Abstract
Hepatitis C virus (HCV) is a positive strand RNA virus that propagates primarily in the liver. We show here that the liver-specific microRNA-122 (miR-122), a member of a class of small cellular RNAs that mediate post-transcriptional gene regulation usually by repressing the translation of mRNAs through interaction with their 3′-untranslated regions (UTRs), stimulates the translation of HCV. Sequestration of miR-122 in liver cell lines strongly reduces HCV translation, whereas addition of miR-122 stimulates HCV translation in liver cell lines as well as in the non-liver HeLa cells and in rabbit reticulocyte lysate. The stimulation is conferred by direct interaction of miR-122 with two target sites in the 5′-UTR of the HCV genome. With a replication-defective NS5B polymerase mutant genome, we show that the translation stimulation is independent of viral RNA synthesis. miR-122 stimulates HCV translation by enhancing the association of ribosomes with the viral RNA at an early initiation stage. In conclusion, the liver-specific miR-122 may contribute to HCV liver tropism at the level of translation.
Keywords: 5′-UTR, HCV, IRES, microRNA, translation
Introduction
Hepatitis C virus (HCV) is one of the major causative agents of human hepatitis (Choo et al, 1989). HCV has a single-stranded RNA genome of about 9 600 nt that contains a single large open reading frame (ORF). The viral RNA genome is of positive polarity, which means that it can serve directly as the template for translation of the viral polyprotein which is then proteolytically processed to yield the virus structural proteins and the non-structural (NS-) proteins required for replication (Appel et al, 2006).
The cis signals that control translation and replication of the viral RNA genome reside in its 5′- and 3′-untranslated regions (UTRs) flanking the polyprotein-coding region. In the 5′-UTR (see Figure 1A), the RNA stem-loop structures I and II are involved in replication. Partially overlapping, the stem-loops II–IV constitute an internal ribosome entry site (IRES). This structured RNA region can bind directly to the ribosome independently of most translation initiation factors (Pestova et al, 1998; Spahn et al, 2001) and governs the cap-independent translation of the viral RNA. Also, the 3′-UTR is involved in RNA replication (Friebe and Bartenschlager, 2002; Yi and Lemon, 2003). Moreover, sequence elements in the 3′-UTR also stimulate in cis the translation directed by the HCV IRES (Ito et al, 1998; McCaffrey et al, 2002; Bradrick et al, 2006; Song et al, 2006), a process that may serve as an inspection for the integrity of the viral RNA to allow efficient translation only of complete viral genomes that have not been attacked by cellular nucleases.
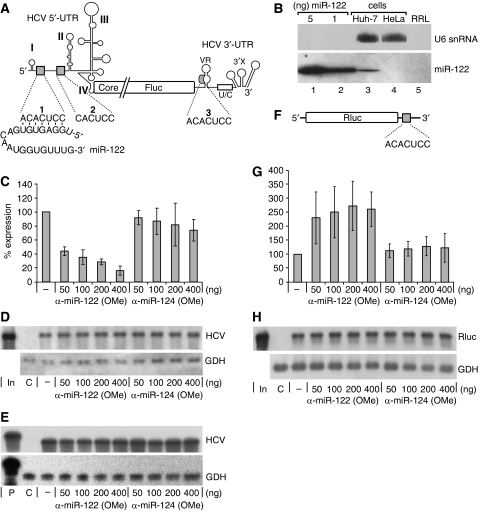
Figure 1.
Sequestration of miR-122 in Huh-7 hepatoma cells decreases HCV translation. (A) The reporter RNA with the HCV 5′-UTR, partial core, ubiquitin (not shown) and firefly luciferase (Fluc) sequences and the HCV 3′-UTR with an authentic 3′-end (Song et al, 2006). miR-122 and its target sequences (grey boxes) are indicated. The core–ubiquitin–Fluc fusion reading frame is drawn interrupted to adjust image scale. (B) Detection of miR-122 in Huh-7 cells, HeLa cells and nuclease-treated rabbit reticulocyte lysate (RRL) by northern blot. U6 snRNA was detected as a control. The indicated amounts of synthetic miR-122 were applied for comparison. (C) Fluc activity after translation of HCV reporter RNA in Huh-7 cells together with 2′-O-methylated antisense oligonucleotides. (D) RNA stability controls by northern blot with antisense RNA probes for HCV Fluc reporter RNA (Fluc 3′-region) and for glyceraldehyde-3-phosphate dehydrogenase (GDH) mRNA. (E) RNA 5′-end integrity controls by RNase protection assay for HCV reporter RNA (using an HCV 5′-UTR 5′-region antisense probe) and for GDH mRNA. In, input RNA; P, RNA probe; C, untransfected cells. (F) Capped and polyadenylated Rluc reporter RNA with the miR-122 target sequence in the 3′-UTR. (G) Translation efficiency of Rluc reporter RNA (F) in Huh-7 cells. (H) Northern blot controls for (G).
HCV replicates preferentially in the liver, suggesting that the viral life cycle depends on factors primarily expressed in liver cells. Besides the fact that cellular surface receptors may contribute to the liver tropism of HCV (Poumbourios and Drummer, 2007), HCV RNA synthesis was recently reported to be stimulated by microRNA-122 (miR-122) (Jopling et al, 2005), a miRNA present preferentially in liver cells and in the liver cell line Huh-7 (Lagos-Quintana et al, 2002; Sempere et al, 2004; Chang et al, 2005; Fu et al, 2005).
miRNAs are small RNAs that function in post-transcriptional gene regulation (Eulalio et al, 2008; Filipowicz et al, 2008). Similar to the small interfering RNAs (siRNAs) involved in RNA interference (Elbashir et al, 2001), miRNAs are processed to ∼22-bp miRNA duplexes with 3′-overhangs (Zhang et al, 2004). Bound in a protein complex containing an Argonaute (Ago) protein, the duplex is unwound starting from its thermodynamically less stable end. The strand that has its 5′-end at the less stable end of the duplex (the guide strand) is retained in the complex, whereas the opposite strand (the passenger strand) is discarded (Khvorova et al, 2003; Schwarz et al, 2003).
The function of this miRNA–protein (miRNP) complex in post-transcriptional gene regulation then depends on the extent of base pairing between the small RNA and its mRNA target (Eulalio et al, 2008; Filipowicz et al, 2008). When the miRNA (or siRNA, respectively) contained in the complex matches perfectly to its target mRNA (usually in the mRNA's 3′-UTR), an RNA-induced silencing complex forms, and Ago cleaves the target mRNA opposite to the miRNA. In contrast, when base pairing between miRNA and target mRNA is imperfect, the interaction of the miRNP complex with the target mRNA results in translation repression (Doench et al, 2003). The action of miRNAs in translation repression appears to involve various molecular mechanisms acting at different steps of the initiation stage or at the elongation stage (reviewed in Filipowicz et al, 2008; Wu and Belasco, 2008). Only very recently, also cases of translation stimulation by miRNAs were reported (Vasudevan et al, 2007; Orom et al, 2008).
In the UTRs of HCV, there are three candidate target sequences for miR-122 (see Figure 1A). One sequence is located in the variable region of the HCV 3′-UTR. Two other sequences are located between stem-loops I and II of the 5′-UTR and thus reside in a region involved in the regulation of both translation and replication. However, in contrast to the action of miRNAs on repression of cellular mRNA translation, HCV uses miR-122 in a way that stimulates the accumulation of the viral RNA genome. Jopling et al (2005) reported that sequestration of miR-122 by antisense oligonucleotides resulted in decreased accumulation of viral genomes.
Here, we show that miR-122 actually enhances the efficiency of HCV translation. These findings not only complement the current knowledge about the stimulation of HCV propagation by the liver-specific miR-122 and contribute to the understanding of cellular determinants involved in HCV liver tropism but also detail a new mode of action of an miRNA in the stimulation of RNA translation in general.
Results
The liver-specific miR-122 stimulates HCV translation
To identify a possible role of miR-122 in translation regulation and tissue tropism of HCV, we focused our analysis on the three potential target sequences for miR-122 in the non-coding regions of HCV, as target sequences in the polyprotein-coding region can be expected to be rendered non-functional by traversing ribosomes during translation. In the 5′-UTR, two miR-122 target sequences are located between stem-loops I and II (Figure 1A). Even if the second sequence displays only six nucleotides homology to miR-122 instead of seven as the first, we considered this sequence to be a candidate miR-122 target site as it matches the miR-122 seed sequence. Also, a conserved 7-nt sequence that could be an miR-122 target is located in the otherwise variable region of the 3′-UTR.
Both the HCV 5′-UTR with the IRES spanning into the core coding sequence and the 3′-UTR were combined in a reporter construct with a firefly luciferase (Fluc) reporter gene (Figure 1A). The resulting RNA contained the 5′- and 3′-terminal HCV cis signals in a monocistronic reporter system including an authentic 3′-end, which is known to be important for 3′-UTR function in translation (Song et al, 2006).
After transfection of this RNA into Huh-7 hepatoma cells that contain endogenous miR-122 (Figure 1B), we found that sequestration of the endogenous miR-122 by complementary 2′-O-methylated anti-miR-122 oligonucleotides significantly decreased HCV IRES-directed translation (Figure 1C; Supplementary Figure S1). In contrast, an 2′-O-methylated oligonucleotide complementary to the brain-specific miR-124 (Figure 1C) or to a randomized sequence (data not shown) did not significantly decrease HCV reporter RNA expression. The same result was obtained using electroporation instead of liposome-mediated transfection (Supplementary Figure S2). All expression values were corrected for variations in cell density or overall transfection efficiency by co-transfected Renilla luciferase (Rluc) mRNA. RNA stability controls were performed by northern blots (Figure 1D) to exclude variations in RNA amounts or stabilities that may account for the differences in translation efficiency. Moreover, RNase protection assays (Figure 1E) show the integrity of the 5′-end of the HCV 5′-UTR with the miR-122-binding sites. In conclusion, miR-122 stimulates HCV translation.
To substantiate that miR-122 functions in the established way when targeting the 3′-UTR of a given mRNA, that is, by repressing translation (Doench et al, 2003), we introduced the miR-122 target sequence ACACUCC into the 3′-UTR of an artificial Rluc mRNA (Figure 1F). After sequestration of the endogenous miR-122 in Huh-7 cells, we observed a significant increase in Rluc translation efficiency (Figure 1G), indicating that the repression of translation by miR-122 binding to the 3′-UTR of a given mRNA works as already known. In this case, the Rluc expression values were corrected for variations using co-transfected Fluc mRNA, and the stability of the mRNA was checked by northern blot (Figure 1H).
miR-122 impacts on the HCV 5′-UTR
The site of interaction of miR-122 with the HCV-Fluc reporter RNA was identified by mutations in the miR-122 target sequences (Figure 2A). With each of the single-site mutant reporter RNAs 5mut1 or 5mut2, the level of expression significantly dropped (Figure 2B), indicating the importance of each of the miR-122-binding sites in the 5′-UTR. Moreover, sequestration of the endogenous miR-122 from the Huh-7 cells resulted in a further moderate decrease of expression with both single-site mutants, indicating that these two miR-122 target sites in the 5′-UTR are involved in translation regulation by miR-122. Consistently, concomitant mutation of both sequences in the 5mut1,2 construct disabled the response of the HCV reporter RNA translation to miR-122 sequestration.
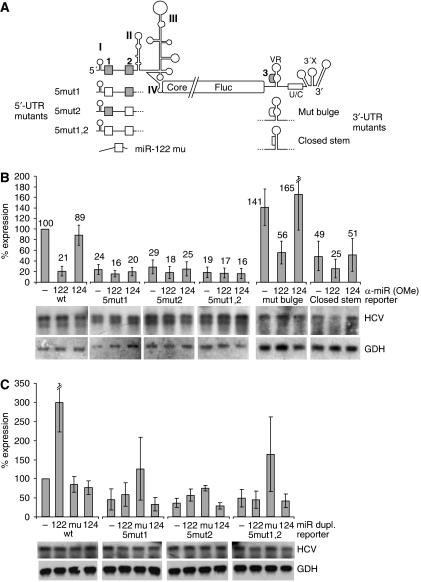
Figure 2.
The miR-122 target sites reside in the HCV 5′-UTR. (A) Substitution mutations (white boxes) of the miR-122 target sequences (grey) in the HCV 5′- and 3′-UTR (see also Supplementary Figure S3) and the miR-122 mu with the compensatory seed mutation. (B) Translation of the 5′- and 3′-UTR mutant HCV reporter RNAs in Huh-7 cells in the presence of anti-miR-122 or anti-miR-124 oligonucleotides. Lower panels, RNA stability controls by northern blot. (C) Translation of the 5′-UTR mutant HCV reporter RNAs in Huh-7 cells in the presence of ectopically added duplex miR-122, miR-122 mu or miR-124, respectively.
In the reporter RNA mut bulge, the primary sequence of the miR-122 complementary sequence in the variable region of the HCV 3′-UTR was altered but the secondary structure was maintained (see Figure 2A and Supplementary Figure S3). This RNA retains its sensitivity to miR-122 sequestration (Figure 2B), indicating that the slight increase in translation efficiency is independent of miR-122. In mutant closed stem, the miR-122 complementary sequence was mutated to change both its primary sequence and its secondary structure by allowing it to largely base pair with the sequence located opposite to it (Figure 2A; Supplementary Figure S3). Also this RNA retained its sensitivity to miR-122 sequestration, whereas the overall translation efficiency was reduced (Figure 2B). Moreover, also the translation efficiency of an RNA with a gross deletion of most of the variable region including the miR-122 complementary sequence (Supplementary Figure S4A) is decreased after miR-122 sequestration (Supplementary Figure S4B). Taken together, these results show that the miR-122 complementary sequence in the 3′-UTR is not required for HCV translation regulation by miR-122, whereas the sequence and the structure of the variable region of the 3′-UTR are important for the overall translation regulation of HCV (Song et al, 2006).
Further addition of ectopic miR-122 on top of the moderate levels of the endogenous miR-122 present in the Huh-7 cells stimulates translation of the wild-type HCV reporter RNA by about three-fold (Figure 2C). Combined with the four-fold reduction after sequestration of the endogenous miR-122, we estimate that miR-122 is able to stimulate HCV translation in liver cells by at least 12-fold.
To verify a direct interaction between the miRNA and its target sequences in the 5′-UTR, we co-transfected miR-122 mu in which the corresponding seed sequence was mutated to restore base pairing with the mutated HCV reporter RNA. This miR-122 mu stimulates translation from the 5mut1 and 5mut2 single-site mutant HCV reporter RNAs by more than two-fold (Figure 2C), whereas the authentic miR-122 also stimulates these mutants. These results show that both miR-122 target sites in the 5′-UTR appear to be involved in translation regulation. As these HCV 5′-UTR single-site mutants contain one mutated and still one wild-type miR-122 target site, it is not to be expected that they are stimulated by the wild type or by the mutant miR-122 as efficiently as the wild-type HCV RNA is stimulated by wild-type miR-122. In contrast, translation of the 5mut1,2 double mutant RNA is stimulated by the mutated miR-122 mu more than three-fold (Figure 2C). This extent of stimulation is similar to that obtained with the wild-type miR-122 and the wild-type HCV RNA, indicating that both miR-122-binding sites are required in combination for translation regulation by miR-122. Moreover, neither the endogenous nor ectopically added wild-type miR-122 is able to stimulate translation of the double mutant 5mut1,2. Considering that this double mutant still contains the miR-122 complementary sequence in the 3′-UTR, the latter result confirms that direct base pairing between miRNA and the two miRNA target sites in the 5′-UTR of the HCV reporter RNA is involved in the stimulation of HCV translation. Moreover, miR-122 also stimulates translation of the HCV reporter RNA lacking the 3′-UTR in Huh-7 cells (Supplementary Figure S4C), confirming that the miR-122 target sites in the HCV 5′-UTR confer the stimulation. In conclusion, the liver-specific miR-122 contributes to HCV liver tropism at the level of translation.
To validate the effect of miR-122 on HCV translation in the biological context, we used the full-length HCV RNA genome (Figure 3A). Translation of this full-length genome in Huh-7 hepatoma cells was stimulated by miR-122 but not by miR-124 (Figure 3B), confirming the role of miR-122 in HCV translation. Even though a significant effect of HCV RNA genome amplification on the overall translation efficiency of the HCV genomes seems unlikely at the early time points analysed (4 h after transfection), we additionally used an NS5B polymerase GND mutant full-length genome (Figure 3C) to exclude that genome amplification contributes to translation efficiency. Moreover, also sequestration of the endogenous miR-122 by antisense oligonucleotides resulted in a decrease of translation with the GND polymerase mutant HCV genome (Figure 3D). These results confirm that HCV translation stimulation by miR-122 occurs at the level of translation. The lower stimulation compared with the shorter reporter RNAs suggests that the miR may be somehow sequestered by the very long RNA or that it could be potentially interesting to search for possible regulatory elements in the HCV genome that may modulate the effect of miR-122 on the 5′-UTR.
Figure 3.
miR-122 stimulates HCV full-length genome translation. (A) The full-length HCV RNA. The polyprotein reading frame is drawn interrupted to adjust image scale. (B) Translation of the full-length HCV RNA genome in Huh-7 cells after co-transfection of ectopically added miR-122 or miR-124. (C) Translation as in (B) but of a replication-deficient full-length HCV RNA genome with an NS5B polymerase GND mutation. (D) Translation of the full-length HCV GND mutant genome in the presence of anti-miR-122. All values represent core antigen amounts in lysates 4 h after transfection. Lower panels: northern blot controls.
Ectopic miR-122 can stimulate HCV translation in non-hepatoma cells
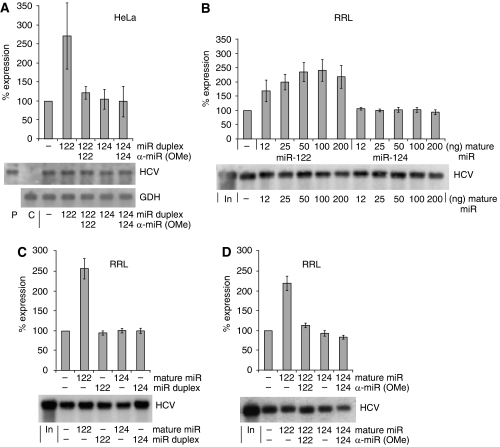
To elucidate whether miR-122 is a decisive determinant of the tissue-specific stimulation of HCV translation, we used the non-hepatoma HeLa cells that do not contain endogenous miR-122 (see Figure 1B). Co-transfection of miR-122 duplex RNA strongly stimulates translation of the HCV reporter RNA (Figure 4A), whereas miR-124 did not show this effect. Also, in the cell-free rabbit reticulocyte lysate (RRL), miR-122 significantly stimulates HCV reporter RNA translation in a dose–response manner (Figure 4B). However, in the reticulocyte lysate only single-stranded mature miR-122 but not duplex miR-122 was able to confer stimulation (Figure 4C), consistent with the observation that cellular components required for loading duplex miRNA into miRNA ribonucleoprotein complexes are missing from reticulocyte lysate (Wang et al, 2006). As a control, sequestration of the mature miR-122 with 2′-O-methylated antisense oligonucleotides blocked the stimulation (Figure 4D). These effects were not observed with miR-124 (Figure 4B–D). Furthermore, miR-122 also stimulates HCV delta 3′-UTR RNA translation in RRL (Supplementary Figure S4D), confirming the importance of the 5′-UTR for miR-122 stimulation.
Figure 4.
miR-122 stimulates HCV translation in HeLa cells and reticulocyte lysate. (A) Translation of the wild-type HCV reporter RNA (see Figure 1A) in HeLa cells in the presence of miR-122 duplex and sequestration control with anti-miR-122. Duplex miR-124 and anti-miR-124 were used as controls. (B) Influence of increasing amounts of mature single-stranded miR-122 on translation of the HCV Fluc reporter RNA (see Figure 1A) in rabbit reticulocyte lysate (RRL). (C) HCV reporter RNA translation efficiency in RRL in the presence of mature single-stranded or duplex miR-122 and miR-124. (D) Sequestration of added single-stranded miR-122 by antisense anti-miR-122 in RRL. Lower panels, northern blot controls.
miR-122 stimulates HCV translation at the stage of formation of ribosomal 48S initiation complexes
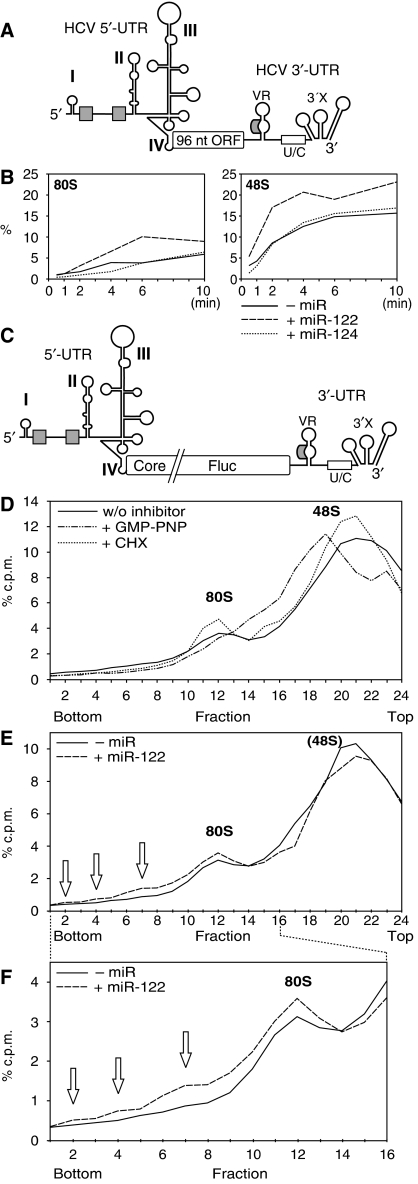
To identify the stage of translation initiation at which miR-122 confers its stimulatory influence on HCV translation, we analysed the formation of ribosomal 48S initiation complexes (including the small ribosomal subunit) and of completely assembled 80S ribosomes associated with the RNA. Therefore, we used a short HCV RNA that contains the regulatory translation signals flanking a short ORF of 96 nt (Figure 5A). miR-122 caused significantly increased association of the small ribosomal subunit with this short HCV RNA to yield ribosomal 48S complexes (Figure 5B), whereas miR-124 had no such effect (Figure 5C). In comparison, miR-122 had no effect on an artificial capped and polyadenylated Fluc mRNA (Supplementary Figure S5).
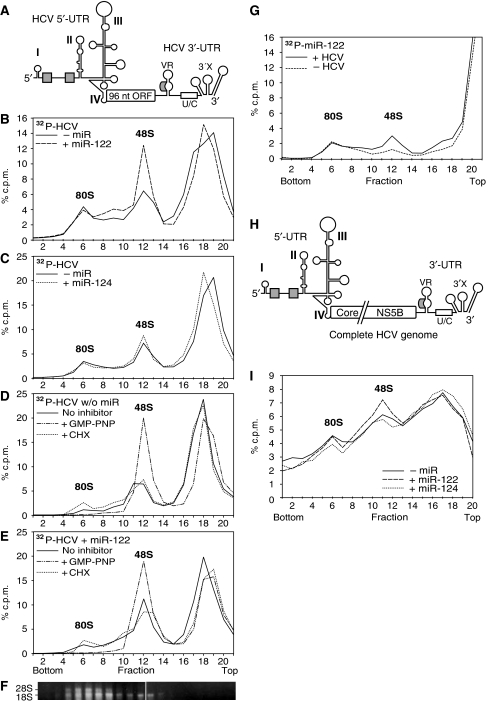
Figure 5.
miR-122 stimulates HCV translation at the stage of ribosomal 48S initiation complex formation. (A) The short RNA used for initiation complex formation contains the HCV IRES fused to an artificial 96 nt ORF that includes partial core coding sequences, followed by the HCV 3′-UTR with a precise 3′-end. (B) Sucrose gradient analysis of initiation complexes formed after 10 min incubation in RRL with the radiolabelled short HCV RNA shown in (A) in the presence or absence of miR-122. Reactions were performed in the presence of the elongation inhibitor anisomycin to ‘freeze' reactions after assembly of complete 80S ribosomes. The gradients were run for 5.5 h to resolve 48S and 80S initiation complexes. (C) Initiation complexes formed in the presence or absence of miR-124. (D, E) Initiation complexes formed with the radiolabelled short HCV RNA in the absence (D) or presence (E) of miR-122 and either in the absence of translation inhibitors, or in the presence of the GTP analogue GMP-PNP or the elongation inhibitor cycloheximide (CHX). (F) Ribosomal RNAs were re-extracted from a gradient run in parallel to localize ribosomal subunits. (G) Incorporation of radiolabelled miR-122 into ribosomal initiation complexes in the presence or absence of non-labelled short HCV RNA. (H) The full-length HCV RNA genome (NS5B polymerase GND mutant). The polyprotein reading frame is drawn interrupted to adjust image scale. (I) Initiation complexes formed after 10 min incubation in RRL with the full-length HCV RNA genome in the presence of miR-122 or miR-124.
To identify the ribosomal 48S initiation complexes, we used the non-hydrolysable GTP analogue GMP-PNP (Anthony and Merrick, 1992) in the absence (Figure 5D) and in the presence (Figure 5E) of miR-122. The peak raised by GMP-PNP (Figure 5D and E) perfectly co-migrates with the peak raised by miR-122 (Figure 5B and E). In turn, a peak of ribosomal 48S initiation complexes already boosted by GMP-PNP to maximal efficiency cannot be further accentuated by miR-122 (compare Figure 5D with 5E), indicating that the ribosomal initiation complexes affected by miR-122 and GMP-PNP are the same. In contrast, the elongation inhibitor cycloheximide enhances the accumulation of 80S ribosomes (Figure 5D and E). Furthermore, ribosomal RNAs were re-extracted from the gradient fractions to identify the ribosomal subunits by their content of 18S and 28S rRNAs (Figure 5F).
To demonstrate the co-migration of HCV RNA and miR-122 in the gradients, we used radiolabelled miR-122 and unlabelled HCV reporter RNA. With the HCV RNA, increased amounts of radiolabelled miR-122 were incorporated into the 48S initiation complexes (Figure 5G), an effect not observed with miR-124 (Supplementary Figure S6).
These results were confirmed with the full-length HCV genome (Figure 5H). In the presence of miR-122 more 48S initiation complexes are formed (Figure 5I), even though the effect was less prominent than with the short HCV RNA, most likely due to the unspecific binding of more RNA-binding proteins to the longer RNA (compare Supplementary Figure S7).
miR-122 accelerates the association of the small ribosomal subunit early in the initiation phase
In a kinetic analysis (Figure 6B; Supplementary Figure S8), we found that miR-122 (but not miR-124) increases the formation of 48S complexes with the short HCV RNA (see Figure 6A) at early time points, that is, already after 0.5 min. This enhancement in 48S complex formation by miR-122 is maximal after 1 and 2 min. Subsequently (after 4–6 min), also the formation of complete 80S ribosomes is increased in the presence of miR-122. After 10 min, the 80S complexes were not further increased but slightly decreased, whereas the 48S complexes increased again, leading to the speculation that the maximal number of complete ribosomes had left the initiation site between 6 and 10 min and made it free for another cycle of ribosome loading.
Figure 6.
miR-122 accelerates an early step of the association of the small ribosomal subunit with the HCV RNA. (A) The short HCV RNA used in (B). (B) Relative amounts of the short HCV RNA present in 48S and 80S complexes at the given times in the presence of miR-122 or miR-124. The data were obtained from the gradients shown in Supplementary Figure S8 by summing up fractions 5–7 for 80S and 11–13 for 48S complexes. (C) The HCV Fluc 3′-UTR RNA used in (D–F). (D) Initiation complexes formed after 5 min incubation with the radiolabelled HCV Fluc 3′-UTR RNA in the presence of GMP-PNP or cycloheximide (CHX), respectively. The gradients in (D, E) were run only for 2.5 h to also resolve polysomes. (E) Initiation complexes formed with the HCV Fluc 3′-UTR RNA in the presence or absence of miR-122. Polysomes are marked by open arrows. Please note that the 48S peak is not resolved from the large peak of free RNA due to the shorter gradient run time. The profile shown represents the means of two experiments; two other experiments showed the same result. (F) Blow-up of fractions 1–16 of the gradient shown in (E).
The formation of polysomes was analysed with the longer HCV-Fluc-3′-UTR RNA (Figure 6C). The gradients were run shorter to resolve polysomes also, and an increase in monomeric 80S ribosomes and in polysomes formed with the HCV Fluc RNA in the reticulocyte lysate was observed after the addition of miR-122 (Figure 6E and F). The translation initiation complexes were again identified using GMP-PNP and cycloheximide (Figure 6D), whereas the 48S complexes could not be resolved from the large peak of ribosome-free RNA in all cases (Figure 6E) due to the lower resolution at the shorter gradient run time.
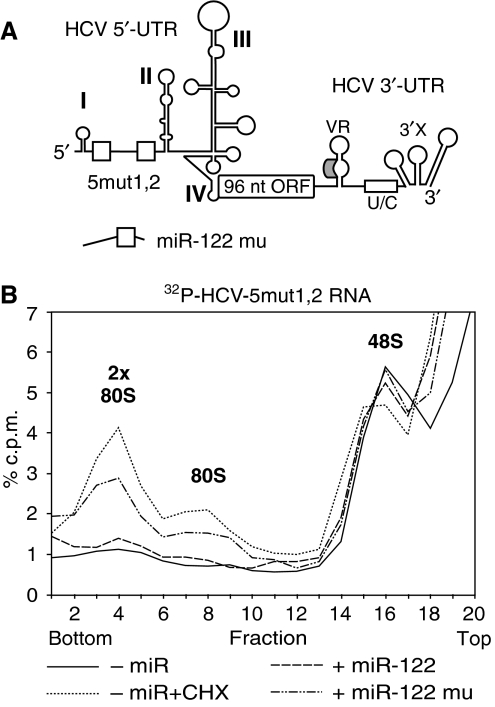
The mutant miR-122 increases the association of ribosomes with the mutant HCV RNA in living cells
To analyse the effect of miR-122 on the formation of ribosomal initiation complexes with the HCV RNA in living cells, and to confirm the direct interaction of miR-122 with its two 5′-UTR-binding sites, we transfected the radiolabelled mutant short HCV 5mut1,2 RNA into HeLa cells, along with either the wild-type or the mutant miR-122 (Figure 7A). The formation of ribosomal initiation complexes was analysed at 2 h after transfection. The results show that the amount of the mutant HCV RNA associated with one or two complete 80S ribosomes is increased after 2 h with the mutant miR-122 mu but not with the wild-type miR-122 (Figure 7B). The ribosomes on the RNA were identified using cycloheximide added 90 min after transfection. These results show that miR-122 increases the association of ribosomes with the HCV RNA in living cells and confirms the direct interaction of the mutant miR-122 with the mutant HCV RNA.
Figure 7.
Mutant miR-122 stimulates the formation of initiation complexes with the mutant HCV RNA in HeLa cells. (A) The short HCV RNA 5mut1,2 with the mutated miR-122 binding sites (open boxes) in the 5′-UTR. (B) Sucrose gradient analysis of translation initiation complexes harvested from HeLa cells 2 h after transfection of the mutant short HCV 5mut1,2 RNA in the absence of microRNA, or in the presence of miR-122 or the mutated miR-122 mu, respectively. As a control, cells transfected with the HCV RNA in the absence of microRNA were treated with 1 mM cycloheximide 30 min before harvesting (dotted line).
Discussion
We show here that miR-122 stimulates HCV RNA translation at an early stage of association of the small ribosomal subunit with the viral RNA. Most experiments were performed with HCV reporter RNAs that do not allow HCV RNA replication in the transfected cells, and also a full-length HCV genome deficient in NS5B polymerase activity shows the same response to miR-122 stimulation.
The experiments with the mutations in the 5′- and 3′-UTR show that those sequences in the 5′-UTR are functional in translation stimulation, whereas that in the 3′-UTR does not significantly confer stimulation by miR-122. The incidental observation that the mutations in the variable region of the 3′-UTR affect translation efficiency confirms our previous finding that the variable region of the 3′-UTR is important for the overall translation regulation of HCV (Song et al, 2006). The expression with the mutant HCV reporter RNAs and the mutated miR-122 showed that compensatory mutations in the miRNA can complement mutations in the miRNA target sites. This demonstrates a direct interaction of the miRNA with its HCV target RNA in translation stimulation. However, even the expression with the double mutant cannot reach the levels obtained with the wild-type HCV reporter in the presence of added wild-type miR-122, as the transfected Huh-7 cells already contain endogenous wild-type miR-122 that stimulates translation with the wild-type but not with the mutant reporter in excess of the ectopically added miR.
In a more mechanistic approach using gradient analysis of the translation initiation complexes formed, we show that miR-122 stimulates the association of the small ribosomal subunit with the HCV RNA. Both HCV RNA and miRNA are contained in the 48S translation initiation complexes identified by the stage-specific translation inhibitor GMP-PNP. The kinetic analysis shows that the stimulation takes place early in the initiation phase by accelerating the association of the small ribosomal subunit with the HCV RNA. At later stages, the formation of polysomes is found enhanced by miR-122 in cell-free extract as well as in living HeLa cells. All these experiments show clearly that miR-122 actually stimulates the translation of HCV RNA.
This finding may complement the result of an earlier study that showed that miR-122 causes the accumulation of HCV replicon RNA (Jopling et al, 2005), and another study followed that experimental system to show that ectopic miR-122 increases the steady-state level of HCV RNA also in a non-hepatic cell line (Chang et al, 2008). Although the effect of miR-122 on overall HCV replication was clearly shown by Jopling and coworkers, we suspected that an effect of miR-122 on HCV translation may have escaped detection in that study, as we know now that monocistronic reporter RNA systems are more appropriate to investigate translation than dicistronic RNAs that contain an additional picornavirus IRES (Song et al, 2006; Jünemann et al, 2007). As our present analysis was focused on the effect of miR-122 on HCV translation, we can only speculate whether or not later steps of the HCV replication cycle, for example, the synthesis of HCV progeny RNA genomes catalysed by the NS5B polymerase, are also modulated by miR-122.
miR-122 may considerably contribute to the liver tropism of HCV on the level of translation regulation. The uptake of HCV into liver cells is obviously related to the metabolism of lipoprotein particles (reviewed in Andre et al, 2005). Cellular surface receptors, such as CD81, scavenger receptor class B type I, claudin-1 and the low-density lipoprotein receptor, are involved in HCV entry into liver cells, whereas DC-SIGN and L-SIGN may help to internalize HCV into endothelial cells on its way to the liver (reviewed in Poumbourios and Drummer, 2007). However, until now miR-122 was found to be expressed almost exclusively in liver cells and in the hepatoma cell line Huh-7 (Lagos-Quintana et al, 2002; Sempere et al, 2004; Chang et al, 2005; Fu et al, 2005). In this scenario, miR-122 can be speculated to provide the decisive increase in translation efficiency that results in more efficient replication of HCV in liver cells compared with other cells and by that finally makes HCV a hepatotropic virus.
The fact that those miR-122 target sequences in the HCV RNA that are functional in translation stimulation reside in the 5′-UTR is surprising only at first glance. Even if it could not be excluded that the coding sequence could also contain miRNA target sequences that are functional in translation regulation, they would most likely be rendered non-functional by traversing ribosomes during translation. In contrast, miRNA target sites that regulate translation of a given capped and polyadenylated ‘standard' mRNA could be expected to be located in the 3′-UTR to avoid interference of the bound miRNP complex with small ribosomal subunits scanning from the mRNA's 5′-end for an initiator AUG. Probably, this is the reason why until now nearly all miRNA-binding sites that function in translation repression have been found in the mRNA's 3′-UTR. However, an miRNA-binding site can be artificially relocated to the 5′-UTR without loss of function when the miRNP complex does not interfere with ribosomal scanning by the use of an IRES downstream of the miRNA-binding site (Lytle et al, 2007). With respect to the location of the miRNA target site, a similar situation is found in HCV: both miR-122-binding sites that function in translation regulation are located upstream of the IRES.
Our results may also have consequences for the general understanding of miRNA function in translation regulation. So far, in the vast majority of cases the action of miRNAs on the 3′-UTRs of cellular mRNAs had been reported to result in the repression of translation or the destabilization of the mRNA (Pillai et al, 2007). Only very recently, a report showed that an miRNA can stimulate translation by acting on the 3′-UTR (Vasudevan et al, 2007), and translation of mRNAs coding for some ribosomal proteins was found to be slightly stimulated by miR-10a binding to the 5′-UTR (Orom et al, 2008).
Why is HCV translation stimulated by the miRNA instead of being repressed? From the mechanistic point of view, we could just ask the reverse question: why not? From transcriptional regulation, we know enhancers and silencers of transcription; the difference in mechanics is just brought along by another kind of influence on the respective target protein complex, the activity of which is to be increased or decreased. And from the functional point of view, it is an advantage for a virus to take the stimulating turn of conformational changes, whereas the activity of cellular mRNAs stored for later use may usually be rather suppressed than stimulated by miRNAs.
Materials and methods
Plasmids
The HCV reporter plasmid pHCV-Fluc-3′-UTR (named pHCV+ 3′-UTR in Song et al, 2006) contains in a pBlueScript SK(+) vector the following elements: the CMV-IE promoter, a T7 promoter fused exactly to the HCV 5′-UTR (nucleotides (nt) 1–341) plus 262 nt core sequence, a ubiquitin sequence, the firefly luciferase (Fluc) gene and the entire HCV 3′-UTR. The corresponding RNA is shown in Figure 1A. In the HCV 5′-UTR, three mutations (Figure 2A) were introduced. In HCV-Fluc-3′-UTR 5mut1, the first miR-122 target site in the 5′-UTR (ACACTCC, nt 22–28) was mutated to CGCGGAA. In mutant 5mut2, the second miR-122 target site (CACTCC, nt 38–43) was mutated to GCGGAA. In the double mutant 5mut1,2, both miR-122 target sites in the 5′-UTR were mutated. In the HCV 3′-UTR, two mutations were generated (Figure 2A; Supplementary Figure S3). In mutant mut bulge, the sequence CTCC in the bulge of the predicted stem-loop in the variable region was mutated to ACGA, leaving the predicted bulge open but mutating its sequence. In mutant closed stem, ACACTCC was mutated to GGAAAA, largely closing the bulge by complementary base pairing to the opposite region of the predicted stem-loop.
The HCV full-length clone pFL-12868 contains a T7 promoter and the complete HCV genome followed by the hepatitis δ virus (HDV) antigenomic ribozyme sequence to allow autocatalytic processing of the authentic HCV 3′-end after transcription. In the GND mutant, the NS5B polymerase active centre residues GDD were mutated to GND (Song et al, 2006).
The construct for transcription of the short HCV RNA used for initiation complex analysis (see Figure 5A) was derived from pHCV-Fluc-3′-UTR by deleting sequences from Aat II in the core sequence to Eco NI in the Fluc 3′-region, leaving a small 96 nt ORF (including the first 61 nt of HCV core and the last 35 nt of Fluc) between the HCV 5′- and 3′-UTR.
Plasmid pM12-Fluc used for transcription of the Fluc 3′-region for northern blot detection of Fluc reporter RNAs was derived from pM12 (Ochs et al, 1999) and contains the FLuc gene followed by 29 nt of the HCV 3′-UTR variable region plus 14 linker nucleotides and a reverse T7 promoter. The plasmid for transcription of the NS5B antisense RNA probe used for the detection of full-length HCV RNAs in northern blots, pNS5B-rev (Song et al, 2006), contains downstream of a T7 promoter 15 linker nucleotides, followed by 29 nt of the variable region and 298 nt of NS5B sequence in antisense orientation. phRL-null (Promega) contains the Renilla luciferase (Rluc) gene downstream of a T7 promoter. In plasmid pRluc-122, the reverse complementary miR-122 target sequence was introduced downstream of the Rluc gene in phRL-null.
Oligonucleotides
Oligonucleotides and 2′-O-methylated RNA oligonucleotides were supplied by CureVac, Thermo Electron, Biomers and Purimex. The sequences were: anti-miR-122, 5′-AGACACAAACACCAUUGUCACACUCCACAGC-3′; anti-miR-124, 5′-UGGCAUUCACCGCGUGCCUUAA-3′; miR-122-mat, 5′-UGGAGUGUGACAAUGGUGUUUG-3′; miR-122*, 5′-AACGCCAUUAUCACACUAAAUA-3′; miR-124-mat, 5′-UUAAGGCACGCGGUGAAUGCCA-3′; miR-124*, 5′-GUGUUCACAGCGGACCUUGAUU-3′; miR-122mu, 5′-UUUCCGCGGACAAUGGUGUUUG-3′; miR-122mu*, 5′-AACGCCAUUAUCCGCGGCCAUA-3′. Duplexes were formed between the guide (mat) and its complementary passenger strand (*).
RNA synthesis
HCV-Fluc reporter plasmids were linearized downstream of the reporter cassette and used to amplify the PCR fragments of interest (Song et al, 2006). RNA delta 3′-UTR contains 15 random nucleotides added to the 3′-end of the Fluc gene. The amplified DNA fragments were gel purified, extracted with phenol/chloroform, ethanol precipitated, dissolved in water and used for transcription using T7 RNA polymerase.
Plasmid pFL-12868 or the GND mutant plasmid were linearized downstream of the HDV ribozyme sequence. The linearized DNA was phenol–chloroform extracted and purified on a G25 column. HCV full-length RNA was in vitro transcribed using T7 RNA polymerase, and authentic 3′-ends were generated by autocatalytic cleavage by the HDV ribozyme.
Plasmid pM12-Fluc-anti was linearized with EcoRV and transcribed with T7 RNA polymerase. The resulting antisense Fluc RNA contains 14 linker nucleotides, 29 nt HCV variable region plus 314 nt of antisense Fluc sequence starting from the Fluc 3′-end. Plasmid pNS5B-rev was linearized with KpnI in the NS5B region. The resulting RNA contains 15 linker nucleotides, 29 nt of the HCV 3′-UTR variable region and 298 nt of NS5B sequence in reverse orientation.
The radioactive RNAs for RNase protection assay analysis of the HCV IRES (Figure 1E) were transcribed from a PCR fragment generated from plasmid pHCV-Fluc-3′-UTR using a DNA oligonucleotide pairing at the very 5′-region of the HCV IRES and a reverse PCR primer in the Fluc sequence introducing a T7 promoter in reverse orientation.
The probe for northern blot analysis of the Rluc RNA (Figure 1H) was transcribed from the Rluc gene inserted downstream of a T3 promoter. To yield digoxygenin-labelled glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) antisense RNA (387 nt), the pTRI-GAPDH-human antisense control template (Ambion) was transcribed with T7 RNA polymerase.
Capped and polyadenylated Rluc RNA used for co-transfections was in vitro transcribed using T7 RNA polymerase from plasmid phRL-null, linearized with BamHI downstream of the Rluc gene. Capped and polyadenylated Fluc RNA used for co-transfections was in vitro transcribed from plasmid pD5 that contains the Fluc gene downstream of a SP6 RNA polymerase promoter. Capped and polyadenylated Rluc-122 RNA used as a control RNA to check miRNA function in the cells (Figure 1F–H) was transcribed with T7 RNA polymerase from plasmid pRluc-122. RNA transcribed from pM12 (Ochs et al, 1999) by SP6 RNA polymerase was used as a co-transfected control RNA to standardize Rluc readings (Figure 1G).
Labelled RNAs were synthesized using T7 RNA polymerase with 1.25 μM [α-32P]UTP (800 Ci/mmol; GE Healthcare or Perkin Elmer) plus 10 μM non-radioactive labelling nucleotide and checked on 8 M urea/6% polyacrylamide gels. Non-radioactive RNAs were transcribed with 500 μM NTPs. Capped RNAs were synthesized with each 500 μM ATP, CTP and UTP, 50 μM GTP and 500 μM m7GTP (Drummond et al, 1985). Capped RNAs were polyadenylated using poly(A)-polymerase (NEB) to a poly(A)-tail length of about 200 nt (Martin and Keller, 1998). After transcription, reactions were treated with RNase-free DNase I, and RNA was purified with RNeasy kits (Qiagen). From RNAs used in sucrose gradient analysis, remaining unincorporated nucleotides were removed on G-25 columns. RNA integrity was checked by denaturing gel electrophoresis. Concentrations of unlabelled RNAs were determined by gel images and photometric analysis, concentrations of labelled RNAs were calculated.
In vitro translation, cells and transfections
In vitro translation reactions were carried out in RRL (Promega) (Ochs et al, 2003; Song et al, 2005). RRL (8 μl) was used in 20 μl reactions, and KCl was supplemented to a final K+ concentration of 135 mM. Reactions were incubated at 30°C for 60 min. Fluc activity was measured according to the supplier (Promega). Liposome-mediated transfection with Lipofectamine 2000 (Invitrogen) and electroporations were performed essentially as described earlier (Song et al, 2006). After 4 h (unless otherwise indicated), cells were washed in phosphate-buffered saline (PBS), PBS was aspirated, 150 μl reporter lysis buffer (Promega) was added to each well and the cells were lysed by gentle agitation. Nuclei were removed by centrifugation, and 20 μl supernatant was used for measuring Fluc activity as described before (Niepmann et al, 1997), or Renilla luciferase activity was measured as described by the supplier (Promega). Core antigen was measured using a chemiluminescence immunoassay (CGS, Abbott).
RNA re-extraction, northern blots and RNase protection assays
At 4 h after transfection, cells were washed in PBS, trypsinized and washed in a medium with 10% FBS. Cells were lysed in lysis buffer (50 mM Tris–Cl pH 8, 140 mM NaCl, 1.5 mM MgCl2, 1 mM DTT and 0.5% NP40) and nuclei were removed by centrifugation. Total RNA from cells was isolated with Trizol (Invitrogen) or RNeasy kits (Qiagen) and eluted in water. RNA from RRL was isolated using phenol–chloroform extraction and RNeasy. Isolation of small RNAs from cells and RRL was carried out by using the miR-Vana kit (Ambion). The RNA was separated on a 20% polyacrylamide gel and blotted to nitrocellulose. Northern blots were performed with dioxygenin-labelled RNAs or oligonucleotides using standard procedures. miR-122 was detected with the dioxygenin-labelled DNA oligonucleotide miR-122 (5′-ACAAACACCATTGTCACACTCCA-3′). U6 snRNA was detected with the oligonucleotide hra-U6 5′-GCAGGGGCCATGCTAATCTTCTCTGTATCG-3′. RNase protection assays were performed as described earlier (Junker-Niepmann et al, 1990).
Initiation complex formation and sucrose gradient sedimentation analysis
Initiation complex formation and gradient analysis were performed essentially as described before (Niepmann et al, 1997). Here, 150 μl binding reactions contained 50 μl RRL, 15 mM Tris–Cl pH 7.5, 0.5 mM MgCl2 and 1 mM DTT. Here, 0.17 mM anisomycin, 2 mM cycloheximide or 2 mM GMP-PNP was added if indicated. K+ was supplemented to a final concentration of 120 mM. Radiolabelled RNA (1 pmol) and 200 ng (26 pmol) miRNA (if indicated) was used to start initiation complex formation. For the binding reaction, samples were incubated at 30°C for 10 min or the times indicated. At all other times vials were kept strictly at 0°C. Reactions were stopped by adding MgCl2 to 30 mM, loaded onto ice-cold 10–35% sucrose gradients containing 50 mM Tris–Cl pH 8.4, 6 mM MgCl2, 60 mM NaCl, 10 mM DTT, and centrifuged in a Beckman SW 40 Ti rotor at 200 000 g and 4°C for 5.5 h (initiation complexes) or 2.5 h (polysomes). Fractions were collected from the bottom and used for scintillation counting.
HeLa cells were transfected with radiolabelled short HCV reporter RNA (see Figure 5A) in the absence or presence of miRNA as indicated. Cycloheximide was added to the medium to 1 mM 90 min after transfection, if indicated. After 120 min, cells were harvested, lysed in lysis buffer and nuclei were removed by centrifugation. The lysates were loaded onto sucrose gradients and analysed as above.
Supplementary Material
Supplementary Information
Acknowledgments
We thank IN Shatsky, R Bartenschlager and P Friebe for discussions. This study was supported by the Deutsche Forschungsgemeinschaft, Germany (DFG; SFB 535, GK 370, IRTG 1384 and Ni 604/1-1).
References
- Andre P, Perlemuter G, Budkowska A, Brechot C, Lotteau V (2005) Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis 25: 93–104 [DOI] [PubMed] [Google Scholar]
- Anthony DD, Merrick WC (1992) Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem 267: 1554–1562 [PubMed] [Google Scholar]
- Appel N, Schaller T, Penin F, Bartenschlager R (2006) From structure to function: new insights into hepatitis C virus RNA replication. J Biol Chem 281: 9833–9836 [DOI] [PubMed] [Google Scholar]
- Bradrick SS, Walters RW, Gromeier M (2006) The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res 34: 1293–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM (2008) Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol 82: 8215–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buenda MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM (2005) miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol 2: 17–24 [DOI] [PubMed] [Google Scholar]
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M (1989) Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244: 359–362 [DOI] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA (2003) siRNAs can function as miRNAs. Genes Dev 17: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Armstrong J, Colman A (1985) The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res 13: 7375–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E (2008) Getting to the root of miRNA-mediated gene silencing. Cell 132: 9–14 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Friebe P, Bartenschlager R (2002) Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol 76: 5326–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Tie Y, Xu C, Zhang Z, Zhu J, Shi Y, Jiang H, Sun Z, Zheng X (2005) Identification of human fetal liver miRNAs by a novel method. FEBS Lett 579: 3849–3854 [DOI] [PubMed] [Google Scholar]
- Ito T, Tahara SM, Lai MMC (1998) The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol 72: 8789–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309: 1577–1581 [DOI] [PubMed] [Google Scholar]
- Jünemann C, Song Y, Bassili G, Goergen D, Henke J, Niepmann M (2007) Picornavirus internal ribosome entry site elements can stimulate translation of upstream genes. J Biol Chem 282: 132–141 [DOI] [PubMed] [Google Scholar]
- Junker-Niepmann M, Bartenschlager R, Schaller H (1990) A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J 9: 3389–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739 [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA (2007) Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA 104: 9667–9672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W (1998) Tailing and 3′-end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA 4: 226–230 [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Ohashi K, Meuse L, Shen S, Lancaster AM, Lukavsky PJ, Sarnow P, Kay MA (2002) Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther 5: 676–684 [DOI] [PubMed] [Google Scholar]
- Niepmann M, Petersen A, Meyer K, Beck E (1997) Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J Virol 71: 8330–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs K, Rust RC, Niepmann M (1999) Translation initiation factor eIF4B interacts with a picornavirus internal ribosome entry site in both 48S and 80S initiation complexes independently of initiator AUG location. J Virol 73: 7505–7514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs K, Zeller A, Saleh L, Bassili G, Song Y, Sonntag A, Niepmann M (2003) Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J Virol 77: 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30: 460–471 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU (1998) A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Filipowicz W (2007) Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 17: 118–126 [DOI] [PubMed] [Google Scholar]
- Poumbourios P, Drummer HE (2007) Recent advances in our understanding of receptor binding, viral fusion and cell entry of hepatitis C virus: new targets for the design of antiviral agents. Antivir Chem Chemother 18: 169–189 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Friebe P, Tzima E, Jünemann C, Bartenschlager R, Niepmann M (2006) The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol 80: 11579–11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Tzima E, Ochs K, Bassili G, Trusheim H, Linder M, Preissner KT, Niepmann M (2005) Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA 11: 1809–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J (2001) Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962 [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318: 1931–1934 [DOI] [PubMed] [Google Scholar]
- Wang B, Love TM, Call ME, Doench JG, Novina CD (2006) Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell 22: 553–560 [DOI] [PubMed] [Google Scholar]
- Wu L, Belasco JG (2008) Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell 29: 1–7 [DOI] [PubMed] [Google Scholar]
- Yi M, Lemon SM (2003) 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol 77: 3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W (2004) Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information