Abstract
The accuracy of ribosomal translation is achieved by an initial selection and a proofreading step, mediated by EF-Tu, which forms a ternary complex with aminoacyl(aa)-tRNA. To study the binding modes of different aa-tRNAs, we compared cryo-EM maps of the kirromycin-stalled ribosome bound with ternary complexes containing Phe-tRNAPhe, Trp-tRNATrp, or Leu-tRNALeuI. The three maps suggest a common binding manner of cognate aa-tRNAs in their specific binding with both the ribosome and EF-Tu. All three aa-tRNAs have the same ‘loaded spring' conformation with a kink and twist between the D-stem and anticodon stem. The three complexes are similarly integrated in an interaction network, extending from the anticodon loop through h44 and protein S12 to the EF-Tu-binding CCA end of aa-tRNA, proposed to signal cognate codon–anticodon interaction to the GTPase centre and tune the accuracy of aa-tRNA selection.
Keywords: cryo-EM, decoding, EF-Tu, ribosome
Introduction
The genetic code in messenger RNA (mRNA) uses 64 nucleotide triplets (codons), exposed in the ribosomal A site and recognized by aminoacyl-tRNAs (aa-tRNAs) or class-1 release factors (RFs). Sixty-one of these, the sense codons, base-pair with cognate aa-tRNA anticodons, whereas the three stop codons interact with release factors (Freistroffer et al, 2000). The match between the mRNA codon and the tRNA anticodon determines whether the aa-tRNA advances into the A site, eventually appending its amino acid to the nascent polypeptide chain, or dissociates from the ribosome. Although the ‘decision' to accept or reject an aa-tRNA is ultimately based on the codon–anticodon interaction, the accuracy of aa-tRNA selection is also contributed to by: (i) stereo-chemical recognition of the codon–anticodon helix (Ogle and Ramakrishnan, 2005) and (ii) proofreading of aa-tRNA initiated by GTP hydrolysis in EF-Tu (Thompson and Stone, 1977; Ruusala et al, 1982; Pape et al, 1999; Gromadski and Rodnina, 2004).
For each codon, the cognate ternary complex with its aa-tRNA in the A/T state is privileged in relation to all its near-cognate competitors by slower dissociation from the ribosome and by faster GTP hydrolysis in EF-Tu, making the probability for GTP hydrolysis close to one in the cognate and close to zero in all the near-cognate cases (Blanchard et al, 2004; Gromadski and Rodnina, 2004; Lee et al, 2007). Although considerable structural differences exist among all kinds of aa-tRNA isoacceptors (Sprinzl and Vassilenko, 2005), their binding with the ribosome might be a conserved feature. In other words, the pre-accommodated A/T state must represent binding features that are universally valid for all types of cognate aa-tRNAs, but other features that are idiosyncratic to any particular type of cognate aa-tRNA, designed to accommodate its particular amino acid, sequence and structure, are also expected.
The only previous case studies focused on pre-accommodated cognate Phe-tRNAPhe in ternary complex with EF-Tu·GDP stalled on the ribosome by the antibiotic kirromycin (Stark et al. 2002; Valle et al, 2002, 2003). These cryo-EM maps provided the first view on the conformation and binding interactions of an aa-tRNA with both EF-Tu and the ribosome. In this complex, the anticodon of the Phe-tRNAPhe (referred to as A/T-tRNA in that state) reaches the codon, whereas its acceptor arm (CCA arm) remains bound with both EF-Tu and the GTPase-associated centre (GAC). This ribosome-bound form of Phe-tRNAPhe is structurally distinct from its unbound form (Jovine et al, 2000) by a kink and twist between its D-stem and anticodon stem, which gives it an overwound conformation akin to that of a loaded spring (Valle et al, 2003).
Here, we have determined the cryo-EM structures of kirromycin-stabilized ribosomal complexes also with pre-accommodated Trp-tRNATrp and Leu-tRNALeuI (Leu-tRNACAGLeu) in ternary complexes with EF-Tu and GDP. To identify those structural similarities and differences between different cognate aa-tRNAs that allow them to efficiently surpass the initial selection step, we have compared the modes of binding of pre-accommodated Trp-tRNATrp and Leu-tRNALeuI with that of pre-accommodated Phe-tRNAPhe (Valle et al, 2003).
Our results show very similarly positioned anticodons and CCA ends of the three tRNAs, and suggest that the differences we observe for the conformations of EF-Tu, the GAC, the D- and T-loops of the aa-tRNAs in the three cases reflect small structural adjustments to ensure that the positioning of anticodons and CCA ends is universally conserved, despite differences in cognate tRNA structures and amino acids. We observe similarly distorted conformations for all three aa-tRNAs, suggesting that the springload mechanism observed earlier for Phe-tRNAPhe (Valle et al, 2003) reflects a universal aspect of the decoding process. Significantly, we are also able to identify a universal bonding network assumed to signal codon–anticodon recognition to the GTPase-associated centre, thereby shedding further light on the process by which the aa-tRNA is selected.
Results
Conformation of cognate aa-tRNA bound with EF-Tu·GDP·kirromycin and the ribosome
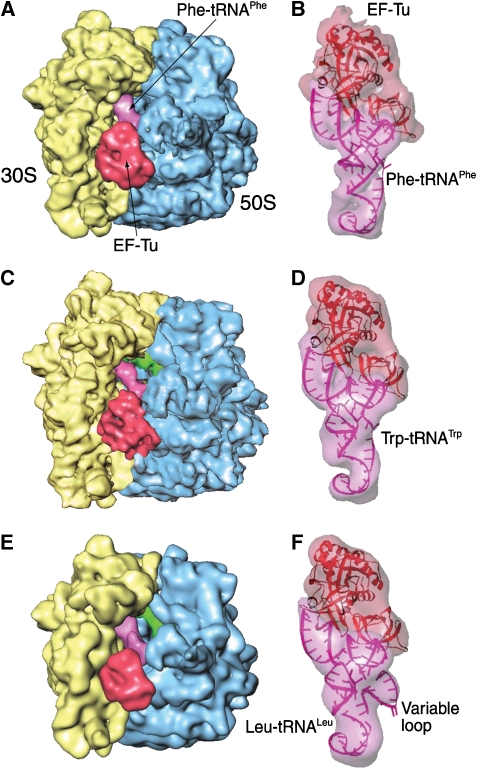
In this study, cryo-EM maps for the complexes of the 70S ribosome with Trp-tRNATrp·EF-Tu·GDP·kirromycin and Leu-tRNALeuI·EF-Tu·GDP·kirromycin have been obtained at resolutions of 9 and 12 Å, respectively. In the presence of kirromycin, the two cognate aminoacyl-tRNAs, similar to the previously observed Phe-tRNAPhe, occupy a common position, referred to as the A/T state, in their respective ribosomal complexes (Figures 1 and 2), where the anticodon reaches the cognate codon in the A site, whereas the CCA arm binds with EF-Tu and the GAC. An fMet-tRNAfMet occupies the P site in all three cryo-EM maps. An unidentified tRNA occupies the E site in both the phenylalanyl and leucyl complexes, but not in the tryptophanyl complex. It should be noted that the Leu complex is formed in the same buffer system as the one previously used for the Phe complex. This buffer is different from the one for the Trp complex (see Materials and methods), as the magnesium concentration is slightly higher (5 versus 3.5 mM). Recent cryo-EM data of pre-translocational ribosomes prepared with 3.5 mM Mg2+ also showed an empty E site (Agirrezabala et al, 2008) and suggested that the higher magnesium concentration may promote the unspecific binding of deacylated tRNAs to the E-site, as the ribosome in the same pre-translocational state, prepared with 5 mM magnesium (Valle et al, 2003), invariably showed an occupied E-site.
Figure 1.
Cryo-EM maps of the E. coli 70S ribosome bound with three aminoacyl-tRNA·EF-Tu·GDP·kir ternary complexes, respectively: yellow: 30S subunit; blue: 50S subunit; purple: A/T-tRNA; red: EF-Tu. (A) Map of the 70S ribosome, at a resolution of ∼10 Å, bearing a Phe-tRNAPhe in the A/T site and a codon UUU in the A site (Valle et al, 2003). (B) Density belonging to the ternary complex (semi-transparent), segmented from the entire ribosome complex shown in (A). Fitting models of tRNA and EF-Tu in ribbon representations. (C) Map of the 70S ribosome, at a resolution of 9 Å, bearing a Trp-tRNATrp in the A/T site and UGG codon in the A site. (D) Map of the ternary complex (semi-transparent), segmented from the ribosome complex shown in (C). Fitting models of EF-Tu (red) and tRNA (pink), both in ribbon representation. (E) The map of the 70S ribosome, at a resolution of 12 Å, bearing a Leu-tRNALeuI in the A/T site and a CUG codon in the A site. (F) Map of the ternary complex (semi-transparent), segmented from the ribosome complex shown in (E). Fitting models of EF-Tu (red) and tRNA (pink), in ribbon representations.
Figure 2.
Stereo views of the cryo-EM maps shown in Figure 1 (semi-transparent), fitted with the atomic models (cartoon respresentations). (A) Phenylalanyl complex, (B) tryptophanyl complex and (C) leucyl complex.
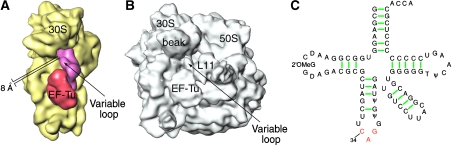
According to its sequence, Escherichia coli Leu-tRNALeuI has a long variable arm comprising 14 nucleotides. In contrast, E. coli Phe-tRNAPhe and Trp-tRNATrp have short variable arms comprising only four nucleotides. The density attributed to the variable loop of Leu-tRNALeuI (Figure 3A) appears to be oriented outwards from the D-loop towards the head of the 30S ribosomal subunit. The stem of the arm shows stronger density than its distal loop, which could be attributed to the latter's mobile positions. The variable loop shows contact neither with EF-Tu nor with the ribosome, but it has a weak connection with the beak of the 30S subunit, visible only at low-density thresholds (Figure 3).
Figure 3.
Visualization of the long variable arm in Leu-tRNALeuI. (A) Cryo-EM map of the 30S subunit and the ternary complex, computationally separated from the map shown in Figure 1E. (B) Cryo-EM map of the 70S ribosomal complex, displayed at low-density threshold to show the connection between the variable loop of the A/T-tRNA and the beak of the 30S subunit. (C) Sequence and secondary structure of E. coli Leu-tRNALeuI. The anticodon nucleotides are shown in red.
The model structure of the Phe-tRNAPhe, derived by fitting of the X-ray coordinates of tRNA into the density map (PDB code: 1QZA; Valle et al, 2003), is distinct from the free tRNAPhe form (PDB code: 1EVV; Jovine et al, 2000) by the presence of a kink and twist at the conjunction between the anticodon stem and the D stem. The conformations of both Trp-tRNATrp and the Leu-tRNALeuI were modelled by using the real-space refinement program RSRef (Chapman, 1995). To provide conformational flexibility, the Phe-tRNAPhe model was split into four rigid pieces, one for each arm of the tRNA. In addition, the long variable arm of leucyl-tRNALeuI (Figure 3), which does not exist either in the Phe-tRNAPhe or in the Trp-tRNATrp, constituted a fifth rigid piece. A fragment from the X-ray structure of the archaeon Pyrococcus horiloshii Leu-tRNALeuI was used to model this arm (PDB code: 1WZ2; Fukunaga and Yokoyama, 2005).
The fitting results show that the kink and twist are formed in both the Trp-tRNATrp and the Leu-tRNALeuI models (Figure 4) at the same position as in the Phe-tRNAPhe model. Furthermore, the anticodon loop and the CCA end in all three tRNAs exhibit common orientations. These results strongly suggest that this conformational distortion in relation to the free structure of tRNA is a universal feature required for cognate aa-tRNA selection. The common orientation of the anticodon loop appears to be of vital importance for optimal testing of codon–anticodon interactions. In addition, differences in the conformations of the three aa-tRNAs are found primarily in the T- and D-loops. The phosphate atom of nt 71 in the CCA arm is positioned about 3.5 Å from one model to another, and so is nt 56 (P). Other significant differences are observed between the D-loops of the three aa-tRNAs, which in the A/T state do not interact directly with the ribosome. The corresponding nucleotides in the three D-loops are separated by distances ranging from 3 to 8 Å. The differences in the T- and D-loops evidently compensate for the different tRNA sequences, thus ensuring that the anticodon ends and the CCA ends bind at the same positions.
Figure 4.
Models of the deformed A/T-tRNAs. (A) Stereo view of superimposed atomic models of the three aminoacyl-tRNAs in ribbon representations. Blue: tRNAPhe; grey: tRNATrp; green: tRNALeuI. (B) Superimposition of the tRNA models shown in (A) with the crystal structure of the A-site tRNA in the ribosome (red; PDB code: 1GIX, Yusupov et al, 2001). The green dashed line designates the position where the A/T-tRNA is kinked and twisted compared with the X-ray structure of the unbound tRNA.
In addition, the fitting model shows that the closest distance between the loop apex and the beak of the 30S subunit, at nt A1051 of 16S rRNA, is 8 Å. It should be noted, however, that the fitting model of the long variable arm uses a fragment of the X-ray structure as a single rigid piece, even though the loop may be quite flexible.
EF-Tu in ternary complex bound with the ribosome
Major differences between the domain rearrangements of free EF-Tu in complex with GTP and GDP have been observed (Kjeldgaard et al, 1993; Parmeggiani and Nissen, 2006). On GTP hydrolysis, domain I changes its orientation relative to domains II and III in free EF-Tu, but in the ribosome complexes that are subject to this study, the major conformational changes associated with GTP hydrolysis on EF-Tu are blocked by the antibiotic kirromycin. As a result, the conformation of EF-Tu remains similar to the GTP-bound form of the factor.
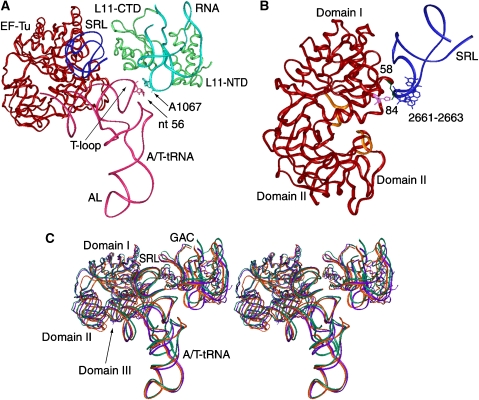
The conformation of EF-Tu in each of the three complexes was modelled from the X-ray structure of E. coli EF-Tu in complex with a GTP analogue, kirromycin, and Phe-tRNAPhe (PDB code: 1OB2; Nilsson and Nissen, 2005), using the real-space refinement program RSRef (Chapman, 1995). Each of the three domains of EF-Tu, individually treated as rigid bodies, was fitted into the cryo-EM maps, allowing the three domains to be reoriented relative to one another (Figure 5). The fitting results show that domain I is positioned similarly in the three ribosome complexes, within a distance of 1 Å, which is below the error margin. In contrast, the positions of domains II and III, which are strongly connected with the CCA arm, linked to the amino acid, vary in the three maps, with the relative positions of the backbones differing by up to 6 Å.
Figure 5.
Stereo view of the superimposed atomic models (ribbon representations) of EF-Tu matching the conformations in the cryo-EM maps, as shown in Figure 1. Blue: EF-Tu in the Phe-tRNAPhe complex; grey: EF-Tu in the Trp-tRNATrp complex; green: EF-Tu in the Leu-tRNALeu I complex.
Early footprinting experiments by Moazed et al (1988) brought evidence that EF-Tu interacts directly with the ribosome through nucleotides around position 2661 of 23S rRNA, located in the apical sarcin–ricin loop (SRL) of LH95. In our three ternary complex maps, amino acid Arg58 in domain I of EF-Tu, located in the GTP-binding region, is consistently seen to be proximal to the apical 2661–2663 loop in the SRL (Figure 6). Another amino acid, His84, is also placed close to the SRL and Arg58 in our fitting model. Mutation of Arg58 of EF-Tu decreased the rate of GTP hydrolysis in the factor, implicating Arg58 as important for its GTPase activity (Knudsen and Clark, 1995; Zeidler et al, 1996). Also, mutagenesis of His84 in EF-Tu was shown to decrease the rate of GTP hydrolysis in the factor (Cool and Parmeggiani, 1991). Accordingly, these pieces of biochemical evidence suggested important functions of both Arg58 and His84 in the ribosome-dependent activation of GTP hydrolysis, as now rationalized by the positioning of these two residues close to the SRL in our three cryo-EM maps.
Figure 6.
Illustration of the contacts involving the ternary complex, the GAC and the SRL. (A) A ribbon representation of the tryptophanyl complex (red: EF-Tu, pink: tRNATrp green: L11; light blue: RNA in the GAC; dark blue: SRL. (B) Illustration of the binding residues of EF-Tu (58 and 84) and the SRL (nts 2661–2663), shown in a different orientation from (A). (C) Stereo view of superimposed three ternary complexes together with the GAC and the SRL. Purple: phenylalanyl complex; orange: tryptophanyl complex; green: leucyl complex.
The dynamic function of the GAC in the process of aa-tRNA selection
The GAC was found to move from an ‘open' position in the factor-unbound ribosome to a ‘closed' position on ternary complex binding (Valle et al, 2003). In the closed position, nucleotide A1067 in the GAC binds with the T-loop of the Phe-tRNAPhe (Valle et al, 2003; Li et al, 2006). This study reveals that the binding between the nucleotide A1067 and the T-loop of the aa-tRNA is conserved among the three complexes.
We have modelled the conformation of the GAC using three rigid pieces from the X-ray structure of the 70S ribosome (PDB code: 2AW4; Schuwirth et al, 2005): the 58 nts of RNA (nts 1054–1101), the N- and the C-termini domains of L11. The fitting models show that nt A1067 is always positioned less than 4 Å away from nt 56 in the T-loop of the aa-tRNAs (Figure 6A), even though in these models the base of nt A1067 remains in its X-ray crystal conformation, where it stacks with base nt 1068. (The size of the pieces used for RSRef and the limited resolution of the map preclude the detection of the change in the orientation of the A1067 base that was predicted by molecular dynamics simulations (Li et al, 2006).)
Protein L11 is found to display great structural variety in the three fitting models. As the position of the N-terminal domain changes by a distance of about 8 Å, the relative orientation between the two domains of L11 varies in the three fitting models, and none of these conformations is similar to the X-ray conformation of L11 (Schuwirth et al, 2005). The loop around residue 62 in the NTD in the three models is about 10 Å away from its X-ray position when the CTD is superimposed on the X-ray structure (Li et al, 2006). The high mobility of the N-terminal domain of L11 is a part of the dynamics of the entire L11 lobe (Valle et al, 2003). The C-terminal domain of L11 exhibits little flexibility among the models, as it is tightly associated with helices 43 and 44 of 23S rRNA. Even though the N-terminal domain of L11 is connected with the mobile L10, it does not bind with the aa-tRNA, and therefore the movement does not disrupt the binding between the aa-tRNA and the GAC (Figure 6C).
The decoding centre and its vicinity
The universally conserved bases of the 16S rRNA in the 30S subunit decoding centre, including nts A1492-A1493 in h44 and G530 in h18, are directly involved in the recognition of aa-tRNAs (Yoshizawa et al, 1999). A1492 and A1493 flip outwards from h44, and G530 rotates from a syn to an anti-conformation, thereby establishing specific interactions with the minor groove formed by the first two base pairs of the codon–anticodon helix. This mechanism allows the mRNA-programmed ribosome to directly recognize the geometry of this groove (Ogle et al, 2001). Specifically, aa-tRNAs with near-cognate codon–anticodon interactions can be efficiently rejected because of the distorted shape of the codon–anticodon helix. This hypothesis invites an exploration of the function that fixed-binding geometry might have in the signalling of the cognate codon–anticodon interaction to the GTPase activation of EF-Tu.
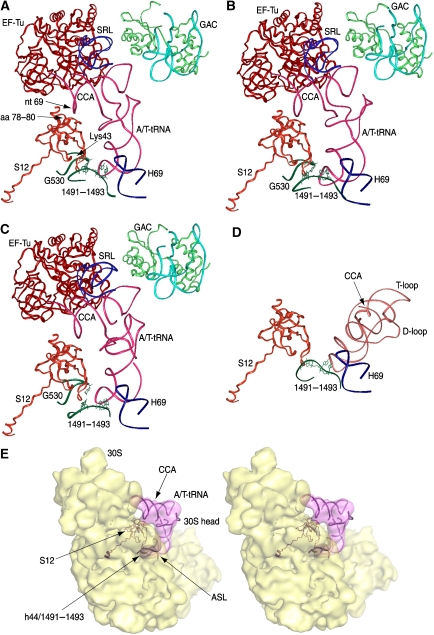
On the basis of the X-ray structure of the empty E. coli 70S ribosome (PDB code: 2AVY; Schuwirth et al, 2005), atomic models of the three ribosomal complexes were produced using RSRef. Protein S12 was treated as a rigid body. Helices H69 and h44 were also individually treated as rigid pieces in the fitting. Bases A1492–A1493 are oriented inwards in the X-ray structure because of the lack of cognate tRNA in the structure (Ogle et al, 2001). To characterize the flipped-out base positions, the fragment from nt 1491 to nt 1497 in h44 was replaced by the corresponding nts in the X-ray structure of the Thermus thermophilus 30S subunit in the presence of a cognate PhetRNAPhe (PDB code: 1IBM, Ogle et al, 2001). The fitting models show that the backbone of nts A1491–A1493 is always associated with their respective aa-tRNA, as well as with the G530 loop in the 16S rRNA (helix 18). The vital importance of bases A1492, A1493 and G530 in aa-tRNA selection was revealed by mutational analysis (Cochella et al, 2007). Mutation in these bases disrupts their interaction with the codon–anticodon helix and therefore impairs cognate aa-tRNA selection.
In line with the preservation of contacts between bases in h44 and the short codon–anticodon helix, we found that nucleotide 1491 of h44 may interact directly with Lys43 of protein S12 in each of the complexes. In addition, amino acids 78–80 of S12 are in close contact with nt 69 of the aa-tRNA, a position near EF-Tu (Figure 7). These common features allow us to define a bonding network connecting the anticodon, the two sites of S12 and the CCA arm. The contact of S12 to the CCA arm of the aa-tRNA is present only in the EF-Tu-bound state, whereas it is absent after aa-tRNA has been accommodated into the A site, as observed in the Phe-tRNAPhe complex (Valle et al, 2003), even though S12 remains in contact with h44 (Figure 7D). This remaining contact has been shown to be important for translocation (Cukras et al, 2003). The relative orientation between A-tRNA and A/T-tRNA has been extensively discussed by Valle et al (2003).
Figure 7.
Molecular network linking the decoding centre and the CCA arm of aa-tRNA, evident in the fitting models. All structural elements are in ribbon representations. (A) Structural elements located around A/T-tRNA in the fitting model of the 70S ribosomal complex bearing the tRNAPhe. Pink: tRNA; red: EF-Tu; blue: SRL; green: L11; light blue: RNA in the GAC; orange: S12; dark blue: H69; dark green: fragments in h44 and h18. (B) Structural elements corresponding to (A), but from the tRNATrp fitting model. (C) Structural elements corresponding to (A), but from the tRNALeuI fitting model. (D) Interactions of tRNA in the A-site, from X-ray structure (PDB code: 1GIX), with its surrounding elements from crystal structure (PDB 2AVY): orange: S12; green: h44 fragment; blue: H69. Protein S12 in PDBs 2AVY and 1GIX are aligned. (E) Stereo view of the cryo-EM map of the 30S subunit and the AT-tRNATrp. The fitting model, a portion of the model shown in (B), includes protein S12, Trp-tRNATrp and a portion of h44 around nts 1491–1493.
Another observation common for the three complexes is that nts 1913–1916 in H69 contact the aa-tRNA at nts 24–26, that is, at the very place where the kink and twist are formed. This finding is in line with the previous conjecture linking this contact with the destabilization of the aa-tRNA required for sterically feasible codon–anticodon pairing (Valle et al, 2003).
Discussion
Common versus distinct features of aa-tRNAs in the A/T state
Our cryo-EM maps show that cognate aa-tRNAs bind with the ribosome with a number of shared characteristics, even though they have distinct identities by different anticodons, aminoacyl groups and other structural features.
In addition to Phe-tRNAPhe characterized in an earlier study, the new cryo-EM maps now enable us to provide quasi-atomic models for Trp-tRNATrp and Leu-tRNALeuI by fitting. These models allow us to identify common as well as distinct features of the three aa-tRNAs, as they are selected by the mRNA-programmed ribosome.
Among the common features are the distortion in aa-tRNA conformation and the conserved binding of the aa-tRNAs to the GAC through nt A1067. A conformational network, linking the decoding information to the site of GTP hydrolysis, is also conserved for the three tRNA structures as described below. Another common feature is that all three aa-tRNAs interact with H69 at the position of the kink between their anticodon stem and D-stem.
Besides the common features, we also found differences among the complexes, in the conformation of EF-Tu, and in the positions of the GAC and the D- and T-loops. The differences among tRNA sequences appear to contribute to the slight variations in the conformation of the D- and T-loops. The variation in the conformation of EF-Tu may serve to accommodate the differences in the amino acids charged to the aa-tRNAs.
EF-Tu domain rearrangements
As shown in our present models, Arg58 and His84 are positioned in the vicinity of the SRL. The residues at these two homologous positions are crucial for hydrolysis of GTP in GTPases and their switch from the GTP state to the GDP state (Coleman et al, 1994). His 84 is highly conserved in EF-Tu across species, and position 58 is occupied by glutamine in other heterotrimeric GTPases (Vetter and Wittinghofer, 2001; Noble and Song, 2008).
At the atomic level, the mechanism of GTP hydrolysis associated with EF-Tu has not been well understood, mainly because of the lack of information on its intermediate structure during GTPase activation. Conformational variability of EF-Tu is evident from a comparison of several crystal structures in the presence of different types of antibiotics. Our structural data show that, owing to the presence of kirromycin, EF-Tu is in a conformation similar to the GTP-bound form of the factor even after the hydrolysis of the nucleotide. One possible interpretation is that a conformational rearrangement of the SRL repositions residues such as Arg 58 and His 84 in the nucleotide-binding pocket as a launch pad to create the transition state. However, a more conclusive definition will come with the visualization of EF-Tu bound to the ribosome in a pre-hydrolysis GTPase-activated state.
LaRiviere et al's (2001) biochemical results show that all types of aminoacyl-tRNAs bind with EF-Tu with similar affinity, in spite of greatly varying contributions to the affinity from different amino-acid side chains. This uniformity is accomplished by reciprocal affinities of the two substrates: a low affinity of the sequence-induced structure of the tRNA to EF-Tu when the amino acid has high affinity to the factor, and vice versa. This affinity calibration requires that the structure of EF-Tu is flexible enough to accommodate all types of amino-acid side chains and tRNA structures. Interestingly, our present fitting models suggest considerable variation in the ribosome-bound EF-Tu conformation with different aa-tRNAs. There are obvious differences in the orientations of domains II and III of EF-Tu that are associated with the accommodation of the different amino acids, as well as in the varying CCA arms in the respective tRNA. Despite the variations in size, shape and charge among the amino acids and in the sequence and structure of tRNA, we suggest that it is the flexibility of EF-Tu's conformation that allows the CCA end of aa-tRNA to be uniformly positioned relative to both EF-Tu and the ribosome. This adaptability of EF-Tu leads to a common starting point for the accommodation of all types of aa-tRNAs into the A-site.
The antibiotic kirromycin was used to maintain the binding between EF-Tu and SRL after GTP hydrolysis, preventing the release of EF-Tu from the ribosome, even though kirromycin does not bind directly with the SRL. As shown by a recent X-ray study (Parmeggiani and Nissen, 2006), kirromycin actually binds with EF-Tu in the pocket between domains I and III so that domain I, including Arg 58, is retained in the binding network composed of the SRL and catalytic residues after GTP hydrolysis. It is most likely that the binding causes only domain I to move towards domain III (Parmeggiani and Nissen, 2006), so that the binding of kirromycin affects neither the binding between aa-tRNA and EF-Tu nor the binding between aa-tRNA and the GAC.
Function of protein S12
Protein S12 is directly involved in virtually every protein factor-binding event during translation: with EF-G during translocation; with initiation factor 2; with release factor 3; and with the ribosomal recycling factor (Frank et al, 2007). Recent mutation analysis of protein S12 indicated that altered residues, including K42A, reduce the rates of both peptide formation and GTP hydrolysis (Sharma et al, 2007). Lys43 is a key link between h44 and S12 in all our present fitting models. In the decoding site, S12 binds with G1491 in h44. This nucleotide and its neighbours, A1492 and A1493, form an important binding network with both anticodon and S12. Mutation of G1491, similar to the experiment on A1492 and A1493, may provide further indication of how this base is involved in tRNA selection. At the other end, S12 binds with the nucleotide at the position of 69 in the aa-tRNA in all three models.
The integrity and dynamics of the tRNA body are important for the rapid and selective promotion of GTPase activation of EF-Tu, in response to a cognate codon–anticodon interaction (Piepenburg et al, 2000; Cochella et al, 2007), and depend on the deformed conformation of the aa-tRNA with the kink and twist. Our present study explains the capacity of S12 to modulate selection fidelity as described by Sharma et al (2007). Thus, a successful signal transmission from the decoding site to the site for GTP hydrolysis requires not only conformational rearrangements in the decoding center of the ribosome, but also a specific conformation of aa-tRNA. We have now provided structural evidence that the aa-tRNA in its deformed conformation binds with the ribosome at both the decoding centre and the protein S12. Thus, in summary, the signal from codon recognition to the GTPase is evidently mediated by a bonding network involving S12.
A refined model of aa-tRNA selection
It was recognized earlier from experiments (Thompson and Stone, 1977; Ruusala et al, 1982) that there are two selection steps for aminoacyl-tRNAs on the mRNA-programmed ribosome: initial selection of ternary complex and proofreading of aa-tRNA (Hopfield, 1974; Ninio, 1975), irreversibly separated by GTP hydrolysis. This double-checking mechanism ensures high accuracy of tRNA selection, with an estimated in vivo error rate of below 3 × 10−4, in conjunction with high kinetic efficiency of cognate aa-tRNA binding and passage through all the steps leading up to peptidyl transfer (Kurland et al, 1996).
Later, a detailed model of the sequential binding events during aa-tRNA selection was postulated and modified as more experimental data became available (Pape et al, 1999; Ogle et al, 2001; Rodnina and Wintermeyer, 2001; Blanchard et al, 2004; Gromadski and Rodnina, 2004; Frank et al, 2005; Ogle and Ramakrishnan, 2005; Cochella et al, 2007; Lee et al, 2007). Essentially, in initial selection, non-cognate and near-cognate ternary complexes dissociate from the ribosome more rapidly, with the GTPase activation of EF-Tu occurring more slowly, than for the cognate ternary complex. Taken together, these features ensure high probability of acceptance of the cognate, but not of a near-cognate, ternary complex. After GTP hydrolysis and release of EF-Tu, the cognate tRNA moves rapidly to A-site accommodation and peptidyl transfer, whereas a near-cognate tRNA that has ‘survived' the initial selection is most likely to dissociate from the ribosome in the proofreading step. The present study, in conjunction with other experimental data (Pape et al, 1999; Ogle et al, 2001; Rodnina and Wintermeyer, 2001; Blanchard et al, 2004; Gromadski and Rodnina, 2004; Frank et al, 2005; Ogle and Ramakrishnan, 2005; Cochella et al, 2007; Lee et al, 2007), suggests how these essential features of tRNA selection are implemented by a sequence of well-defined structural rearrangements of ribosomal complexes.
Initial selection of ternary complex. The ternary complex selection process starts with a post-initiation state, referred to as state 0 in the studies by Blanchard et al (2004) and Lee et al (2007). Here, the highly mobile L7/L12 stalk is able to recognize EF-Tu in the aa-tRNA-bound complex (Helgstrand et al, 2007). Once EF-Tu is recognized and has loosely bound with the ribosome (state 1), it causes the ternary complex of aa-tRNA·EF-Tu·GTP to interact reversibly with the decoding site for initial codon recognition (state 2; Blanchard et al, 2004). The interaction between the ternary complex and the ribosome induces conformational changes in the head of the 30S subunit, the GAC, and the aa-tRNA itself (state 3; Pape et al, 1999; Ogle et al, 2002; Frank et al, 2005; Cochella and Green, 2005). The resultant conformation tightens the binding of the aa-tRNA with the ribosome (‘induced fit') and deforms the conformation of aa-tRNA with a kink and twist around the anticodon stem. Subsequently, the GAC moves to a ‘closed' position, and EF-Tu reaches the SRL in H95. The ternary complex binds with the ribosome through the following sites: (i) the A1067 base in the GAC, including protein L11, and helices 43 and 44 (H43, H44) of the 23S rRNA, (ii) the SRL in H95, (iii) H69, (iv) nucleotides A1492–A1493 of helix 44 (h44) of the 16S rRNA, as well as G530 in h18 in the decoding centre and (v) S12. These interactions are ensured by the deformed conformation of the aa-tRNA. In case of an unsuccessful recognition, for non-cognate aa-tRNA, the pseudo-activated GTPase state (state 3′) has a short lifetime and is reversible (Lee et al, 2007). In contrast, a successful recognition transfers EF-Tu into the GTPase-activated state (state 4) that results in GTP hydrolysis (Blanchard et al, 2004). If, and only if, there is a cognate codon–anticodon interaction, the function of the bonding network that we observe is to promote (i) a tight contact between the minor groove of the anticodon–codon helix and the flipped bases of h44; and (ii) binding of nt 1491 with Lys43 of S12. There exists no bond between these atoms in the X-ray structure (PDB code: 2AVY), but the two residues are brought into a bonding distance (about 2 Å) in the fitting model. By pivot action, this binding event is relayed to the interaction between S12 and nt 69 in the CCA arm of aa-tRNA at the other end, where the arm binds with domains II and III of EF-Tu. This interaction, in turn, effects conformational changes in EF-Tu.
The effect of mutations of S12 on tRNA selection fidelity (Sharma et al, 2007) could be explained by the fact that a non-cognate aa-tRNA cannot be stabilized in the deformed conformation, as the initial codon–anticodon contact does not lead to the conformational rearrangement essential for formation of the complete network involving S12.
Proofreading of aa-tRNA. The GTPase-activated state is followed by the next step in the sequential model, the release of inorganic phosphate (Gromadski and Rodnina, 2004). EF-Tu changes its conformation from the GTP- to the GDP-bound form, breaks its contact with the SRL and the GAC and is released from the ribosome (state 5; Nilsson and Nissen, 2005). Then, the GAC moves back to the open position, freeing up the T-loop of the aa-tRNA. Subsequently, the second rejection step takes place (Pape et al, 1999). Although a cognate aa-tRNA is fully accommodated with high probability, and moves its CCA end into the A site (state 6), a near-cognate tRNA dissociates from the ribosome with high probability. Finally, the CCA end of the tRNA moves to the peptidyl-transferase centre, leading to the formation of the peptide bond (going back to its undistorted low-energy state 7).
Materials and methods
Preparation and image processing of the ternary complexes
The leucyl and tryptophanyl complexes were prepared using in vitro translation systems in polymix and HiFi buffers, respectively (Zavialov et al, 2001; Gromadski and Rodnina, 2004), as described earlier (Valle et al, 2003, Cochella and Green, 2005). For the preparation of the leucyl complex, the tRNALeuI was overexpressed and a bulk tRNA prep containing 80% tRNALeuI was used. The fMet-Leu Stop mRNA had CUG codon cognate for tRNALeuI (Leu-tRNACAGLeu). Kirromycin at a concentration of 0.25 mM was added to the respective elongation mixtures to stall the ternary complex on the ribosome. To prepare samples for cryo-EM, aliquots of the ribosomal complexes were thawed separately on ice and diluted with the respective buffer to 30 nM concentration of ribosome. Quantifoil 2/4 grids were used for cryo-EM and prepared following standard procedures (Grassucci et al, 2007).
For the leucyl ternary complexes, film images were manually acquired on an FEI Tecnai F20 at 200 kV and a nominal magnification of × 50,000. The film plates were scanned using a Zeiss Imaging scanner (Z/I Imaging Corporation) with a step size of 14 μm, resulting in a pixel size of 2.82 Å. About 80 000 particles were included in the final reconstruction.
In the case of the tryptophanyl complexes, the automated acquisition program AutoEMation (Lei and Frank, 2005) was used to collect CCD images on an FEI Tecnai Polara at 300 kV and a nominal magnification of × 59 000. The microscope is equipped with a single-port 4K × 4K CCD camera (TVIPS TemCam-F415), and the effective CCD magnification for an EM setting of 59 000 × is 100 000 × , corresponding to a pixel size of 1.5 Å. The latest version of AutoEMation is now extended to TVIPS CCD cameras supported by FEI software and has features with enhanced productivity and stability (J Lei, X Agirrezabala and J Frank, in preparation). The AutoEMation session lasted 68.5 h and was left completely unattended, including overnight periods, except for refilling of liquid nitrogen 1–2 times a day. For imaging, 1125 holes were automatically selected from 19 grid squares, and 3375 high-magnification CCD images were acquired because the system can automatically collect multiple high-magnification low-dose images in each hole. After visual inspection and evaluation of micrographs and their power spectra, 3116 CCD images were selected from which the total number of particles used was about 290 000.
The 3D reconstruction followed the standard SPIDER protocols for reference-based reconstruction (Frank, 1996; Shaikh et al, 2008). The resolution for the maps is 12 and 9.2 Å for the leucyl and tryptophanyl ternary complexes, respectively, using a cutoff of 0.5 in the Fourier Shell Correlation.
Fitting and modelling
The initial atomic model of the 70S ribosome was based on the X-ray structure of an empty E. coli 70S ribosome (PDB codes: 2AVY, 2AW4; Schuwirth et al, 2005). The P-tRNA from the X-ray structure of the T. thermophilus 70S ribosome in complex with mRNA and tRNAs (1GIX; Yusupov et al, 2001) was added to our model. The A/T-tRNA models were built as described earlier (Valle et al, 2003; see details in text). These models were docked into cryo-EM maps using the RSRef program (Chapman 1995) as described earlier (Gao et al, 2003). The maps and the models were visualized using programs IRIS Explorer 5 and InsightII (Accelrys Inc., San Diego, CA).
Accession numbers
The cryo-EM maps have been deposited in the 3D-EM database, EMBL-European Bioinformatics Institute, Cambridge: EMD-1565 (70S·fMet-tRNAfMet·Trp-tRNATrp·EF-Tu·GDP·kir); EMD-1564 (70S·fMet-tRNAfMet·Leu I-tRNALeuI·EF-Tu·GDP·kir). Atomic models have been deposited in the Protein Data Bank under the following ID codes: 3EP2 (Phe-tRNAPhe·EF-Tu·S12·SRL·GAC·h44); 3EQ3(Trp-tRNATrp·EF-Tu·S12·SRL·GAC·h44); 3EQ4 (Leu-tRNALeuI·EF-u·S12·SRL·GAC·h44).
Acknowledgments
We thank Richard Gursky for collecting the images of the leucyl complex in the electron microscope. We also thank Michael Watters for assistance with the preparation of the illustrations. This work was supported by HHMI and NIH grants R01 GM55440 and R37 GM29169 (to JF), NIH grant GM70768 and the Swedish Research Council grants (to ME and SS), Göran Gustafsson Stiftelse and LEO foundation grants (to SS) and NIH grant R01 GM059425 (to RG).
References
- Agirrezabala X, Lei J, Brunelle J, Ortiz-Meoz RF, Green R, Frank J (2008) Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell 32: 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD (2004) tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11: 1008–1014 [DOI] [PubMed] [Google Scholar]
- Chapman MS (1995) Restrained real-space macromolecular atomic refinement using a new resolution-dependent electron density function. Acta Crystallogr A51: 69–80 [Google Scholar]
- Cool RH, Parmeggiani A (1991) Substitution of histidine-84 and the GTPase mechanism of elongation factor Tu. Biochemistry 30: 362–366 [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R (2005) An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308: 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L, Brunelle JL, Green R (2007) Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol 14: 30–36 [DOI] [PubMed] [Google Scholar]
- Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR (1994) Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science 265: 1405–1412 [DOI] [PubMed] [Google Scholar]
- Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R (2003) Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol Cell 12: 321–328 [DOI] [PubMed] [Google Scholar]
- Frank J (1996) Three-dimensional Electron Microscopy of Macromolecular Assemblies. San Diego, CA: Academic Press [Google Scholar]
- Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M (2005) The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett 579: 959–962 [DOI] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ (2007) The process of mRNA–tRNA translocation. Proc Natl Acad Sci USA 104: 19671–19678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Kwiakowski M, Buckingham RH, Ehrenberg M (2000) The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci USA 97: 2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Yokoyama S (2005) Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol 10: 915–922 [DOI] [PubMed] [Google Scholar]
- Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, Chapman MS, Frank J (2003) Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113: 789–801 [DOI] [PubMed] [Google Scholar]
- Grassucci RA, Taylor DJ, Frank J (2007) Preparation of macromolecular complexes for cryo-electron microscopy. Nat Protoc 2: 3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV (2004) Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 13: 191–200 [DOI] [PubMed] [Google Scholar]
- Helgstrand M, Mandava CS, Mulder FA, Liljas A, Sanyal S, Akke M (2007) The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J Mol Biol 365: 468–479 [DOI] [PubMed] [Google Scholar]
- Hopfield JJ (1974) Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA 10: 4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovine L, Djordjevic S, Rhodes D (2000) The crystal structure of yeast phenylalanine tRNA at 2.0 A resolution: cleavage by Mg(2+) in 15-year-old crystals. J Mol Biol 301: 401–414 [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M, Nissen P, Thirup S, Nyborg J (1993) The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1: 35–50 [DOI] [PubMed] [Google Scholar]
- Knudsen CR, Clark BF (1995) Site-directed mutagenesis of Arg58 and Asp86 of elongation factor Tu from Escherichia coli: effects on the GTPase reaction and aminoacyl-tRNA binding. Protein Eng 8: 1267–1273 [DOI] [PubMed] [Google Scholar]
- Kurland CG, Hughes D, Ehrenberg M (1996) Limitations of translational accuracy. In Escherichia coli and Salmonella, In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds), pp 979–1004. Washington DC: ASM Press [Google Scholar]
- LaRiviere FJ, Wolfson A.D, Uhlenbeck OC (2001) Uniform binding of aminoacyl tRNAs to elongation factor Tu by thermodynamic compensation. Science 294: 165–168 [DOI] [PubMed] [Google Scholar]
- Lee T, Blanchard SC, Kim HD, Puglisi JD, Chu S (2007) The role of fluctuations in tRNAselection by the ribosome. Proc Natl Acad Sci USA 104: 13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Frank J (2005) Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J Struct Biol 150: 69–80 [DOI] [PubMed] [Google Scholar]
- Li W, Sengupta J, Rath BK, Frank J (2006) Functional conformations of the L11-ribosomal RNA complex revealed by correlative analysis of cryo-EM and molecular dynamics simulations. RNA 12: 1240–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Robertson JM, Noller HF (1988) Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334: 362–364 [DOI] [PubMed] [Google Scholar]
- Nilsson J, Nissen P (2005) Elongation factors on the ribosome. Curr Opin Struct Biol 3: 349–354 [DOI] [PubMed] [Google Scholar]
- Ninio J (1975) Kinetic amplification of enzyme discrimination. Biochimie 57: 587–595 [DOI] [PubMed] [Google Scholar]
- Noble CG, Song H (2008) Structural studies of elongation and release factors. Cell Mol Life Sci 65: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V (2002) Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V (2005) Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Parmeggiani A, Nissen P (2006) Elongation factor Tu-targeted antibiotics: four different structures, two mechanisms of action. FEBS Lett 580: 4576–4581 [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV (1999) Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18: 3800–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O, Pape T, Pleiss JA, Wintermeyer W, Uhlenbeck OC, Rodnina MV (2000) Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry 39: 1734–1738 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W (2001) Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem 70: 415–435 [DOI] [PubMed] [Google Scholar]
- Ruusala T, Ehrenberg M, Kurland CG (1982) Is there proofreading during polypeptide synthesis? EMBO J 1: 741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD (2005) Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias F, Boisset N, Leith A, Frank J (2008) SPIDER image-processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc, (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Cukras AR, Rogers JE, Southworth DR, Green R (2007) Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol 374: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS (2005) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 33: D139–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, Zemlin F, Wintermeyer W, van Heel M (2002) Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat Struct Mol Biol 9: 849–854 [DOI] [PubMed] [Google Scholar]
- Thompson RC, Stone PJ (1977) Proofreading of the codon-anticodon interaction on ribosomes. Proc Natl Acad Sci USA 74: 198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J (2002) Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J 21: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J (2003) Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
- Yoshizawa S, Fourmy D, Puglisi JD (1999) Recognition of the codon-anticodon helix by ribosomal RNA. Science 285: 1722–1725 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 29: 883–896 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M (2001) A posttermination ribosomal complex is the guanine exchange factor for peptide release factor RF3. Cell 107: 1–20 [DOI] [PubMed] [Google Scholar]
- Zeidler W, Schirmer NK, Egle C, Ribeiro S, Kreutzer R, Sprinzl M (1996) Limited proteolysis and amino acid replacements in the effector region of Thermus thermophilus elongation factor Tu. Eur J Biochem 239: 265–271 [DOI] [PubMed] [Google Scholar]