Abstract
The PAX2 gene encodes a transcription factor expressed during development. In humans, PAX2 mutations cause the renal-coloboma syndrome, whereas homozygous mutations are lethal, causing severe organ malformation, notably in the brain and kidney. Wilms tumor (WT) of the kidney results from a failure in the mesenchymal-epithelial transition, a crucial step partly controlled by PAX2. Downstream target genes regulated by PAX2 are still undefined. We therefore hypothesized that identification and characterization of the genes regulated by PAX2 may improve our understanding of developmentally related malignancies including WT. We used nickel agarose chromatin enrichment, chromatin immunoprecipitation, and the human embryonic kidney-derived cell line HEK293 to identify regulatory elements responding to PAX2. Among others, we identified WNT5A as a gene potentially regulated by PAX2. Here, we demonstrate that WNT5A is a direct target of PAX2 in HEK293 cells, using both transactivation and electrophoretic mobility shift assays. We were unable to find any WNT5A disease-associated mutations after screening a panel of 99 WT samples. However, quantitative reverse transcription-polymerase chain reaction in human favorable-histology WT revealed that ∼66% of the cases expressed significantly less WNT5A than human fetal kidney. Moreover, the WiT9 WT cell line revealed a weak expression of the WNT5A gene. A correlation of decreased WNT5A expression with predominant blastemal histology tumors suggests a possible inhibitory role in WT pathogenesis. This study underlines the importance of PAX2 in the regulation of WNT5A. Further in vivo study is necessary to determine whether the PAX2 and WNT5A are truly involved in WT pathogenesis.
Introduction
Wilms tumor (WT) is the most common renal cancer affecting young children. The origin of this tumor is in the pluripotent embryonic kidney precursor cells that fail to differentiate properly during early renal development [1,2]. Pediatric kidney tumors arise in the early stem cells of the kidney and are generally responsive to chemotherapy. In contrast, cancers affecting adult kidney arise from the tubular cells and are usually highly resistant to therapy.
The association of WT with persistent foci of embryonic tissue called nephrogenic rests reflects the link with early kidney development [2]. Nephrogenic rests consist mainly of blastemal cells with varying degrees of differentiation. Interestingly, these lesions share some genetic alterations with their adjacent tumors and suspected to be precursor lesions to WT [3,4]. It is likely that genes involved in early kidney development are also involved in the initiation and progression of WT [5]. Thus, it is important to elucidate the entity and nature of the genes and mechanisms governing the differentiation of embryonic kidney stem cells.
Two transcription factors PAX2 and WT1 are among such genes [6–9]. PAX2 belongs to the paired box gene family, characterized by a conserved domain, the paired box that binds specifically to DNA-binding sites to initiate regulation [10]. The functions attributed to PAX2 in different developmental stages, including activation and/or repression of downstream genes in different cell lineages, are complex and not completely understood. In developing mammalian kidney, PAX2 is one of the earliest markers expressed within the nephric duct and seems to orchestrate the branching of the ureteric bud through activation of glial cell line-derived neurotrophic factor (GDNF) and its corresponding receptor [11,12]. In mice, Pax2 null mutations cause abnormal kidney development with a marked absence of ureteric bud outgrowth [13,14]. Moreover, pax2 plays a crucial role in the mesenchyme surrounding the ureteric bud and during the early stage of epithelial differentiation [15–17]. During the apoptotic suppression process, PAX2 seems to play an important role, particularly during the fixation of nephron number and branching morphogenesis [18–21]. Mice hemizygous for Pax2 have reduced numbers of nephrons, less developed branching, and increased apoptosis in ureteric bud cells [19,22]. Delivery of Bax, a proapoptotic gene, to developing ureteric buds also reduces branching [23]. Conversely, blocking the expression of the antiapoptotic gene bcl2 or caspases reverses the branching defects observed in mice heterozygous for the pax2 gene [24]. At early stages of kidney development, the branching ureteric buds signal the neighboring metanephric mesenchymal cells, inducing them to aggregate and express Pax2 while simultaneously undergoing profound changes leading to the transition from mesenchyme to epithelium. It is believed that Pax2 expression regulates the mesenchymal-epithelial transition; however, its precise role and the downstream target genes controlled by Pax2 are not yet known [13]. To search for evidence that PAX2 has a direct role in WT pathogenesis, we previously used denaturing high-performance liquid chromatography (DHPLC) and sequencing analysis to screen the entire PAX2 coding sequence for mutations. We found no evidence for disease-causing mutations, suggesting that the direct involvement of PAX2 in WT is uncommon [25]. However, these data do not exclude the possibility that the pathway downstream from PAX2 may be involved. Therefore, we used the nickel agarose chromatin enrichment (NACE) and chromatin immunoprecipitation (ChIP) techniques to identify genes regulated by PAX2 [26].
In this study, we describe a possible functional role for the PAX2 target gene, WNT5A. We have validated WNT5A as a PAX2 target by using electrophoretic mobility shift assay (EMSA) and by polymerase chain reaction (PCR)-amplifying an enriched chromatin generated by means of ChIP technique, where an anti-Pax2 polyclonal antibody was used. We have also demonstrated that WNT5A is underexpressed in many favorable-histology Wts relative to fetal kidney. These data suggest a pathway where the PAX2 gene regulates renal nephron development through WNT signaling pathway with the participation of WNT5A gene.
Materials and Methods
Cell Culture
Human fetal kidney cell line HEK293 was purchased from the American Type Culture Collection (Manassas, VA) and was propagated in minimum essential medium supplemented with 10% fetal bovine serum and 1% glutamine. The WT cell line WiT49, obtained recently as a kind gift from Dr. Herman Yeger (Hospital for Sick Children, Toronto, Ontario, Canada), was propagated in Dulbecco's modified Eagle's medium, containing 15% fetal bovine serum and 1% insulin. Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2, and their growth was monitored until reaching 60% to 70% confluence, at which stage the medium was changed and cells were transfected.
Transfection
HEK293 cells were transiently transfected with 4 µg of pcDNA-4/His/Max-PAX2 DNA vector using the transfection reagent Effectine (Qiagen Inc., Valencia, CA). HEK293 cells were also transfected with an empty vector (pcDNA-4/His/Max) as a negative control. After 24 and 36 hours of incubation at 37°C in a humidified incubator containing 5% CO2, cells were subjected to NACE or ChIP [26].
Polymerase Chain Reaction Amplification
All primers used for the PCR amplification were designed using the PRIMER3 software package (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and their sequences are listed in Table 1. Amplification products were resolved on 1% agarose gel stained with ethidium bromide.
Table 1.
Sequence of Primers Used for PCR Assays.
| Forward | Reverse |
| 5′-GAG GAG AAG CGC AGT CAA TC-3′ | 5′-GAA GAG GAA GAA CAC GCA CA-3′ |
| 5′-CCA CCT CGA GAT TTC ACG TA-3′ | 5′-TTT TCC TAC CTA TCT GCA TCA CC-3′ |
| 5′-GCC AGT GAT CCC TTG TCC T-3′ | 5′-GGG CAA GGT GAG AAG ACA GA-3′ |
| 5′-CAT CTG GCT CCT CCA TGA AT-3′ | 5′-GGT GGC ACC CAC TAC TTG-3′ |
| 5′-ATG AGT CCA ACA TGC AGA GA-3′ | 5′-GTA ACT GTA TCA GTG AAA AT-3′ |
| 5′-ATG AGT CCA ACA TGC AGA GA-3′ | 5′-GTT GGC AGG CAC AGC TGG TT-3′ |
| 5′-AAC CAG CTG TGC CTG CCA AC-3′ | 5′-GTA ACT GTA TCA GTG AAA AT-3′ |
The PCR conditions were optimized to amplify a specific band of expected size using DNA samples as templates. The NACE-enriched WNT5A 5′-upstream element (AB) was amplified totally or partially (subfragments A and B) before cloning.
Nickel Agarose Chromatin Enrichment and ChIP
Both NACE and ChIP analyses were performed as described earlier, with slight changes in sonication time and washing stringency [26,27]. For NACE assays, HEK293 cells were fixed in their culture medium and lysed to extract and sonicate DNA five times for 20 seconds each to obtain sizes ranging between 200 and 1000 bp. Purified chromatin was then incubated with Ni2+ agarose beads for 6 hours and washed, and DNA/protein complexes were purified as described earlier [26]. After reversing the cross-linking of the complexes formed by DNA/proteins, DNA was purified through Qiagen columns after a complete digestion of proteins. We recently succeeded to optimize conditions for a specific anti-Pax2 antibody used in ChIP that was carried out exactly as described above for NACE with the exception of replacing Ni2+ with a polyclonal Pax2 antibody and a final purification through phenol chloroform extraction [26,28]. A mouse IgG polyclonal antibody was used as a negative control for ChIP assays.
Cloning and Sequencing of the NACE-Enriched Chromatin
Five hundred nanograms of the enriched DNA was end-filled and ligated to the EcoRV-digested pBluescript II SK(-) vector (Stratagene, La Jolla, CA). Ligation products were used to transform XL-Blue competent cells, and DNA from putative-positive clones was extracted. Enzymatic digestions confirmed the presence of the insert, and positive clones were sequenced on an automated sequencer (CEQ8000 Genetic Analysis System; Beckman Coulter, Mississauga, Ontario, Canada) using dye terminator cycle sequencing chemistry (Beckman Coulter).
Construct Preparation
The PAX2 cDNA encoding the entire open reading frame was subcloned into EcoRI-XbaI sites in the pcDNA4-His/Max plasmid (Invitrogen, Carlsbad, CA). The WNT5A 5′-upstream enriched fragment (AB) of ∼300 bp was amplified and cloned first into pGEM-T-easy vector (Promega, Madison, WI) and subsequently subcloned upstream of the luciferase gene in the pGL3 vector (Promega). The AB fragment was subdivided by PCR amplification into units, A and B, of ∼155 and ∼145 bp, respectively. The fragments were subcloned upstream to the luciferase reporter in pGL3 vector as described above. A synthesized fragment (P2BS1) of 54 bp, containing a confirmed PAX2 binding site [29], was end-filled, cloned upstream to the cytomegalovirus promoter of the pGL3 vector, and used as a positive control for transactivation.
Transactivation Assays
We amplified fragment AB covering the total ∼300 bp as well as overlapping fragments A and B, which, together, covered the total region of ∼300 bp. The three amplicons A, B, and AB were each cloned into a TA vector-sequenced and subcloned into pGL3-CMV-luciferase reporter vector. HEK293 cells were cultured in 12-well plates at a density of 1 x 105 cells/ml and cotransfected 36 hours later with PAX2-pcDNA4-His/Max or pcDNA4-His/Max and 300 ng of pGL3 vector containing either the PAX2 binding site or fragments A, B, and AB placed upstream of the Luciferase gene in pGL3. Twenty-four hours later, cells were harvested and subjected to luciferase activity-monitoring according to the manufacturer's guidelines (Promega). A reporter construct containing one PAX2 binding site [29] (P2BS-pGL3, see sequence in Table 2) was used as positive control. For each set of experiments, transfections were performed in triplicates and repeated three times for confirmation.
Table 2.
Sequence of Oligonucleotides Used for EMSA.
| Oligos | Sense—Biotin-Labeled | Antisense |
| P2BS | 5′-TTC GTG GTC ATG TAA GTT GTT TCA CTT GGA TGA AAT CCC TCT TCA GGA GAT GTT-3′ | 5′-AAC ATC TCC TGA AGA GGG ATT TCA TCC AAGTGA AAC AAC TTA CAT GAC CAC GAA-3′ |
| α | 5′-ATG AGT CCA ACA TGC AGA GAG AAG CAG AGA-3′ | 5′-TCT CTG CTT CTC TCT GCA TGT TGG ACT CAT-3′ |
| β | 5′-GAA GCA GAG AAG AGA GAT GAA TGC AAC AGG-3′ | 5′-CCT GTT GCA TTC ATC TCT CTT CTC TGC TTC-3′ |
| γ | 5′-ATG CAA CAG GGG TGG GAG AGA TGG AAA GAT-3′ | 5′-ATC TTT CCA TCT CTC CCA CCC CTG TTG CAT-3′ |
| δ | 5′-ATG GAA AGA TAG AGA GAT AGA CAG AGA CAG-3′ | 5′-CTG TCT CTG TCT ATC TCT CTA TCT TTC CAT-3′ |
| ε | 5′-ACA GAG ACA GAG GCA GAG AGA TCT GAG AAC-3′ | 5′-GTT CTC AGA TCT CTC TGC CTC TGT CTC TGT-3′ |
| ζ | 5′-ATC TGA GAA CCA GCT GTG CCT GCC AAC-3′ | 5′-GTT GGC AGG CAC AGC TGG TTC TCA GAT-3′ |
| γ′ | 5′-TAC GTT GTC CCC ACC CTC TCT ACC TTT CTA-3′ | ATG CAA CAG GGG TGG GAG AGA TGG AAA GAT-3′ |
Electrophoretic Mobility Shift Assays
In addition to the P2BS oligonucleotides containing the PAX2 binding site, six overlapping oligonucleotides (α, β, γ, δ, ε, and ζ) covering the total sequence of the A fragment were synthesized (Table 2) with one strand conjugated to biotin. HEK293 cells expressing endogenous and recombinant Pax2 proteins were lysed, and protein extracts were resolved on 6% polyacrylamide Tris-glycine-EDTA. Five micrograms of protein extracts was incubated with 1.3 mM DTT, 5 µg of sheared salmon sperm DNA, 1 µg of poly dIdC (Sigma, St. Louis, MO), and biotin-labeled oligonucleotides (α, β, γ, δ, ε, and ζ) as well as P2BS fragments. Samples were incubated at room temperature for 20 minutes to form protein-DNA complexes. After 45 minutes of a prerun of the gel, samples were analyzed by electrophoresis. Protein-DNA complexes were subsequently transferred to a membrane, and signals were revealed by using a chemiluminescent nucleic acid detection module (Pierce, Rockford, IL).
Supershift Assay
Ten micrograms of HEK293 protein extract was incubated overnight with 2 µg of anti-Pax2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) in 12 µl of 5% skim milk. Biotin-labeled probes were subsequently added and incubated for another 20 minutes at room temperature. Samples were resolved on a 5% acrylamide gel using 1x TGE buffer (50 mM Tris, 380 mM glycine, and 2 mM EDTA, pH 8.2) at 4°C for 1.5 hour using 20 mA. The transfer and the detection of the signal were performed as above for EMSA.
Silencing of PAX2 and WNT5A
Two oligonucleotides designed for each of the two genes PAX2 and WNT5A to specifically knock down their expression, were purchased from Qiagen and their sequences are as follow: PAX2-siRNA1 5′-CACAGCTACACGCCCATTAAA-3′; PAX2-siRNA2 5′-CCCGTAGTTGCTCTTTCGGTA-3′; WNT5A-siRNA2 5′-CCGGATAACCTTGTAACATAT-3′; WNT5A-siRNA6 5′-TTGGTGGTCGCTAGGTATGAA-3′. siRNA duplexes were transfected into WiT49 cells using HiPerFect (Qiagen) according to the manufacturer's instructions. Cells were plated into 60 mm and/or six-well dishes at a density of 5 x 104 cells/ml 24 hours before transfections. A total of 5 µl of 20 µM siRNA solution and 20 µl of the transfection reagent HiPerFect (Qiagen) were incubated with 0.5 ml of serum-free Dulbecco's modified Eagle's medium for 10 minutes before adding the resulting mixture to the cells dropwise. For data confirmation, each siRNA transfection was processed in duplicate.
Protein Extraction and Western Blot
Twenty-four hours of incubation after transfection, cells were rinsed twice with cold phosphate-buffered saline and lysed in protein lysis buffer (20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid pH 7.6, 75 mM NaCl, 2.5 mM MgCl2, 0.1 mM ethylenediaminetetraacetic acid, 0.1% Triton X-100, 1 µg/ml leupeptin, 1 µg/ml aprotinin, 1 µg/ml pepstatin, and 1 mM phenylmethylsulfonyl fluoride). The cell lysates were kept on ice for 10 minutes, rotated at 4°C for 20 minutes, and centrifuged at 10,000 rpm for 15 minutes, and the supernatant was subjected to Bradford protein assay using a BioRad kit (Bio-Rad, Hercules, CA) to determine protein concentrations using bovine serum albumin as a standard control. Equal amounts of proteins from each set of experiments were loaded on a 6% acryl/bisacrylamide gel. Because WNT5A is a secreted protein, its detection required the collection of the medium at 24 hours of incubation after transfection, then concentration through a “centricon” concentrator column and quantification.
The protein extracted from WiT49 cells or concentrated medium were resolved on 10% polyacrylamide gel and transferred to a nitrocellulose membrane. Immunoblot analyses for Pax2 and Wnt5a proteins were performed with rabbit polyclonal and monoclonal antibodies respectively (cell signaling, dilution of 1:150). Immunoblots were incubated overnight at 4°C and followed by 2 hours of incubation at room temperature with an HRP-conjugated antirabbit IgG secondary antibody (dilution 1:5000; Cell Signaling, Danvers, MA). Generated signals were revealed by chemiluminescent protein detection kit (Pierce).
In Silico Analysis
Both Ensembl (http://www.ensembl.org/index.html) and National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) Web sites were used for the BLAST analysis of the relevant clones, for the screening of the nucleotide databases for other species, and for the localization of regions similar to the WNT5A upstream element [30]. Similar sequences from human and mouse were aligned using the NCBI Web site.
Mutation Analysis
DNA preparation and mutational analysis were performed as described earlier [25]. DNA samples from 99 favorable-histology WT cases were obtained from the Renal Tumors Bank of the Children's Oncology Group [31] after approval from our institutional research ethics committee. Exons 1 to 4 of the human WNT5A were subjected to PCR amplification using the panel of primers listed in Table 1. For each WT sample, a wild type DNA control was amplified in parallel under the same conditions. The specificity of the PCR products was monitored by electrophoresis on a 1.5% agarose gel.
For DHPLC analysis, 10 µl of PCR product from each WT and from the wild type were mixed together, heated to 95°C for 5 minutes, and cooled to 25°C at a rate of 1.5°C/min. Denaturing high-performance liquid chromatography analysis was performed using the WAVE nucleic acid fragment system (Transgenomics, Omaha, NE) equipped with a L7300+ column oven using aDNA Sep column as described earlier [25].
Quantitative Reverse Transcription-Polymerase Chain Reaction and Statistical Analysis
Thirty-eight frozen tissue samples of favorable-histology WT were subjected to total RNA extraction using TRIzol reagent (Life Technologies, Gaithersburg, MD). After DNase treatment, 5 µg of each RNA sample was run on a denaturing agarose gel, and the quality of RNA was monitored under UV light using ethidium bromide staining of the gel. cDNA was produced using 0.5 µg of RNA with the SuperScript II Reverse Transcriptase (Invitrogen). Quantitative PCRs were carried out using the 7900HT Fast Real-Time PCR systems (Applied Biosystems, Foster City, CA) and performed following the guidelines provided by the company (Applied Biosystems) using the TaqMan Gene Expression Assay Kit. All cDNA samples were assayed in triplicate for each primer set. Primers to amplify ∼140-bp fragments of WNT5A and GAPDH genes together with specific probe for each gene were included in the 20x TaqMan Gene Expression Assay Mix (Applied Biosystems). Expression data normalized to GAPDH and fetal kidney controls were analyzed using the ΔΔCt method [32]. The Children's Oncology Group Renal Tumors Reference Pathologist categorized tumor histology.
Results
Nickel Agarose Chromatin Enrichment Generates Potential Candidate Genes Regulated by PAX2
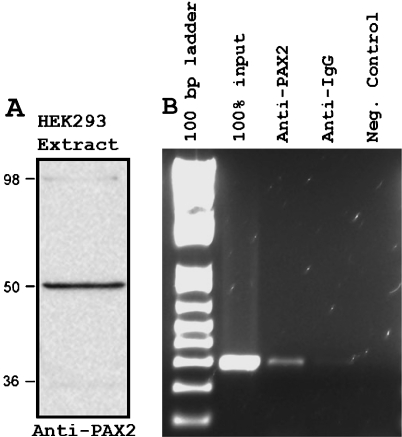
At the start of this project, we were unable to find a highly specific Pax2 antibody as required in ChIP analysis. Thus, we used the NACE technique, an antibody-free affinity-enrichment method based on Ni2+-NTA affinity chromatography, a variant of ChIP that can be used as an alternative to overcome the lack of a specific antibody and nonspecific cross-linking [26]. We have used the human embryonic kidney cell line HEK293 as a model system to identify gene targets of PAX2 and its regulation of gene expression. Pax2 protein expression was detectable by Western blot in HEK293 cells (Figure 1A), making it an appropriate model to isolate and test the predicted PAX2-target regulatory elements.
Figure 1.
Endogenous PAX2 binds to WNT5A 5′-upstream region. (A) Western analysis of Pax2 protein expression in HEK293 cells nuclear extract. (B) Chromatin immunoprecipitation was performed using a polyclonal antibody directed against Pax2. The enriched chromatin was amplified using primers flanking the NACE enriched WNT5A 5′-upstream region. A specific band of an expected size (∼300 bp) was obtained in the sample treated with the anti-Pax2 antibody but not in samples treated with an anti-IgG or mock controls.
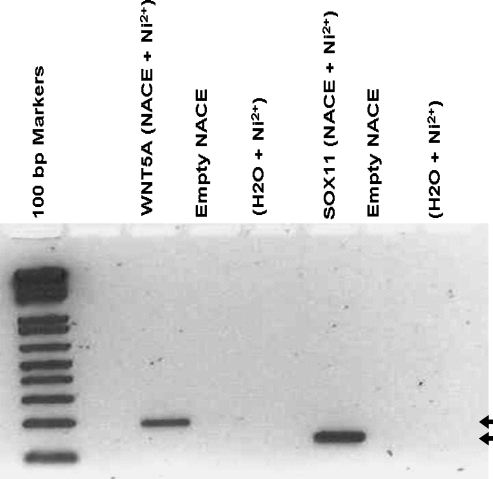
DNA fragments from 140 PAX2 NACE clones were sequenced and BLAST-searched in public sites at NCBI (http://www.ncbi.nlm.nih.gov/) and Ensembl (http://www.ensembl.org/index.html). BLAST analysis revealed DNA sequences related to 1 adhesion molecule, 23 novel genes, 5 proto-oncogenes, and 11 transcription factors including WNT5A, WT1, WIT1, and the SOX11 genes. The remaining 76 clones were repetitive sequences or could not be sequenced and were excluded from further investigations. Table 3 lists the NACE-generated putative regulatory elements binding to PAX2. Whereas the identification of genes such as WT1, previously described as a PAX2-target gene supports the specificity of the NACE procedure, validation experiments were, nevertheless, undertaken for candidate genes, which we intended to characterize (Figure 2). Owing to the known central role of the WNT pathway in the development of the kidney, we chose to further analyze the WNT5A gene first.
Table 3.
Recapitulation of NACE-Generated Regulatory Elements Presumably Interacting with PAX2.
| No. and Gene Symbol | Locus | Category | No. and Gene Symbol | Locus | Category |
| 01. ABLIM3 | 5q33.1 | Adaptor | 33. BIRC6 | 2p22.21 | Protooncogene |
| 02. CIB1 | 15q25.3-q26 | Adhesion | 34. HRB2 | 12q21.1 | Protooncogene |
| 03. CD4 | 12p13.31 | Antigen | 35. EIF2C4 | 1p34.3 | Protooncogene |
| 04. VRK2 | 2p16-p15 | Enzyme | 36. NOT4 | 7q33 | Transcription factors |
| 05. MAP2K5 | 15q22.2-31 | Enzyme | 37. RNF39 | 6P21.3 | Transcription factors |
| 06. GUCY1B2 | 13q14.2-14.3 | Enzyme | 38. SOX11 | 2P25.3 | Transcription factors |
| 07. DPP10 | 2q14.1 | Enzyme | 39. ZNF141 | 4p16.3 | Transcription factors |
| 08. ? | Chromosome #9 | Novel/hypothetical | 40. BTG4 | 11q23.2–3 | Transcription factors |
| 09. ? | 1q41 | Novel/hypothetical | 41. N6AM1 | 21q22.1 | Transcription factors |
| 10. ? | XP11.22 | Novel/hypothetical | 42. GTF2B1 | 1p22.1–3 | Transcription factors |
| 11. ? | Chromosome #17 | Novel/hypothetical | 43. ARHGAP8 | 22q13.31 | Transcription factors |
| 12. ? | 9p24.1 | Novel/hypothetical | 44. WT1 and WIT1 | 11p13 | Transcription factors |
| 13. ? | Chromosome #20 | Novel/hypothetical | 45. TCF4 | 18q21.1 | Transcription factors |
| 14. ? | Chromosome #12 | Novel/hypothetical | 46. MAFK | 7p22 | Transcription factors |
| 15. ? | Chromosome #22 | Novel/hypothetical | 47. ELL2 | 5q15 | RNA Associated |
| 16. ? | 1p32.1 | Novel/hypothetical | 48. NXT1 | 20p12 | RNA Associated |
| 17. ? | Chromosome #7 | Novel/hypothetical | 49. SRRM2 | 16P13.3 | RNA Associated |
| 18. ? | Chromosome #1 | Novel/hypothetical | 50. HTR1E | 6q14–15 | Signaling/receptor |
| 19. ? | Chromosome #4 | Novel/hypothetical | 51. MAPKAP1 | 9q34.11–12 | Signaling/receptor |
| 20. ? | 18q21.1 | Novel/hypothetical | 52. DCAMKL1 | 9q34.11–12 | Signaling/receptor |
| 21. ? | Chromosome #2 | Novel/hypothetical | 53. DLG2 | 11q | Signaling/receptor |
| 22. ? | 11q14.1 | Novel/hypothetical | 54. CCKAR | 4p15.2 | Signaling/receptor |
| 23. ? | 22q122.1 | Novel/hypothetical | 55. AKAP13 | 15q24–25 | Signaling/receptor |
| 24. ? | 21q21 | Novel/hypothetical | 56. TNFSF4 | 1q24–25 | Signaling/receptor |
| 25. ? | 15q23 | Novel/hypothetical | 57. GEF6 | Chromosome X | Signaling/receptor |
| 26. ? | 3q26.31 | Novel/hypothetical | 58. PTPRM | 18P11.2 | Signaling/receptor |
| 27. ? | Chromosome #4 | Novel/hypothetical | 59. HARP | Chromosome #16 | Signaling/receptor |
| 28. ? | Chromosome #7 | Novel/hypothetical | 60. TNFRSF9 | 1p36 | Signaling/receptor |
| 29. ? | Chromosome #9 | Novel/hypothetical | 61. ACTRT1 | X26.1 | Structural protein |
| 30. ? | 16q21–22.1 | Novel/hypothetical | 62. SH120 | 1P36.13 | Structural protein |
| 31. WNT5A | 3p21.1 | Protooncogene | 63. ABLIM3 | 5q33.1 | Structural protein |
| 32. SEMA3E | 7q21.11 | Protooncogene | 64. FAM36A | 1q44 | ? |
Figure 2.
Validation of NACE specificity. A representative example of PCR amplification using NACE product as template and primers designed to amplify the 5′upstream region of WNT5A and SOX11 genes. Both WNT5A and SOX11 were confirmed to be specifically present in the Pax2 NACE product. Bands of the expected sizes were obtained (arrows) specifically in PAX2-transfected samples enriched by NACE. Material enriched from HEK293 cells transfectd by an empty vector was used as a control along with water and Ni2+ agarose beads (mocks).
Confirmation of WNT5A As a Target Regulated by PAX2
To determine whether endogenous PAX2 binds to the WNT5A promoter in vivo, we performed ChIP analysis using HEK293 cells, and an antibody, which conditions for specific binding to endogenous PAX2, were recently optimized. Western blot analysis, using HEK293 cells, protein extracts showed a specific band of an expected size of ∼50 kDa, consistent with the molecular weight of PAX2 (Figure 1A). As shown in Figure 1B, a ∼300-bp fragment located upstream of WNT5A containing the putative PAX2 binding site was amplified from PAX2 immunoprecipitates, indicating that endogenous PAX2 binds to the putative WNT5A regulatory region. Additionally, amplification of the ∼300-bp fragment using the ChIP product as template confirms the specificity of the NACE-enriched fragment, corresponding to a regulatory element mapped ∼82 kb upstream of the WNT5A gene.
Transactivation Assays
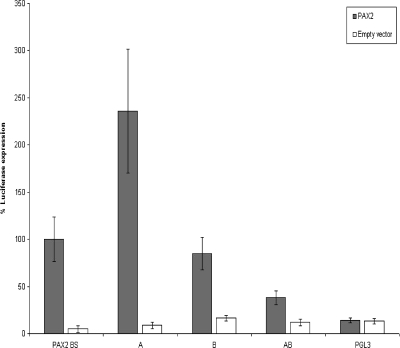
To prove whether the NACE- and ChIP-derived ∼300-bp WNT5A upstream fragment possessed a PAX2-responsive element, a transactivation assay was performed. Monitoring luciferase activity revealed that PAX2 significantly enhanced the expression from all WNT5A upstream element luciferase reporters, A, B, and AB (Figure 3). PAX2-enhanced luciferase expression was greatest when cotransfected with fragment A, whereas AB fragment expression was similar to the P2BS control. The pGl3 empty vector was used as a negative control.
Figure 3.
Transactivation assays of the WNT5A 5′-upstream region. The ∼300-bp NACE-enriched fragment (AB) and subfragments A and B were cloned into the pGL-3 CMV luciferase vector. The three vectors were cotransfected with pcDNA-4/His/Max-PAX2 expression vector and pRL-CMV internal control plasmid into HEK293 cells. Dual-luciferase activity ratios were calculated relative to express-tagged PAX2 activation of P2BS, the PAX2 binding site reporter. Error bars, SEM.
Electrophoretic Mobility Shift Assays
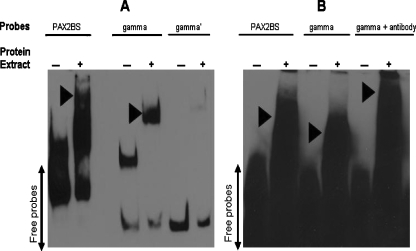
Based on data obtained with the transactivation assays, fragment A was suspected to contain the binding site of PAX2. Six overlapping biotin-labeled oligonucleotides (α, β, γ, δ, ε, and ζ), covering fragment A as well as the PAX2 binding site P2BS, were synthesized (Table 2). A protein extract from HEK293 cells was incubated with each oligo, and reactions were resolved on a 10% polyacrylamide gel. In addition to that observed with the P2BS oligo, a clear shift was obtained only with the gamma-oligo (γ) (Figure 4A) indicating that the binding site is likely located within this 30-bp sequence. The synthesis of an oligo gamma′ (γ′) where pyrimidines were replaced by purines and purines by pyrimidines eliminated the observed shift (Figure 4A).
Figure 4.
PAX2 binding to sequences in the WNT5A 5′-upstream region. Electrophoretic mobility shift assay to assess the direct binding of PAX2 to potential sequences including a synthetic control binding site (P2BS), the PAX2 binding site from the PAX2 5′-upstream region (gamma-γ), and the same sequence altered by changing purines to pyrimidines and vice versa (gamma′-γ′). (▸) Arrowheads indicate shifts caused by proteins-probes complex formation (A). An anti-Pax2 antibody was added to the complex formed between protein extracts and biotin-labeled gamma oligos to validate the specificity of the binding. A supershift migrating at higher molecular weight was observed (B).
To validate the specificity of the binding, we optimized conditions for a supershift assay using a specific antibody to Pax2 protein. A detectable label migrating at higher apparent molecular weight was observed as a result of the anti-Pax2 antibody binding with the biotinlabeled probes/protein extracts' heterocomplexes (Figure 4B).
In Vitro siRNA-Mediated Silencing of PAX2 and WNT5A
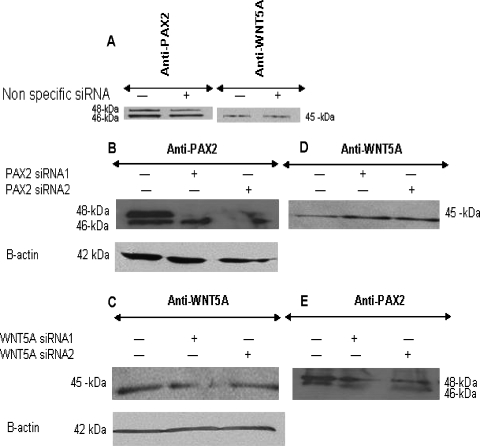
To knock down PAX2 and WNT5A genes, we used siRNA formulations commercialized by Qiagen and optimized to efficiently shut down the expression of PAX2 and WNT5A. WiT49 cells were transfected by either PAX2-siRNA1/PAX2-siRNA2 or WNT5A-siRNA2/WN5A-siRNA6 sets. Cotransfection with both sets (PAX2-siRNA1 and PAX2-siRNA2 or WNT5A-siRNA2 and WN5A-siRNA6) was highly toxic to the cells. An siRNA with no specific binding to human, mouse, and rat (sequence not revealed by the company) was used as a negative control (Figure 5A). siRNA-treated samples were subjected to Western analysis using antibodies directed against Pax2, Wnt5a, and β-actin as loading controls. All the four siRNA significantly contributed to a successful knock down of PAX2 and WNT5A expression (Figure 5, B and C). For PAX2 knock down, siRNA2 looks slightly more efficient than siRNA1 (Figure 5B), whereas for WNT5A gene, siRNA2 seems more effectual than siRNA6 (Figure 5C). β-Actin monitoring of the treated samples showed that the observed knock down was not caused by an eventual loss of the material but by siRNA treatment (Figure 5, B and C).
Figure 5.
siRNA-based knock down of PAX2 and WNT5A genes. Two locations within PAX2 and WNT5A genes were specifically targeted to knock down their expression using commercialized siRNA (Qiagen). A nonspecific siRNA was used as a negative control where protein expression is not altered in treated samples compared to nontreated samples (A). It is noteworthy that WNT5A expression is weak when compared to the PAX2 expression in the WiT49 WT cell line. Both PAX2-siRNA1 and PAX2-siRNA2 were efficient to reduce drastically PAX2 expression (B), whereas WNT5A expression was clearly detectable in the same samples (D). Culture medium where WiT49 cells were propagated and treated with WNT5A-siRNA2 and WNT5A-siRNA6 was concentrated and subjected to Western blot analysis using an anti-Wnt5A antibody. Like PAX2-siRNA, both WNT5A-siRNA2 and WNT5A-siRNA6 reduced the WNT5A expression (C). The presence of PAX2 expression was detected in these WNT5A knocked down samples (E).
Mutation Screenings of WNT5A in WT Patients
The four exons covering the entire coding sequence of WNT5A were amplified separately using primers listed in Table 1 and were subjected to mutation analysis using DHPLC. None of the 99 WT samples displayed a chromatogram with abnormal pattern when compared to their control DNA samples, providing no evidence for exon-specific mutations in WNT5A in WTs.
Quantitative Reverse Transcription-Polymerase Chain Reaction
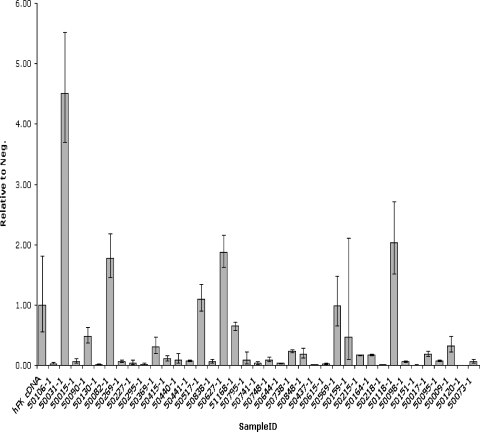
To assess the expression profile of WNT5A in WT samples, we performed quantitative reverse transcription-polymerase chain reaction (RT-PCR) using the TaqMan Gene Expression Assay from Applied Biosystems on RNA isolated form favorable-histology WT samples. Of the 38 cases tested, 25 showed significantly less expression of WNT5A compared to the human fetal kidney control (Figure 6 and Table 4). Exceptionally, one sample highly overexpressed WNT5A relative to fetal kidney, whereas the expression levels in the remaining 12 cases were not significantly different from fetal kidney.
Figure 6.
Quantitative RT-PCR to assess the expression levels of WNT5A in WT samples of favorable-histology. Gene expression assay (Applied Biosystems) was performed on 38 WT samples. Human fetal kidney was included as a control to which all data were normalized. Error bars, relative SE.
Table 4.
Expression Analysis and Histology Classification for the 38-Sample Experimental Set.
| Sample | ID No. | ΔCt | σ | Test Statistic | P | Significance [α < 0.05] | Histology Classification |
| Control | HFK cDNA | 10.27 | 0.86 | 0 | 1 | No | - |
| 1 | 50106-1 | 15.30 | 0.69 | 7.919 | .02 | Yes | Epithelial |
| 2 | 50031-1 | 8.09 | 0.29 | 4.155 | .05 | Yes* | Epithelial |
| 3 | 50015-1 | 14.23 | 0.74 | 6.047 | .03 | Yes | Blastemal |
| 4 | 50090-1 | 11.33 | 0.38 | 1.954 | .19 | No | Stromal |
| 5 | 50130-1 | 16.94 | 1.24 | 7.657 | .02 | Yes | Blastemal |
| 6 | 50082-1 | 9.44 | 0.29 | 1.582 | .25 | No | Stromal |
| 7 | 50269-1 | 14.30 | 0.40 | 7.369 | .02 | Yes | Stromal |
| 8 | 50227-1 | 14.93 | 1.17 | 5.565 | .03 | Yes | Blastemal |
| 9 | 50295-1 | 17.01 | 2.12 | 5.106 | .04 | Yes | Stromal |
| 10 | 50369-1 | 11.98 | 0.61 | 2.825 | .11 | No | Stromal |
| 11 | 50415-1 | 13.42 | 0.50 | 5.505 | .03 | Yes | Stromal |
| 12 | 50440-1 | 13.81 | 1.21 | 4.147 | .05 | Yes | Stromal |
| 13 | 50441-1 | 13.99 | 0.28 | 7.143 | .02 | Yes | Stromal |
| 14 | 50517-1 | 10.13 | 0.29 | 0.266 | .82 | No | Stromal |
| 15 | 50838-1 | 14.27 | 0.71 | 6.223 | .02 | Yes | Blastemal |
| 16 | 50627-1 | 9.36 | 0.20 | 1.776 | .22 | No | Stromal |
| 17 | 51168-1 | 10.89 | 0.15 | 1.25 | .34 | No | Stromal |
| 18 | 50795-1 | 13.89 | 1.44 | 3.742 | .06 | No | Blastemal |
| 19 | 50741-1 | 15.24 | 0.92 | 6.842 | .02 | Yes | Blastemal |
| 20 | 50748-1 | 13.73 | 0.63 | 5.648 | .03 | Yes | Blastemal |
| 21 | 50644-1 | 15.20 | 0.33 | 9.291 | .01 | Yes | Blastemal |
| 22 | 50738-1 | 12.38 | 0.14 | 4.21 | .05 | Yes | Blastemal |
| 23 | 50848-1 | 12.69 | 0.58 | 4.035 | .06 | No | Blastemal |
| 24 | 50437-1 | 17.07 | 0.13 | 13.57 | .01 | Yes | Blastemal |
| 25 | 50615-1 | 15.92 | 0.71 | 8.768 | .01 | Yes | Blastemal |
| 26 | 50569-1 | 10.30 | 0.59 | 0.048 | .97 | No | Stromal |
| 27 | 50159-1 | 11.36 | 2.17 | 0.813 | .5 | No | Stromal |
| 28 | 50215-1 | 12.85 | 0.10 | 5.183 | .04 | Yes | Stromal |
| 29 | 50164-1 | 12.78 | 0.11 | 5.039 | .04 | Yes | Stromal |
| 30 | 50218-1 | 17.60 | 0.04 | 14.789 | 0 | Yes | Blastemal |
| 31 | 50118-1 | 9.24 | 0.42 | 1.854 | .2 | No | Stromal |
| 32 | 50098-1 | 14.34 | 0.26 | 7.871 | .02 | Yes | Stromal |
| 33 | 50151-1 | 17.99 | 0.44 | 13.88 | .01 | Yes | Stromal |
| 34 | 50017-1 | 12.71 | 0.37 | 4.533 | .05 | Yes | Stromal |
| 35 | 50095-1 | 14.04 | 0.16 | 7.494 | .02 | Yes | Blastemal |
| 36 | 50009-1 | 11.88 | 0.56 | 2.733 | .11 | No | Stromal |
| 37 | 50120-1 | 17.83 | 0.09 | 15.176 | 0 | Yes | Stromal |
| 38 | 50073-1 | 14.28 | 0.62 | 6.547 | .02 | Yes | Stromal |
This sample showed significant over expression (Figure 1).
We subsequently examined the relationship between WNT5A expression and tumor histology. Samples were categorized in WNT5A expression as either aberrant or normal and compared to histologic classification based on predominant histologic finding; that is, blastemal, epithelial, or stromal as determined on central pathology review on Children's Oncology Group study AREN03B2 (Table 4). Fifteen samples showed predominant blastemal histology, two samples showed predominant epithelial histology, and the remainder showed predominantly stromal histology. Using the McNemar exact test for nonparametric data, a significant correlation between WNT5A underexpression and predominant blastemal histology was observed (P = .000122, n = 14/38).
Discussion
We used the HEK293 cell line to identify target genes regulated by PAX2, a transcription factor known for its role in kidney development [33]. Our preliminary experiments revealed that this cell line expresses PAX2, making it an appropriate model to isolate and test the predicted target regulatory elements. In contrast to most cancers where both animal models and cell lines are available, the study of WT still suffers from the lack of good cell line models. The few WT lines that have been reported, including WiT49 and HWFT, usually harbor a p53 mutation, characteristic of the anaplastic variant of WT and likely not representative of the much more common favorable-histology WT (95% of WTs are favorable histology), which rarely includes mutation of the p53 gene [34,35]. We chose HEK293 cells as a very well characterized line, which is easily used for transfection/transactivation assays, especially for its human embryonic kidney origin. However, findings generated by using this model remain speculative because a good cell line representing favorable-histology WTs have not yet been established.
BLAST sequence analysis of the PAX2 NACE-derived sequences revealed responsive elements from a variety of genes including adhesion proteins, proto-oncogenes, transcription factors, and some novel genes (Table 3). Approximately 54% of the clones (76/140) were either repetitive sequences or could not be sequenced and were excluded from further analysis.
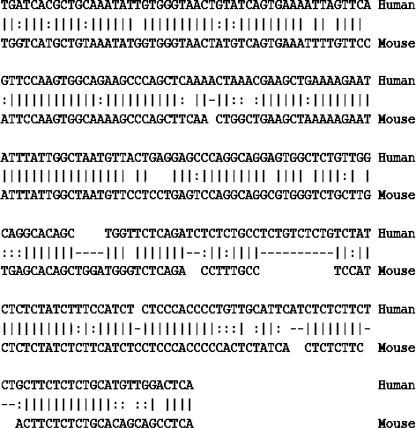
Among the potential PAX2 targets obtained using the NACE and ChIP techniques, a ∼300-bp WNT5A 5′-upstream element was particularly interesting. WNT genes are expressed in a variety of human tissues, including bladder and fetal kidney, and, therefore, play a major role in kidney development [33] and is implicated in cancer [36]. The NACE-enriched fragment, localized ∼82 kb upstream of the WNT5A gene, is conserved between human and mice and represents one of the upstream noncoding putative regulatory regions (Figure 7). Likewise, the identified ∼300 bp lies within an area belonging to other genes located in the vicinity, notably within the last intron of ERC2 gene, upstream the WNT5A. Owing to its central position in several developmental pathways and its involvement in the control of different steps in certain organs' formation including the kidney [13], PAX2 can adopt polyvalent functions and has the capacity to interact simultaneously with different entities.
Figure 7.
Alignment of the human ∼300-bp WNT5A-responsive element with its counterpart in mouse 73% sequence identity. Human genomic upstream region of WNT5A was compared to the mouse counterpart and a percent identity plot (PIP) was generated. There are several regions of homologous sequence in the 100-kb 5′-upstream region. None of these homologous regions match known genes or expressed sequence tags and are therefore assumed to be noncoding regulatory sequences. The NACE-enriched ∼300-bp fragment (bar) overlaps one of these homologous regions that share more than 70% identity with the mouse.
The results of our transactivation experiments demonstrated that the WNT5A upstream element was sufficient to provide PAX2-dependent activation of gene expression and led to the identification of an authentic PAX2 responsive element within the 155-bp subfragment of the WNT5A 5′-upstream element (Figure 3). The fragment contains a DNA sequence to which PAX2 binds specifically as shown by EMSA and confirmed by the supershift assay (Figure 4B). Moreover, mutations of (γ), the fragment harboring the putative PAX2 binding site, resulted in a complete removal of the observed shift (Figure 4A). Surprisingly, the subfragment A displayed the highest luciferase expression activity compared to the positive control (PAX2BS) and particularly to the total fragment AB of ∼300 bp (Figure 3). This discrepancy might be caused by the presence of an inhibitory element in fragment AB, which is not present in the smaller fragment A (Figure 3).
Because we found that WNT5A is a direct target of the PAX2 gene, which itself interacts reciprocally with WT1, and because WT1 is known to be involved in approximately 15% of sporadic WTs [37], we tested whether mutations of WNT5A could be detected in WTs. The analysis of a panel of 99 WT samples revealed no disease-associated mutations, however, indicating that mutation of WNT5A itself is unlikely to be a significant cause of WT.
Interestingly, however, the expression levels of WNT5A in favorable histology WTs, assessed by quantitative RT-PCR, showed that approximately two-thirds of the cases displayed significantly less WNT5A expression than human fetal kidney control (Figure 6). Furthermore, there seemed to be a significant correlation between low WNT5A expression levels and categorization of the tumors as having predominantly blastemal histology.
The development of the metanephric kidney is at least a two-step process in which the undifferentiated metanephric mesenchyme is rescued from apoptosis by the developing ureteric bud and subsequently triggered into a mesenchymal-epithelial conversion to form the mature nephron [38]. Tissue that has been rescued from the apoptotic fate, but is to receive the second transition signal, is often termed the metanephric blastema. In general, the first transition occurs in cells proximal to the invading ureteric bud, whereas subsequent transitions are observed after primary and secondary branchings have occurred. Thus, the topography of the developing kidney is reflective of its chronological maturity, with the “earliest” developmental stages occurring near the outer edges around the buds of the invading ureters and the “mature” tissue appearing at the core.
Some WTs are characterized by a histologic pattern where proliferation of undifferentiated metanephric blastemal cells is predominant [39] suggesting that errors in the second stage of nephrogenesis may be involved in Wilms tumorigenesis. In this context, the correlation between the underexpression of WNT5A and predominantly blastemal tumor histology could represent a cause for the absence of a secondary transition (i.e., mesenchymal-epithelial transition). However, given that WNT signaling pathway genes are affected by the promoter methylation status [40], WNT5A underexpression in WT samples could be the result of its promoter hypermethylation that hampers the normal transcription of the gene.
As a signal transducer, WNT5A might be suited for a key role in the complex cellular cross-talk between the developing ureter and the developing epithelia. Interestingly, up-regulation of WNT5A has previously been shown to suppress tubule branching during lung development [41]. WNT5A has also been implicated in regulating the mesenchymal-epithelial transition in uterine tissue [42].
In addition to its role in the regulation of branching morphogenesis [43,44], PAX2 is also up-regulated during early kidney development and repressed inmature renal tissue [15,45]. Furthermore, PAX2 is constitutively expressed in a variety of renal tumors including WT [45]. The PAX2 knock down using specific siRNA and the WiT5AWT line showed a detectable level of WNT5A protein expression in samples treated with PAX2-siRNA1 and PAX2-siRNA2 (Figure 5, B and D). WNT5A expression increased slightly in both samples treated with PAX2-siRNA1 and PAX2-siRNA2 compared with nontreated samples (Figure 5, A, B, and D). Likewise, a detectable level of PAX2 protein expression was detected in samples that had undergone WNT5A-siRNA2 and WNT5A-siRNA6 transfection (Figure 5, C and E). Based on this in vitro siRNA data, it is likely that when PAX2 is highly expressed (i.e., in WT), the WNT5A is down-regulated (Figure 5A) and vice versa, although an in vivo study would be more suitable to confirm this hypothesis.
Reconciling these data with the expression analysis in Figure 6, it is tempting to speculate that PAX2 and WNT5A share an inhibitory-like cross-relationship. PAX2 tends to be overexpressed in Wilms tumors, whereas WNT5A tends to be underexpressed, and given that we now demonstrate that PAX2 plays a specific regulatory role involving a cis-region of the WNT5A gene, an inhibitory relationship is entirely plausible.
During early renal development, up-regulated PAX2 is suppressed by WT1 after the completion of the first transitional stage [45], in more mature tissue, a dichotomy that is crucial to proper nephrogenesis [46]. PAX2 inhibition of WNT5A would then be relieved, promoting the second transitional step. When PAX2 is constitutively up-regulated, as it is in certain cases of WT, this second stage would not occur as reflected in the tumor histology (i.e., predominately blastemal).
A very recently discovered tumor suppressor gene WTX, operating through β-catenin, was shown to suppress the canonical WNT signaling pathway by enhancing β-catenin degradation [47]. Likewise, WNT5A was also shown to antagonize canonical Wnt/β-catenin signaling pathway [48] and regulate cell proliferation [49]. Given that both WTX and WNT5A genes block the Wnt/β-catenin signaling pathway, it is therefore likely that both genes operate through the same chain of command line [47,48]. This is the first report indicating a direct relationship between PAX2 and WNT5A in the human embryonic kidney cells (HEK293) and indicating the importance of PAX2 in the regulation of this transcription factor and member of the WNT pathway. Further study is necessary to identify other intermediary molecules involved in these pathways and to determine whether the PAX2 and WNT5A are truly involved in WT pathogenesis.
Acknowledgments
The authors thank Elizabeth Perlman for review and classification of tumors' histology and Weizheng Guo and Ana Milanovic for their technical support.
References
- 1.Beckwith JB. National Wilms Tumor Study: an update for pathologists. Pediatr Dev Pathol. 1998;1(1):79–84. doi: 10.1007/s100249900010. [DOI] [PubMed] [Google Scholar]
- 2.Beckwith JB, Kiviat NB, Bonadio JF. Nephrogenic rests, nephroblastomatosis, and the pathogenesis of Wilms' tumor. Pediatr Pathol. 1990;10(1–2):1–36. doi: 10.3109/15513819009067094. [DOI] [PubMed] [Google Scholar]
- 3.Charles AK, Brown KW, Berry PJ. Microdissecting the genetic events in nephrogenic rests and Wilms' tumor development. Am J Pathol. 1998;153(3):991–1000. doi: 10.1016/S0002-9440(10)65641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park S, Bernard A, Bove KE, Sens DA, Hazen-Martin DJ, Garvin AJ, Haber DA. Inactivation of WT1 in nephrogenic rests, genetic precursors to Wilms' tumour. Nat Genet. 1993;5(4):363–367. doi: 10.1038/ng1293-363. [DOI] [PubMed] [Google Scholar]
- 5.Van Heyningen V, Hastie ND. Wilms' tumour: reconciling genetics and biology. Trends Genet. 1992;8(1):16–21. doi: 10.1016/0168-9525(92)90019-z. [DOI] [PubMed] [Google Scholar]
- 6.Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, Haber DA, Rose EA, Kral A, Yeger H, Lewis WH, et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms' tumor locus. Cell. 1990;60(3):509–520. doi: 10.1016/0092-8674(90)90601-a. [DOI] [PubMed] [Google Scholar]
- 7.Dahl E, Koseki H, Balling R. Pax genes and organogenesis. Bioessays. 1997;19(9):755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- 8.Haber DA, Buckler AJ, Glaser T, Call KM, Pelletier J, Sohn RL, Douglass EC, Housman DE. An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms' tumor. Cell. 1990;61(7):1257–1269. doi: 10.1016/0092-8674(90)90690-g. [DOI] [PubMed] [Google Scholar]
- 9.Huang A, Campbell CE, Bonetta L, McAndrews-Hill MS, Chilton-MacNeill S, Coppes MJ, Law DJ, Feinberg AP, Yeger H, Williams BR. Tissue, developmental, and tumor-specific expression of divergent transcripts in Wilms tumor. Science. 1990;250(4983):991–994. doi: 10.1126/science.2173145. [DOI] [PubMed] [Google Scholar]
- 10.Treisman J, Harris E, Desplan C. The paired box encodes a second DNA-binding domain in the paired homeo domain protein. Genes Dev. 1991;5(4):594–604. doi: 10.1101/gad.5.4.594. [DOI] [PubMed] [Google Scholar]
- 11.Brophy PD, Ostrom L, Lang KM, Dressler GR. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development. 2001;128(23):4747–4756. doi: 10.1242/dev.128.23.4747. [DOI] [PubMed] [Google Scholar]
- 12.Dressler GR, Wilkinson JE, Rothenpieler UW, Patterson LT, Williams-Simons L, Westphal H. Deregulation of Pax-2 expression in transgenic mice generates severe kidney abnormalities. Nature. 1993;362(6415):65–67. doi: 10.1038/362065a0. [DOI] [PubMed] [Google Scholar]
- 13.Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development. 1995;121(12):4057–4065. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- 14.Torres M, Gomez-Pardo E, Gruss P. Pax2 contributes to inner ear patterning and optic nerve trajectory. Development. 1996;122(11):3381–3391. doi: 10.1242/dev.122.11.3381. [DOI] [PubMed] [Google Scholar]
- 15.Dressler GR. Pax-2, kidney development, and oncogenesis. Med Pediatr Oncol. 1996;27(5):440–444. doi: 10.1002/(SICI)1096-911X(199611)27:5<440::AID-MPO9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 16.Dressler GR, Douglass EC. Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc Natl Acad Sci USA. 1992;89(4):1179–1183. doi: 10.1073/pnas.89.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eccles MR, Yun K, Reeve AE, Fidler AE. Comparative in situ hybridization analysis of PAX2, PAX8, and WT1 gene transcription in human fetal kidney and Wilms' tumors. Am J Pathol. 1995;146(1):40–45. [PMC free article] [PubMed] [Google Scholar]
- 18.Dziarmaga A, Hueber PA, Iglesias D, Hache N, Jeffs A, Gendron N, Mackenzie A, Eccles M, Goodyer P. Neuronal apoptosis inhibitory protein (NAIP) is expressed in developing kidney and is regulated by PAX2. Am J Physiol Renal Physiol. 2006;291(4):F913–F920. doi: 10.1152/ajprenal.00004.2006. [DOI] [PubMed] [Google Scholar]
- 19.Porteous S, Torban E, Cho NP, Cunliffe H, Chua L, McNoe L, Ward T, Souza C, Gus P, Giugliani R, et al. Primary renal hypoplasia in humans and mice with PAX2 mutations: evidence of increased apoptosis in fetal kidneys of Pax2(1Neu)+/- mutant mice. Hum Mol Genet. 2000;9(1):1–11. doi: 10.1093/hmg/9.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Sanyanusin P, McNoe LA, Sullivan MJ, Weaver RG, Eccles MR. Mutation of PAX2 in two siblings with renal-coloboma syndrome. Hum Mol Genet. 1995;4(11):2183–2184. doi: 10.1093/hmg/4.11.2183. [DOI] [PubMed] [Google Scholar]
- 21.Sanyanusin P, Schimmenti LA, McNoe LA, Ward TA, Pierpont ME, Sullivan MJ, Dobyns WB, Eccles MR. Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat Genet. 1995;9(4):358–364. doi: 10.1038/ng0495-358. [DOI] [PubMed] [Google Scholar]
- 22.Torban E, Eccles MR, Favor J, Goodyer PR. PAX2 suppresses apoptosis in renal collecting duct cells. Am J Pathol. 2000;157(3):833–842. doi: 10.1016/S0002-9440(10)64597-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dziarmaga A, Clark P, Stayner C, Julien JP, Torban E, Goodyer P, Eccles M. Ureteric bud apoptosis and renal hypoplasia in transgenic PAX2-Bax fetal mice mimics the renal-coloboma syndrome. J Am Soc Nephrol. 2003;14(11):2767–2774. doi: 10.1097/01.asn.0000094082.11026.ee. [DOI] [PubMed] [Google Scholar]
- 24.Clark P, Dziarmaga A, Eccles M, Goodyer P. Rescue of defective branching nephrogenesis in renal-coloboma syndrome by the caspase inhibitor, Z-VAD-fmk. J Am Soc Nephrol. 2004;15(2):299–305. doi: 10.1097/01.asn.0000111248.23454.19. [DOI] [PubMed] [Google Scholar]
- 25.Tamimi Y, Dietrich K, Stone K, Grundy P. Paired box genes, PAX-2 and PAX-8, are not frequently mutated in Wilms tumor. Mutat Res. 2006;601(1–2):46–50. doi: 10.1016/j.mrfmmm.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 26.Tamimi Y, Lines M, Coca-Prados M, Walter MA. Identification of target genes regulated by FOXC1 using nickel agarose-based chromatin enrichment. Invest Ophthalmol Vis Sci. 2004;45(11):3904–3913. doi: 10.1167/iovs.04-0628. [DOI] [PubMed] [Google Scholar]
- 27.Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26(1):37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 28.Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16(2):235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phelps DE, Dressler GR. Identification of novel Pax-2 binding sites by chromatin precipitation. J Biol Chem. 1996;271(14):7978–7985. doi: 10.1074/jbc.271.14.7978. [DOI] [PubMed] [Google Scholar]
- 30.Birney E, Andrews D, Bevan P, Caccamo M, Cameron G, Chen Y, Clarke L, Coates G, Cox T, Cuff J, et al. Ensembl 2004. Nucleic Acids Res. 2004;32(Database issue):D468–D470. doi: 10.1093/nar/gkh038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundy PE, Breslow NE, Li S, Perlman E, Beckwith JB, Ritchey ML, Shamberger RC, Haase GM, D'Angio GJ, Donaldson M, et al. Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable-histology Wilms tumor: a report from the National Wilms Tumor Study Group. J Clin Oncol. 2005;23(29):7312–7321. doi: 10.1200/JCO.2005.01.2799. [DOI] [PubMed] [Google Scholar]
- 32.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270(1):41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 33.Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 34.Sredni ST, de Camargo B, Lopes LF, Teixeira R, Simpson A. Immunohistochemical detection of p53 protein expression as a prognostic indicator in Wilms tumor. Med Pediatr Oncol. 2001;37(5):455–458. doi: 10.1002/mpo.1229. [DOI] [PubMed] [Google Scholar]
- 35.Szuhai K, Ijszenga M, Tanke HJ, Rosenberg C, Hogendoom PC. Molecular cytogenetic characterization of four previously established and two newly established Ewing sarcoma cell lines. Cancer Genet Cytogenet. 2006;166(2):173–179. doi: 10.1016/j.cancergencyto.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Saitoh T, Katoh M. Expression and regulation of WNT5A and WNT5B in human cancer: up-regulation of WNT5A by TNFalpha in MKN45 cells and up-regulation of WNT5B by beta-estradiol in MCF-7 cells. Int J Mol Med. 2002;10(3):345–349. [PubMed] [Google Scholar]
- 37.Park S, Schalling M, Bernard A, Maheswaran S, Shipley GC, Roberts D, Fletcher J, Shipman R, Rheinwald J, Demetri G, et al. The Wilms tumour gene WT1 is expressed in murine mesoderm-derived tissues and mutated in a human mesothelioma. Nat Genet. 1993;4(4):415–420. doi: 10.1038/ng0893-415. [DOI] [PubMed] [Google Scholar]
- 38.Barasch J, Pressler L, Connor J, Malik A. A ureteric bud cell line induces nephrogenesis in two steps by two distinct signals. Am J Physiol. 1996;271(1 Pt 2):F50–F61. doi: 10.1152/ajprenal.1996.271.1.F50. [DOI] [PubMed] [Google Scholar]
- 39.Hastie ND. The genetics of Wilms' tumor—a case of disrupted development. Annu Rev Genet. 1994;28:523–558. doi: 10.1146/annurev.ge.28.120194.002515. [DOI] [PubMed] [Google Scholar]
- 40.Martin V, Agirre X, Jimenez-Velasco A, Jose-Eneriz ES, Cordeu L, Garate L, Vilas-Zornoza A, Castillejo JA, Heiniger A, Prosper F, et al. Methylation status of Wnt signaling pathway genes affects the clinical outcome of Philadelphiapositive acute lymphoblastic leukemia. Cancer Sci. 2008;99:1865–1868. doi: 10.1111/j.1349-7006.2008.00884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li C, Hu L, Xiao J, Chen H, Li JT, Bellusci S, Delanghe S, Minoo P. Wnt5a regulates Shh and Fgf10 signaling during lung development. Dev Biol. 2005;287(1):86–97. doi: 10.1016/j.ydbio.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 42.Mericskay M, Kitajewski J, Sassoon D. Wnt5a is required for proper epithelial-mesenchymal interactions in the uterus. Development. 2004;431(9):2061–2072. doi: 10.1242/dev.01090. [DOI] [PubMed] [Google Scholar]
- 43.Grote D, Souabni A, Busslinger M, Bouchard M. Pax2/8-regulated Gata3 expression is necessary for morphogenesis and guidance of the nephric duct in the developing kidney. Development. 2006;133(1):53–61. doi: 10.1242/dev.02184. [DOI] [PubMed] [Google Scholar]
- 44.Narlis M, Grote D, Gaitan Y, Boualia SK, Bouchard M. Pax2 and pax8 regulate branching morphogenesis and nephron differentiation in the developing kidney. J Am Soc Nephrol. 2007;18(4):1121–1129. doi: 10.1681/ASN.2006070739. [DOI] [PubMed] [Google Scholar]
- 45.Ryan G, Steele-Perkins V, Morris JF, Rauscher FJ, Dressler GR. Repression of Pax-2 by WT1 during normal kidney development. Development. 1995;121(3):867–875. doi: 10.1242/dev.121.3.867. [DOI] [PubMed] [Google Scholar]
- 46.Beland M, Bouchard M. PAX gene function during kidney tumorigenesis: a comparative approach. Bull Cancer. 2006;93(9):875–882. [PubMed] [Google Scholar]
- 47.Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, Maccoss MJ. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007;316(5827):1043–1046. doi: 10.1126/science/1141515. [DOI] [PubMed] [Google Scholar]
- 48.Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman M, Shibuya H, Moon RT, Ninomiya-Tsuji J, Matsumoto K. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca(2+) pathway to antagonize Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23(1):131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126(6):1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]