Abstract
Heterochromatin is defined by distinct posttranslational modifications on histones, such as methylation of histone H3 at lysine 9 (H3K9), which allows heterochromatin protein 1 (HP1)–related chromodomain proteins to bind. Heterochromatin is frequently found near CENP-A chromatin, which is the key determinant of kinetochore assembly. We have discovered that the RNA interference (RNAi)–directed heterochromatin flanking the central kinetochore domain at fission yeast centromeres is required to promote CENP-ACnp1 and kinetochore assembly over the central domain. The H3K9methyltransferase Clr4 (Suv39); the ribonuclease Dicer, which cleaves heterochromatic double-stranded RNA to small interfering RNA (siRNA); Chp1, a component of the RNAi effector complex (RNA-induced initiation of transcriptional gene silencing; RITS); and Swi6 (HP1) are required to establish CENP-ACnp1 chromatin on naïve templates. Once assembled, CENP-ACnp1 chromatin is propagated by epigenetic means in the absence of heterochromatin. Thus, another, potentially conserved, role for centromeric RNAi-directed heterochromatin has been identified.
Metazoan centromeres are mostly composed of repetitive DNA upon which the kinetochore assembles to mediate chromosome segregation. Epigenetic factors contribute to the establishment and maintenance of kinetochores at particular sites, which are composed of CENP-A chromatin (1-4). However, the primary signals specifying the site of CENP-A chromatin, and thus kinetochore assembly, are unknown.
In Drosophila, human, and fission yeast, kinetochores are embedded in heterochromatin (5-7). Fission yeast centromeres have two distinct domains: outer repeats (otr) that flank the central kinetochore domain composed of innermost repeats (imr) and central core (cnt/cc) DNA (3, 8) (Fig. 1A).
Fig. 1.
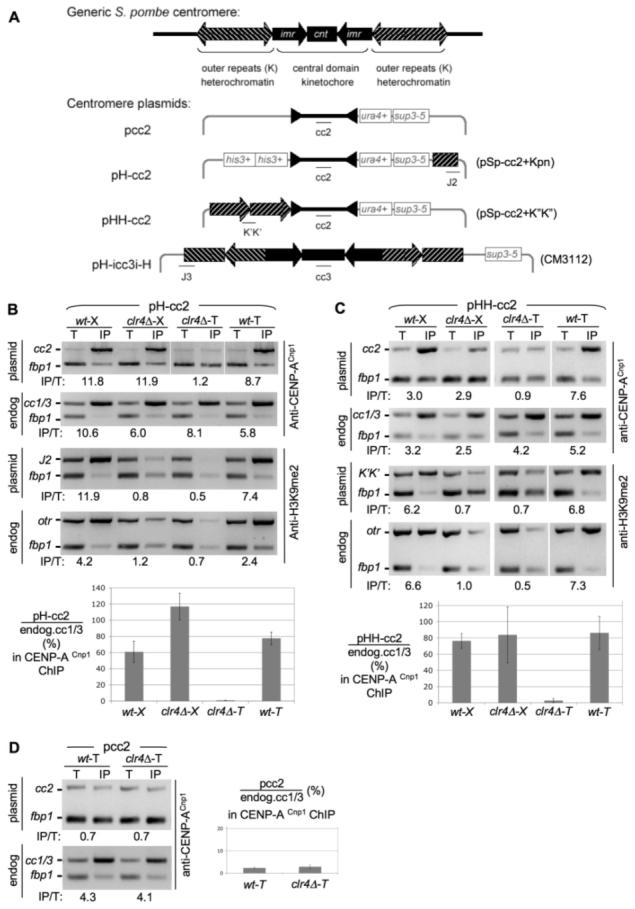
Testing roles for Clr4 in establishment of CENP-ACnp1 chromatin at centromeres. (A) Fission yeast centromere DNA and minichromosomes used (20). Association of CENP-ACnp1 and H3K9me2 with pH-cc2 (B), pHH-cc2 (C), or pcc2 (D) introduced into wild-type (wt) or clr4 cells by crossing (X) or transformation (T) is shown (see also fig. S1). Polymerase chain reaction (PCR) product positions on minichromosomes are indicated (A). fbp1 is the control noncentromeric locus. In CENP-ACnp1 ChIP, enrichment of pcc2 product was compared with input DNA (T) relative to fbp1. Enrichment of endogenous cc1/3 relative to fbp1 was also assessed. In H3K9me2 ChIP, enrichment at plasmid–outer repeats J2 (B) or K″K″ (C) products was compared with the PCR of input DNA relative to the fbp1 product. Enrichment of endogenous otr sequences was also assessed (B and C). Quantitative PCR confirms these results [lower panels (B) and (C) and right (D) and fig. S3] (20). Histograms show percent enrichment on plasmid relative to endogenous cc1/3.
Heterochromatin, containing Swi6 (heterochromatin protein 1 or HP1) bound to histone H3 dimethylated on lysine 9 (H3K9me2), forms over the otr, whereas CENP-ACnp1 replaces histone H3 in the central domain (5, 7, 9-11). Centromeric heterochromatin contributes to centromere function by recruiting cohesin and mediating cohesion between sister centromeres (12, 13). Here, we test the idea that heterochromatin marks sites for CENP-A chromatin assembly (14)
Fission yeast minichromosomes must contain at least part of an otr and a large portion of central domain (imr-cc) DNA to allow kinetochore assembly, segregation function, and minichromosome retention (3, 14-17). Small interfering RNAs (siRNAs) originating from noncoding otr transcripts direct H3K9 methylation and heterochromatin formation on these repeats (8, 18). Defects in various components cause loss or reduction in H3K9 methylation, Swi6, and centromeric cohesin (5, 9, 13). Endogenous chromosomes segregate without centromeric heterochromatin, because sister chromatids remain tethered by arm cohesin. However, mitotic stability of small circular minichromosomes is obliterated (19). Minichromosomes containing otr and cc DNA have centromere activity, resulting in a low loss rate compared with acentrics (15, 16). Here, two minichromosomes were initially used (Fig. 1A) [described in (15, 20)]: pH-cc2 and pHH-cc2 (H denotes an otr heterochromatic element, cc denotes central domain DNA). pcc2 lacks otr repeats. These plasmids carry ura4+ and sup3-5 (suppressor of ade6-704) selection systems. Formation of a functional centromere on transformation of naked DNA into cells requires both otr and cc sequences (3, 14). Thus, the acentric pcc2 plasmid is mitotically unstable and rapidly lost but is maintained in selective media, whereas pH-cc2 and pHH-cc2 are mitotically stable and segregate in wild-type cells (15). Heterochromatin is not essential for cell viability; to determine whether it is required to establish CENP-ACnp1 chromatin on cc DNA, we compared the association of CENP-ACnp1 with minichromosomes after establishment in wild type and transmission, by crossing, into a clr4 background with minichromosome DNA transformed directly into clr4 cells (fig. S1).
Wild-type cells containing pH-cc2 and pHH-cc2 minichromosomes were crossed (indicated by X) to clr4 (clr4::LEU2) cells, and wild-type (leu−) or clr4 (leu+) progeny were selected that contained the minichromosomes. In addition, pcc2, pH-cc2, and pHH-cc2 plasmids were transformed into wild-type and clr4 strains, and primary ura+ ade+ transformants (indicated by T) with intact minichromosome DNA were selected for further analyses (Fig. 1A and fig. S2) (20). Chromatin immunoprecipitation (ChIP) showed the presence of H3K9me2 on the otr of pH-cc2 and pHH-cc2 in wild-type cells derived from crosses or primary transformants (Fig. 1, B and C). However, this heterochromatic modification is lost in clr4. CENP-ACnp1 is associated with endogenous cen1/cen3 central core DNA (cc1/3) relative to the euchromatic fbp1+ locus in all strains (Fig. 1, B and C, and fig. S3). However, no CENP-ACnp1 was detected on pcc2 in either wild type or clr4 (Fig. 1D and fig. S3). Thus, CENP-ACnp1 cannot associate with cc DNA when introduced alone on a plasmid, and Clr4 does not act through cc DNA to recruit CENP-ACnp1. This is consistent with the formation of normal nucleosomal ladders on pcc2 rather than the unusual cc chromatin seen at active centromeres (16, 17).
In contrast, CENP-ACnp1 is enriched on cc of pH-cc2 or pHH-cc2 transformed into wild type (wt-T) or crossed into wild type (wt-X) or clr4 (clr4-X). However, no CENP-ACnp1 is detected on pH-cc2 or pHH-cc2 when transformed directly into clr4, even though enrichment of endogenous cc1/3 is seen (Fig. 1, B and C, and fig. S3). Thus, the presence of CENP-ACnp1 on identical minichromosomes in genetically identical clr4 cells is dependent on whether the minichromosome entered these cells in a preassembled or naked state. This suggests that Clr4-dependent centromeric heterochromatin influences establishment of CENP-ACnp1 within the central domain.
Small pH-cc2 or pHH-cc2 minichromosomes with partial centromeric DNA elements and otr heterochromatin on only one side of a central core may be particularly sensitive to the absence of heterochromatin in such CENP-ACnp1 establishment assays. Therefore, we used a 36-kb minichromosome containing two nearly complete otr repeats flanking an entire cen3 central domain [pH-icc3i-H (Fig. 2A)] (16, 20). Compared with cen1 structure, this minichromosome contains an almost complete centromere with 32.5 kb of contiguous cen3 DNA. pH-icc3i-H was crossed into wild type or clr4, and colonies retaining pH-icc3i-H were selected. In addition, total DNA from wild-type cells containing pH-icc3i-H was used to transform wild type and clr4 to select cells that directly received pH-icc3i-H. Using specific primer pairs to distinguish minichromosome from endogenous sequences, we observed the same pattern of H3K9me2 and CENP-ACnp1 association seen for the smaller minichromosomes (Figs. 1 and 2A). Thus, the association of CENP-A with pH-icc3i-H in genetically identical clr4 cells depended on its route of introduction. This indicates that the Clr4 H3K9 methyltransferase and intact otr heterochromatin are required to establish CENP-ACnp1 chromatin on centromeric cc DNA even on pH-icc3i-H which resembles a complete cen1 (16, 19).
Fig 2.
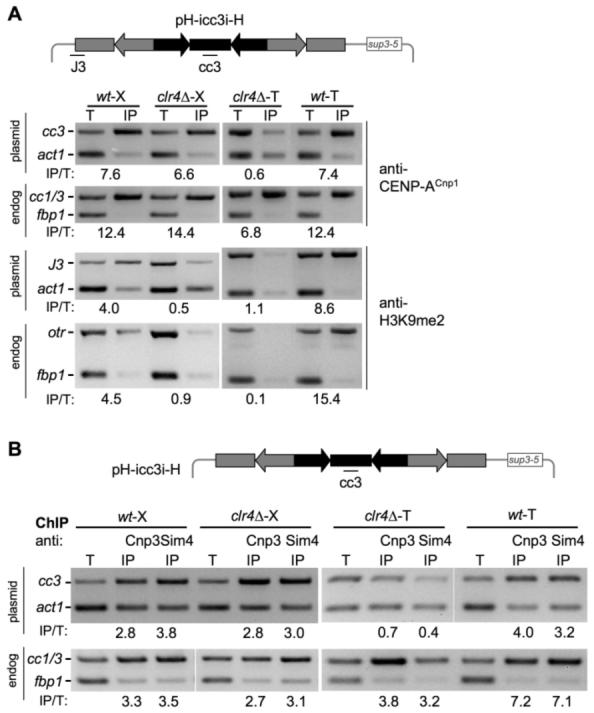
Clr4 is required to establish, but not to maintain CENP-ACnp1 and kinetochore proteins on the large cen3 minichromosome. Association of CENP-ACnp1 and H3K9me2 in (A), and CENP-CCnp3 and Sim4 in (B) were determined by ChIP in wild-type and clr4 strains containing pH-icc3i-H introduced by crossing (X) or transformation (T). fbp1 or act1 are control noncentromeric loci. In CENP-ACnp1, CENP-CCnp3, and Sim4 ChIPs enrichment of plasmid cc3 product was compared with input DNA (T) relative to the act1 product. Positive control: Enrichment of endogenous centromeric cc1/3 was assessed. H3K9me2 ChIP: Enrichment of plasmid–outer repeat J3 product was compared with PCR of input DNA (T) relative to act1. Positive control: Enrichment of endogenous otr centromeric repeats was assessed.
Use of different primers on minichromosomes indicates that CENP-ACnp1 and H3K9me2 are generally found only at cc and otr sequences, respectively, with no evidence of intermingling (fig. S4). A relative reduction in CENP-ACnp1 levels at endogenous centromeres in clr4 cells can be detected, but only in selective minimal media (fig. S5). This provides some indication that intact heterochromatin may affect CENP-ACnp1 recruitment at centromeres in their normal context. Heterochromatin may become critical for establishing CENP-ACnp1 chromatin under particular conditions, for example, on germination of spores that have remained dormant for prolonged periods. Heterochromatin-directed CENP-ACnp1 chromatin assembly may act as a backup system that is redundant with other mechanisms of CENP-ACnp1 deposition and is only revealed by establishment assays.
The CENP-ACnp1 detected on unstable minichromosomes in clr4-X cells might be a remnant of a once-functional kinetochore, but other kinetochore proteins might be lost from such minichromosomes, regardless of their derivation. CENP-C is a conserved inner kinetochore component (21). Sim4 is a component of a conserved multisubunit kinetochore complex (10, 22). CENP-CCnp3 and Sim4 are detected at endogenous cc1 and cc3 (cc1/cc3), relative to a control locus (act1) (Fig. 2B), by ChIP and were also present on pH-icc3i-H derived by crossing or transformation. CENP-CCnp3 and Sim4 were retained on cc DNA of pH-icc3i-H when crossed into clr4, but neither was recruited to pH-icc3i-H on transformation into clr4. Thus, these genetically identical cells (clr4–X, clr4–T) differ in the association of two additional kinetochore proteins with cc3, depending on whether the DNA molecule was initially propagated in wild type before transmission into cells lacking Clr4.
These analyses demonstrate that, once CENP-ACnp1 chromatin and kinetochore proteins assemble on centromeric DNA, they are propagated without flanking heterochromatin. However, heterochromatin influences the establishment of CENP-ACnp1 chromatin and, consequently, a kinetochore on naked centromeric DNA. The addition of functional Clr4 to clr4 cells containing a minichromosome derived by DNA transformation should induce heterochromatin assembly on otr sequences and, thus, CENP-ACnp1 assembly. To test this, we reintroduced the clr4+ gene into clr4 cells propagating pH-icc3i-H derived by crossing or DNA transformation (clr4+clr4–X, clr4+clr4–T; Fig. 3A). H3K9me2 appears on endogenous outer repeats and the plasmid-otr junction (J3) of pH-icc3i-H after clr4+ reintroduction. Moreover, CENP-ACnp1 is now detected on the cc of pH-icc3i-H [compare clr4–T with clr4+clr4–T (Fig. 3A)]. To determine whether Clr4 reintroduction concomitantly restored full centromere-kinetochore function, we assessed minichromosome stability. In clr4–X and clr4–T cells, pH-icc3i-H does not segregate and is rapidly lost. clr4+ reintroduction allowed the minichromosome to acquire centromere activity and to segregate normally. clr4+clr4–X and clr4+clr4–T cells are no longer sensitive to thiabendazole (TBZ), which indicates that full centromere function has been restored (Fig. 3B). Thus, centromeric epialleles that lack or retain CENP-ACnp1 can be created depending on the history of the centromeric DNA; a nonfunctional centromeric epiallele without CENP-ACnp1 can switch to the functional state, which contains CENP-ACnp1, upon provision of Clr4 methyltransferase.
Fig 3.
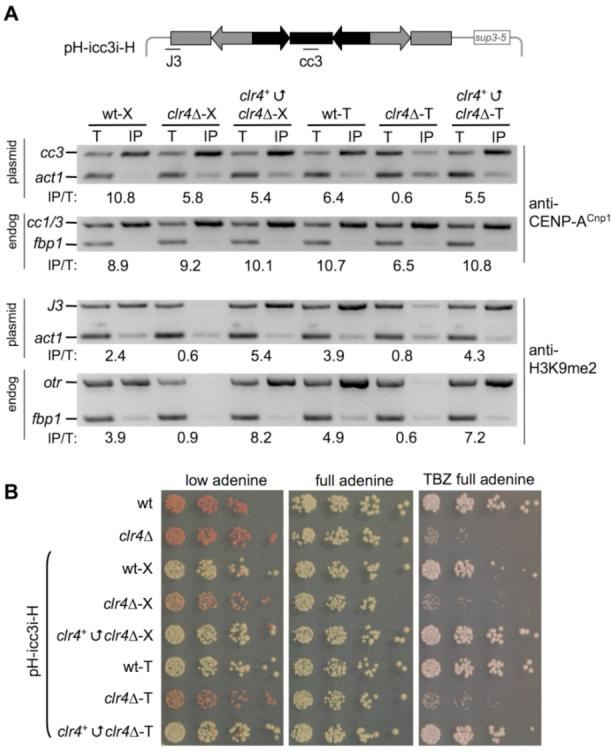
clr4+ reintroduction triggers CENP-ACnp1 recruitment to minichromosome and minichromosome stability. (A) Association of H3K9me2 and CENP-ACnp1 with pH-icc3i-H in clr4 before and after clr4+ reintroduction (clr4+clr4). Primers and conditions as in Fig. 2. (B) Wild-type (wt) and clr4+clr4 cells containing pH-icc3i-H, are indistinguishable. Serial dilution assay on 1/10th adenine assesses minichromosome stability [red colonies (lost); white colonies (retained)] or full adenine ± 10 g/μl TBZ.
Intact otr heterochromatin requires a functional RNAi pathway (8, 23-25). To further dissect the process of CENP-ACnp1 establishment, we transformed pHH-cc2 into cells lacking Swi6 or RNAi components Chp1 or Dcr1. Consistent with other analyses (26), no H3K9me2 was established on pHH-cc2 in chp1 or dcr1 cells (Fig. 4). Moreover, CENP-ACnp1 did not associate with cc2 DNA on these minichromosomes even though CENP-ACnp1 is associated with endogenous centromeres (Fig. 4 and fig. S3). Thus, an active RNAi pathway is required to establish CENP-ACnp1 chromatin on naked centromere DNA. The RNAi pathway is still active in the absence of Swi6; swi6 cells produce siRNAs and retain H3K9me2 on centromere repeats (26, 27). However, although otr chromatin of pHH-cc2 becomes H3K9-methylated when introduced into swi6, no CENP-ACnp1 is detected on its cc2 region. Thus, H3K9me2, and presumably the targeting of RITS to otr chromatin, is not sufficient to induce the assembly of CENP-ACnp1 within the central domain of minichromosomes when introduced as naked DNA. We conclude that a patch of fully intact centromeric heterochromatin is required to establish, but not to maintain, CENP-ACnp1 on the nearby central kinetochore domain of fission yeast centromeres [see our model (fig. S6)]. It is possible that centromeric cohesin, which is intimately associated with heterochromatin, may also play a role in establishing CENP-A chromatin.
Fig 4.
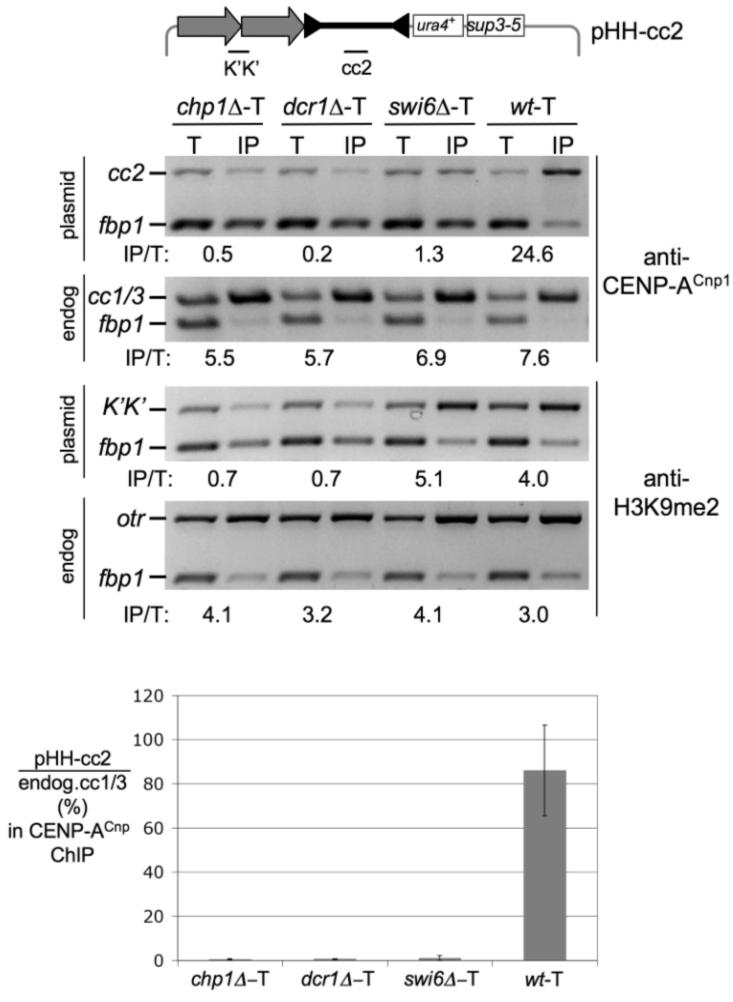
Swi6 and RNAi components are required for CENP-A establishment. CENP-ACnp1 and H3K9me2 ChIP on pHH-cc2 after direct transformation with plasmid DNA and selection in dcr1, chp1, swi6, and wild-type (wt) strains. Primers and conditions as in Fig 1C. Quantitative PCR confirms these results (lower panel and fig. S3) (20).
These analyses demonstrate that, in fission yeast, heterochromatin integrity is essential for establishment of the CENP-A chromatin that underlies kinetochores. It is noteworthy that HP1 heterochromatin is formed 800 kb from the CENP-A chromatin of a human neocentromere (28); thus, it is conceivable that heterochromatin might play a similar role at metazoan centromeres.
Supplementary Material
Supporting Online Material www.sciencemag.org/cgi/content/full/319/5859/94/DC1 Materials and Methods / SOM Text / Figs. S1 to S6 / Tables S1 and S2 / References
Acknowledgments
We thank A. Bird, A. Buscaino, E. Choi, W. Earnshaw, and M. Vogelauer for comments; M. Baum, D. Moazed, O. Niwa, J. Partridge, and M. Yanagida for materials; and the Allshire lab for discussions. The Epigenome Network of Excellence (NoE) is acknowledged for support (EC-FP6). H.D.F. and A.L.P. are funded by Wellcome Trust program grant 065061/Z. R.C.A. is a Wellcome Trust Principal Research Fellow.
References and Notes
- 1.Cleveland DW, Mao Y, Sullivan KF. Cell. 2003;112:407. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan BA, Blower MD, Karpen GH. Nat. Rev. Genet. 2001;2:584. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 3.Morris CA, Moazed D. Cell. 2007;128:647. doi: 10.1016/j.cell.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Mythreye K, Bloom KS. J. Cell Biol. 2003;160:833. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Partridge JF, Borgstrom B, Allshire RC. Genes Dev. 2000;14:783. [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan BA, Karpen GH. Nat. Struct. Mol. Biol. 2004;11:1076. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Chen ES, Yanagida M. Science. 2000;288:2215. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 8.Grewal SI, Jia S. Nat. Rev. Genet. 2007;8:35. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Science. 2001;292:110. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 10.Pidoux AL, Richardson W, Allshire RC. J. Cell Biol. 2003;161:295. doi: 10.1083/jcb.200212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saitoh S, Takahashi K, Yanagida M. Cell. 1997;90:131. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- 12.Bernard P, et al. Science. 2001;294:2539. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 13.Nonaka N, et al. Nat. Cell Biol. 2002;4:89. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 14.Pidoux AL, Allshire RC. Philos. Trans. R. Soc. London B Biol. Sci. 2005;360:569. doi: 10.1098/rstb.2004.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baum M, Ngan VK, Clarke L. Mol. Biol. Cell. 1994;5:747. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi K, et al. Mol. Biol. Cell. 1992;3:819. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marschall LG, Clarke L. J. Cell Biol. 1995;128:445. doi: 10.1083/jcb.128.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verdel A, Moazed D. FEBS Lett. 2005;579:5872. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 19.Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Genes Dev. 1995;9:218. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 20.Materials and methods are available as supporting material on Science Online.
- 21.Saitoh H, et al. Cell. 1992;70:115. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, McLeod I, Anderson S, Yates JR, 3rd, He X. EMBO J. 2005;24:2919. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volpe TA, et al. Science. 2002;297:1833. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 24.Verdel A, et al. Science. 2004;303:672. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noma K, et al. Nat. Genet. 2004;36:1174. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 26.Sadaie M, Iida T, Urano T, Nakayama J. EMBO J. 2004;23:3825. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buhler M, Verdel A, Moazed D. Cell. 2006;125:873. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 28.Saffery R, et al. Mol. Cell. 2003;12:509. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


