, Potten CS, Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 1997;57:4987–4991. [PubMed] [Google Scholar]
- 33.Russo J, Ao X, Grill C, Russo IH. Pattern of distribution of cells positive for estrogen receptor alpha and progesterone receptor in relation to proliferating cells in the mammary gland. Breast Cancer Res Treat. 1999;53:217–227. doi: 10.1023/a:1006186719322. [DOI] [PubMed] [Google Scholar]
- 34.Mallepell S, Krust A, Chambon P, Brisken C. Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc Natl Acad Sci USA. 2006;103:2196–2201. doi: 10.1073/pnas.0510974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy A, Savage K, Gabriel A, Naceur C, Reis-Filho JS, Ashworth A. A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol. 2007;211:389–398. doi: 10.1002/path.2124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.