Abstract
A wheat germ cell-free extract was used to perform in vitro translation of human stearoyl-CoA desaturase in the presence of unilamelar liposomes, and near complete transfer of the expressed integral membrane protein into the liposome was observed. Moreover, co-translation of the desaturase along with human cytochrome b5 led to transfer of both membrane proteins into the liposomes. A simple, single step purification via centrifugation in a density gradient yielded proteoliposomes with the desaturase in high purity as judged by capillary electrophoresis. After in vitro reconstitution of the non-heme iron and heme active sites, the function of the reconstituted enzyme complex was demonstrated by conversion of stearoyl-CoA to oleoyl-CoA. This simple translation approach obviates the use of detergents or other lipids to stabilize and isolate a catalytically active integral membrane enzyme. The applicability of cell-free translation to the assembly and purification of other integral membrane protein complexes is discussed.
Although integral membrane proteins account for almost 25% of open reading frames in fully sequenced genomes, progress on understanding their structure and function has lagged behind their soluble counterparts. In part, this is due to the difficulty in obtaining sufficient quantities of homogenous protein for in vitro studies using traditional expression systems. For example, the available space in cellular membranes, the toxic effects of competition for the membrane insertion machinery, and incorrect lipid composition for proper folding may limit the utility of Escherichia coli for eukaryotic membrane protein production [1]. Efforts to study the enzymology and structure of membrane proteins have been hindered by these difficulties over several decades.
As one example, stearoyl-CoA desaturases are integral membrane proteins thought to have four trans-membrane sequences [2]. They have a conserved motif consisting of 8 His residues hypothesized to provide at least some of the ligands to a catalytically essential diiron center [2]. In 1974, Strittmatter and colleagues published a preparation of the stearoyl-CoA desaturase from the livers of starved and then fed rats [3]. This achievement ultimately permitted a number of important properties of the enzyme complex to be elucidated [4–6]. However, no comparable reports on the successful purification of mammalian stearoyl-CoA desaturase have arisen in the ensuing four decades.
Human SCD isoform 1 (hSCD1) from liver and adipose tissue catalyzes a critical step in de novo lipid biosynthesis [7]; the NAD(P)H and O2-dependent desaturation of stearoyl-CoA to form oleoyl-CoA. This product is an important precursor for phospholipids, triglycerides, and cholesterol esters. The function of stearoyl-CoA desaturase is linked to hypertension, non-insulin dependent diabetes, cardiovascular disease, obesity, and other significant health issues [8]. Human desaturases also require the presence of two other membrane-anchored proteins, NADH cytochrome b5 reductase (cytb5 reductase) and cytochrome b5 (cytb5), for catalytic turnover [9].
Studies of tissue cultures [10], mouse, and rat livers [11] have shown that mammalian SCD activity is tightly regulated at the transcriptional, translational, and post-translational levels. Transcription is under control of the PPARγ signaling pathway [12], and may also respond to various secondary messengers such as polyunsaturated fatty acids. Translational regulation is mediated through mRNA stability by endoplasmic reticulum membrane proteins, like Mga2p [13], and, as recently suggested, the 15-lipoxygenase-differentiation control element, a conserved feature in mammalian 3′-untranslated regions [14]. Other studies in murine microsomes [11] and differentiated mouse 3L3-L1 adipocytes [15] showed post-translational proteolytic processing of the N-terminus of the enzyme, subsequent inactivation, and proteosome-directed degradation. Interestingly, DesA3, part of the stearoyl-CoA desaturase complex from Mycobacterium tuberculosis [16, 17], is also targeted for rapid proteolytic degradation, but in this case by a prokaryotic degradation complex with specificity for the C-terminus [18]. The multiple layers of regulation of desaturase activity, in diverse organisms, emphasize the importance of this enzyme in cellular function [7], but also complicate recombinant expression using living hosts.
As an alternative, cell-free protein translation may potentially circumvent the regulatory issues associated with expression of membrane proteins in living systems [19, 20]. Here we report translation of the hSCD1 and cytb5 complex using wheat germ extract, purification of the complex, and metal and heme reconstitution to create an active form of the complex entirely from in vitro reactions. The approaches described at each step are simple, and have potential applicability to studies of many other integral membrane proteins.
MATERIALS AND METHODS
Materials
Routine reagents were of the highest grade available from standard vendors. Distilled and deionized water was used for all reagent preparations. The genes for human hSCD1 (BC005807) and human cytb5 (BC015182) were obtained from Open Biosystems (http://www.openbiosystems.com/). The cDNAs for the mouse desaturase genes were prepared from total RNA isolated from mouse liver, brain, and pancreas, and were the generous gift of Dr. F.E. Gomez (University of Wisconsin, Madison). The hSCD5 gene was a generous gift from Drs. J.M. Ntambi and M. Miyazaki (University of Wisconsin, Madison). The desA3 gene was amplified from Mycobacterium tuberculosis H37Rv genomic DNA (TB Research Materials Facility at Colorado State University, Prof. J. Belisle, Director, NIH NIAD NO1AI75320). Reagents for FlexiVector cloning were from Promega (Madison, WI). The bacteriorhodopsin gene was amplified from genomic DNA purified from Halobacterium salinarum generously provided by Dr. J. Escalante-Semerena (University of Wisconsin-Madison), and the enhanced green fluorescent protein gene was from the Center for Eukaryotic Structural Genomics [21]. Care must be taken to exclude ribonucleases from all reagents and equipment used for preparation of the cell-free translation reactions, including micropipeters.
Cloning Vector
The vector pEU optimized for wheat germ cell-free translation [22] was modified for FlexiVector (Promega, Madison, WI) cloning to contain 5′-SgfI and 3′-PmeI restriction sites and a toxic selection cassette in the multi-cloning site [23]. One modified vector, named pEU-His-FV, is available from the NIH Protein Structure Initiative Material Repository (http://www.hip.harvard.edu/PSIMR/index.htm), and produces a protein with an N-terminal His6 purification tag. Another vector, named pEU-FV, produces a protein with no purification tag. Desaturase and cytb5 genes were amplified by PCR using the primers indicated in Table 1 and transferred by FlexiVector cloning [23] into pEU-His-FV and pEU-FV. All PCR-amplified genes were sequenced to confirm their fidelity. Plasmid DNA for transcription reactions was purified using Marligen maxi-prep kits (Marligen Biosciences, Ijamsville, MD).
Preparation of Liposomes
Liposomes were prepared from a soybean tissue extract (Avanti Polar Lipids, Alabaster, AL). The lipid powder was dissolved in chloroform and dried for 30 min under vacuum after removal of the bulk organic solvent by evaporation under a stream of N2 gas. The dried lipid film was re-hydrated with 25 mM HEPES, pH 7.5, containing 100 mM NaCl at a concentration of 5 mg/mL. The lipid solution was vortexed for 5 min and subjected to 3 freeze-thaw cycles. An Avanti mini-extruder was used to form unilamelar liposomes by 11 passes through a 100 nm track-etch polycarbonate membrane (Nucleopore, Pleasanton, CA). The liposomes were stored at −80 °C.
Transcription
The transcription reaction has a total volume of 50 μL, and contains 4 μg of purified plasmid DNA, 80 mM HEPES, pH 7.5, 16 mM magnesium acetate, 2 mM spermidine, 10 mM dithiothreitol, 2.5 mM of each nucleotide triphosphate (ATP, UTP, GTP, CTP), 25 units of RNasin (Promega), 30 units of Sp6 RNA polymerase (Promega). The remainder of the total volume was from deionized and sterilized water. The reaction was incubated at 37 °C for 3 h and then centrifuged at 15,000 rpm in an Allegra 21R centrifuge (Beckman Coulter, Fullerton, CA) and F2402H rotor. The supernatant, containing mRNA, was transferred to a 1.7 mL centrifuge tube containing 10 μL of 6 M ammonium acetate. For co-translation, supernatants from transcription reactions for both genes were added to the ammonium acetate. After addition of 150 μL of 100% ethanol, the tubes were mixed, incubated on ice for 5 min and centrifuged at 15,000 rpm for 20 min at 4 °C. The mRNA pellet was washed with 600 μL of 70% ethanol and centrifuged, and after careful removal of the supernatant, allowed to air dry.
Cell-free Translation
The translation mixture has a total volume of 50 μL, and contains 30 mM HEPES, pH 7.8, 100 mM potassium acetate, 2.7 mM magnesium acetate, 1.2 mM ATP, 0.25 mM GTP, 16 mM creatine phosphate, 0.4 mM spermidine, 0.3 mM of each amino acid, 0.8 mg/mL of creatine kinase, 24 units of RNasin, and 60 μg of liposomes. Wheat-germ extract (15 μL, Cell Free Sciences, Yokohama, Japan) was added from a concentrated commercial preparation to a final OD600 of 60 and the remainder of the total volume was from deionized and sterilized water. The purified mRNA pellet was dissolved in the translation mixture and the reaction was placed into a 12 MWCO dialysis cup (Biotech International, Perth, Australia) suspended in a buffer reservoir containing all of the above reagents except creatine kinase, RNasin, liposomes, and wheat germ extract. The reaction was incubated at 26 °C for 16 h. Protein levels were determined by Caliper Lab Chip 90 analysis (Caliper Life Sciences, Hopkinton, MA) by comparison with creatine kinase as an internal standard.
Proteoliposome Purification
A 45-μL aliquot of the completed translation mixture was mixed with a buffer containing 25 mM Hepes, pH 7.4, 100 mM NaCl and 30% (v/v) glycerol to give final volume of 75 μL and 10% (v/v) glycerol; the remaining 5 μL of the translation reaction was retained for analysis by denaturing electrophoresis. The 75-μL sample was mixed with 75 μL of 80% (w/v) Accudenz (Accurate Chemical and Scientific, Westbury, NY) prepared in 25 mM Hepes, pH 7.4, containing 100 mM NaCl and 10% (v/v) glycerol. The mixture was transferred to an Ultra-Clear centrifuge tube (Beckman Coulter, City ST) and sequentially overlaid with 350 μL of 30% Accudenz and then 100 μL of 25 mM Hepes, pH 7.4, containing 100 mM NaCl. The mixture was centrifuged for 4 h at 45,000 rpm (189,000g) and 4 °C in an L-60 ultracentrifuge (Beckman Coulte r) and SW-50.1 rotor with adaptors. After centrifugation, 60-μL fractions were collected from the top of the gradient. Proteoliposomes containing hSCD1 and/or cytb5 were found at the buffer/Accudenz interface (fractions 2–3). The remaining, unbound proteins were found in fractions 7–10. Protein levels were determined by LabChip90 analysis using total detected protein-dye complex fluorescence.
Hemin and Ferrous Iron Preparations
Hemin chloride [24] was added to a 50% (v/v) ethanol/water solution, allowed to stand for a few minutes before 100-fold dilution into 10 mM Tris, pH 8.0, containing 1 mM EDTA. The absorbance was measured at 385 nm using an Agilent 8453 UV-visible spectrophotometer (Agilent Scientific, Santa Clara, CA). To the heme solution, an aliquot of 1 M NaOH (100 μL) was added, mixed, and allowed to stand prior to measuring the absorbance at 385 nm. This process was repeated until no further increase in absorbance at 385 nm was observed, which indicated saturation of heme in the ethanol/water mixture. The solution was filtered through a 0.2-μm filter and the concentration of heme was determined from optical spectrometry (ε385 = 56,000 cm−1). The heme solution was stored at 4 °C.
A solution of Fe2+ was prepared immediately before use by dissolving Fe(NH3)2(SO4)2 and a 10-fold molar excess of ascorbate in degassed 60 mM HEPES, pH 7.4.
Desaturase Assay
The 60-μL fractions obtained from the proteoliposome floatation were added to 140 μL of assay mixture containing 25 mM Hepes, pH 7.2, 180 mM NaCl, 6 nmol stearoyl-CoA, 0.03 μCi [1-14C] stearoyl-CoA, and 2 mM NADH. Unless noted, the heme solution and the Fe2+ solution were added to the assay mixture to give final concentrations of 5.8 μM and 29 μM, respectively. The reaction was initiated by addition of the soluble domain of human cytb5 reductase to a final concentration of 230 nM. The desaturase reaction was incubated at 37 °C and quenched by addition of 200 μL of 2.5 M KOH in ethanol and the fats were saponified at 80 °C. The fatty acids were pro tonated by addition of 280 μL of formic acid. Fatty acids were extracted with 700 μL of hexane, and 300 μL of the hexane layer was evaporated to dryness and re-suspended in 50 μL of hexane. Fatty acids were separated on a thin-layer chromatography plate impregnated with 10% AgNO3 and developed with a chloroform/methanol/acetic acid/water mixture (90:8:1:0.8). Radioactive decay was detected using a Packard Instant Imager (Packard, Meriden, CT).
RESULTS
Construction of Expression Vectors
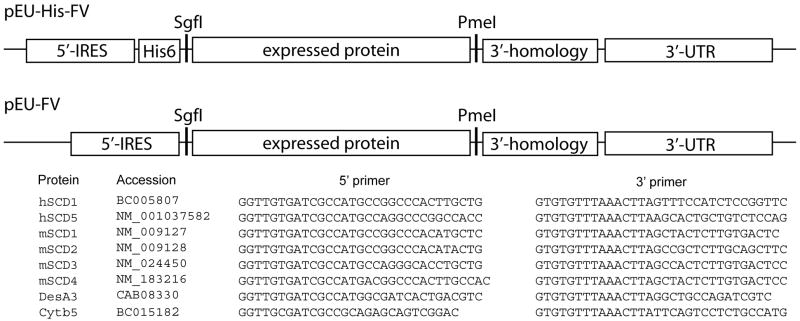
Fig. 1 shows the desaturase and cytb5 genes studied here, the primers used, and location of the 5′-internal ribosome entry site, start codon, His6 purification tag, stop codon, and 3′-untranslated region in the expression vectors used. The 5′-internal ribosome entry site and 3′-untranslated region are required for effective translation in the wheat germ extract [25] while the 3’ homology region is required for efficient transfer of genes between pairs of FlexiVectors [23]. Desaturase genes were cloned into pEU-His-FV, which added an N-terminal His6 tag to the translated protein, and pEU-FV, which contained all the above features except for the His6 purification tag.
Fig. 1.
Schematic representations of the vectors pEU-His-FV and pEU-FV. Each vector contains a 5′-internal ribosome entry sequence, a 3′-untranslated region for translation, and a 3′ homology region to enhance cloning efficiency. pEU-His-FV also contains an N-terminal His6 purification tag. The desaturases and cytb5 genes, their respective accession numbers, and the PCR primers that were used in this study are also indicated.
Cell-Free Translation
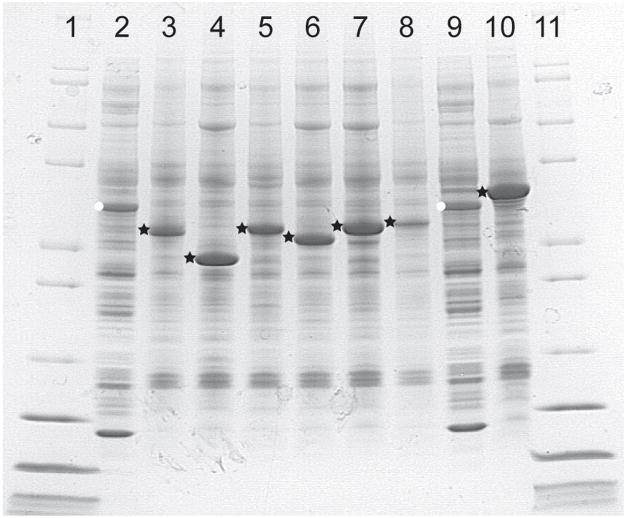
Fig. 2 shows the results of cell-free translation of isoforms of stearoyl-CoA desaturase from human, mouse, and Mycobacterium tuberculosis. Lanes 1 and 8 show the wheat germ extract. The polypeptide of ~43 kDa marked with a white dot is creatine kinase, which is added to the extract at ~700 μg/mL to serve as part of an ATP regeneration system and a relative marker of the level of protein translation. Lanes 2 through 7 and lane 9 were prepared from the pellet fractions obtained by centrifugation of the extract after completion of the translation reactions by re-suspension in an equal volume of SDS-containing electrophoresis buffer. Lanes 2 and 3 contain human hSCD1 and hSCD5, while lanes 4 through 7 contain mouse mSCD1, mSCD2, mSCD3, and mSCD4, respectively. Lane 9 contains DesA3, the mycobacterial stearoyl-CoA desaturase homolog [17]. Except for mSCD4, which does not complement a yeast ole1 auxotroph, the different desaturases accumulated to levels between 1–2 mg/mL of translation mixture as determined by capillary electrophoresis.
Fig. 2.
Expression of human, mouse and mycobacterium desaturases in cell-free translation as visualized by denaturing electrophoresis and Coomassie staining. Translation of pelleted desaturases was estimated by comparison to exogenous soluble creatine kinase (white stars, 0.7 mg/mL, lanes 2 and 9), and verified by capillary electrophoresis. Black circles indicate pellet fractions consisting of human hSCD1 (lane 3, 1.1 mg/mL) and hSCD5 (lane 4, 1.6 mg/mL), mouse mSCD1 (lane 5, 1.3 mg/mL), mSCD2 (lane 6, 1.2 mg/mL), mSCD3 (lane 7, 1.3 mg/mL) and mSCD4 (lane 8, 0.3 mg/mL) and mycobacterial DesA3 (lane 10, 2.3 mg/mL).
Unlike results for bacterial cell-free translation of membrane transporters [20], the desaturases expressed in the as-purchased wheat germ extract formed dense precipitates that could not be re-suspended or solubilized except with aggressive detergents such as SDS. Moreover, a polypeptide from the extract of ~71 kDa co-precipitated with hSCD5, mSCD2, mSCD3, and DesA3, which were the most abundantly expressed desaturases in these experiments. A tryptic digest mass spectral analysis revealed that this protein had high sequence identity with Triticum aestivum Hsp70 [1], a eukaryotic chaperone protein that participates in folding of nascent proteins emerging from the ribosome.
Table 1 shows the lipid and metal composition of the wheat germ extract. Desaturases require non-heme iron for catalytic function, so it was relevant to determine the content of iron. Since the amount of iron detected was ~50-fold lower than the amount of desaturase protein expressed, iron seemed likely to be a limiting reagent for assembly of a functional complex. For reconstitution of cytb5 activity, heme was necessary, but the absence of the characteristic Soret band at ~410 nm in the optical spectrum of the extract suggested that only a trace level of total heme might be available for incorporation. Furthermore, it was of interest to determine the lipid composition of the extract in order to understand whether natural membranes, endogenous substrates, or products of the reaction might be present. The different fatty acids detected were consistent with the composition of natural plant membranes, but like iron, were present in only low amounts relative to the level of protein translation obtained from the wheat germ extract. These findings indicated that the availability of essential metals and lipids required for desaturase function and stability might limit assembly of a functional complex.
Influence of Detergents
Table 2 shows the results of expression of hSCD1, bacteriorhodopsin, and enhanced GFP in the presence of detergents. Based on previous work [26, 27], a small subset of non-inhibitory detergents, Brij-35, Triton X-100 and CHAPS were investigated. This study and others [26] showed that cell-free translation in the presence of detergents gave variable results that were dependent on the identities of both protein and detergent. For example, Brij-35 solubilized the desaturases, but did not completely solubilize bacteriorhodopsin, a well-characterized homologue of proteorhodopsin, that is often used as a control for the effectiveness of cell-free translation of membrane proteins [28] and spontaneous incorporation into liposomes [29]. Due to the complexities of handling detergent-stabilized membrane proteins [30], further studies were deemed undesirable.
Purification of Proteoliposome Complex
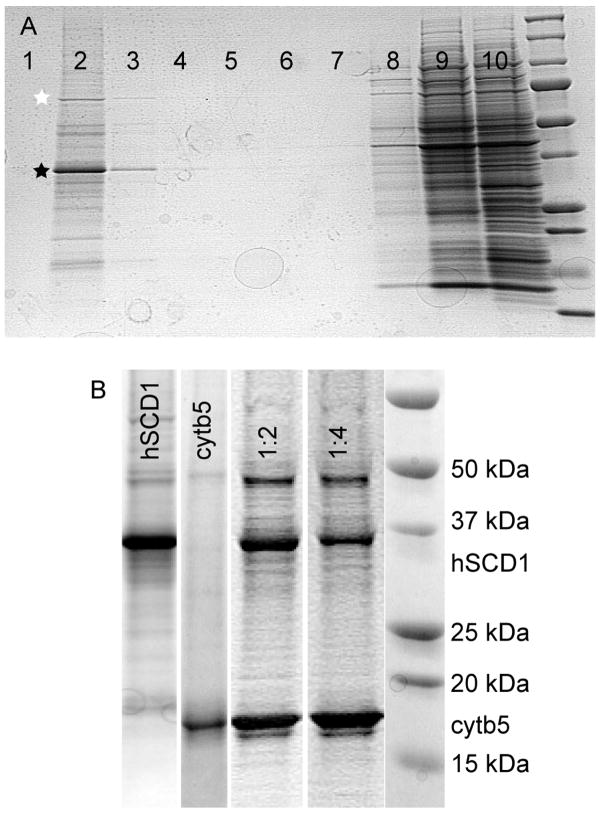
In contrast to the variable results with detergents, Table 2 shows that cell-free translation in the presence of liposomes gave comparable expression to that obtained with no supplementation for each of the proteins tested. After translation, the buoyant proteoliposomes were floated through a density gradient by ultracentrifugation (Fig. 3A, also see Fig. 7 of [31] for purification of cytb5 using this approach). The electrophoresis of Fig. 3A revealed several important results. In the presence of liposomes, essentially all of the translated hSCD1 (marked with a black star and verified by in-gel trypsin-digest mass spectrometry) was captured by the liposomes, as shown by comparing lane 3 and lane 8. The proteoliposomes were remarkably pure, and hSCD1 accounted for >90% of the total protein present as determined by capillary electrophoresis. Among the most frequently observed contaminants, the Hsp70 protein, elongation factor 1α [32], and a 16.9 kDa heat shock protein [33] were identified by mass spectrometry. While cytb5 has been shown to spontaneously insert into liposomes [34], the requirements for insertion of desaturases into lipids are not understood. It was therefore encouraging that both high level translation and effective transfer of the membrane protein were observed in the presence of liposomes.
Fig. 3.
A, Coomassie-stained denaturing gel electrophoresis showing near complete incorporation of hSCD1 (black star) into synthetic liposomes from wheat germ cell-free translation. Proteoliposomes were loaded underneath a discontinuous Accudenz gradient and floated by ultracentrifugation. Bound protein, including hsp70 (white star) and hSCD1 floated (lanes 2 and 3), while unbound protein remained at the bottom (lanes 8–10). B, a comparison of proteoliposomes from floated fraction 2 containing hSCD1 (lane 1), cytb5 (lane 2), and a co-translation of hSCD1 and cytb5 (lane 3, 1 eq of hSCD1 mRNA with 2 eq of cytb5 mRNA; lane 4, 1 eq of hSCD1 mRNA with 4 eq of cytb5 mRNA).
In an important variation of the cell-free translation reaction, hSCD1 and cytb5 were co-translated by adding mRNAs encoding both proteins to the translation reaction (Fig. 3B) The amounts of the two proteins were found to roughly correspond to the size-normalized amounts of mRNA added to the reaction mixture. This result demonstrates a simple approach to vary the relative amounts of proteins that assemble into a macromolecular complex.
Reconstitution of Desaturase Complex and Catalytic Activity
Previous studies have shown that the soluble domain of cytb5 reductase was sufficient to reconstitute the catalytic activity of a membrane-bound complex of rat SCD1 and rat cytb5 [4]. For the present studies, human cytb5 reductase was expressed as previously described [31] and purified from an Escherichia coli cell extract.
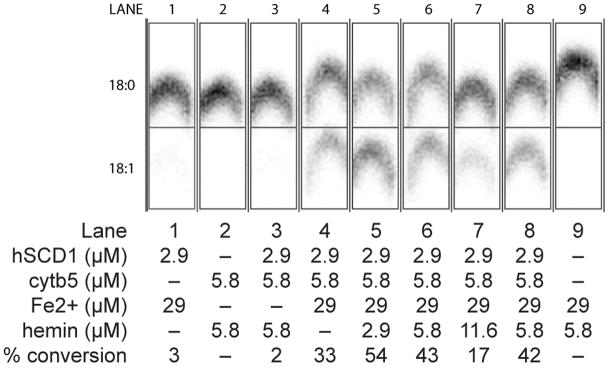
Fig. 4 shows the results of catalytic assays of the purified proteoliposomes. In these reactions, NADH, stearoyl-CoA (18:0), and a tracer amount of [U-14C]-stearoyl-CoA were added to liposome samples, and the reaction was initiated by the addition of cytb5 reductase. The product of the reaction, [U-14C]-oleoyl-CoA, was detected using a phosphoimager after separation by thin-layer chromatography. Control reactions showed that the extract alone (lane 9), Fe2+-treated hSCD1 alone (lane 1) and heme-treated cytb5 (lane 2) alone gave no catalytic activity. Likewise, no catalytic activity was observed from the as-isolated hSCD1 and cytb5 proteoliposomes upon addition of cytb5 reductase, NADH and stearoyl-CoA (lane 3), as the extract lacked substantial amounts of essential iron (Table 2). In order to reconstitute the hSCD1 diiron active site, an Fe2+ and ascorbate solution was added to the proteoliposomes prior to initiation of the reaction (lane 4). In proteoliposomes treated in this manner, 33% of substrate was converted to product, which also revealed that the wheat germ extract contained sufficient heme to partially reconstitute the translated cytb5. The addition of heme (~3 μM) to the translation reaction increased the conversion of substrate to product to 54%, while further 2- and 4-fold increases in the heme concentration inhibited the desaturation reaction (lanes 5, 6, and 7). Furthermore, when hSCD1 and cytb5 were independently translated and purified, the proteoliposomes could be mixed to recover comparable activity to that obtained from co-translation of hSCD1 and cytb5 (lanes 6 versus 8), offering another possibility for assembly of enzyme complexes.
Fig. 4.
Demonstration of catalytic activity from cell-free co-translatation of hSCD1 and cytb5. Concentrations of hSCD1, cytb5, Fe2+, and hemin present in each lane are indicated. Lane 4 contains the Fe2+-activated complex of hSCD1 and cytb5. Lanes 5, 6 and 7 contain the complex treated with increasing amounts of exogenous heme. For comparison, lane 8 shows a desaturation reaction assembled by mixture of Fe2+- and heme-activated proteoliposomes originally containing only hSCD1 and only cytb5.
Table 3 provides an accounting of the two-step translation and purification of hSCD1. In the wheat germ extract, hSCD1 accounted for ~4% of the total protein present after translation in the presence of liposomes. The density gradient separation of the proteoliposomes yielded 24 μg of hSCD1 from a 50 μL translation reaction with greater than 80% purity. The density gradient separation also gave a 25-fold increase in the specific activity of the enzyme, and provided near complete recovery of the enzyme activity from the extract.
The activity of the proteoliposomes obtained from cell-free translation was also compared to a rat liver microsome preparation known to contain active SCD. Thus the rat liver microsome preparation had specific activity for oleate production of ~2 U/mg in a 15 min stopped-time assay, while proteoliposomes obtained from co-translation of hSCD1 and cytb5 at 1:2 ratio of mRNA gave a specific activity for oleate production of ~10 U/mg in the same assay. The desaturation reaction was not impacted by the presence of the N-terminal His6 purification tag.
DISCUSSION
Cell-Free Translation
Cell-free translation has been used in biochemical research for a considerable time [35]. The earliest studies used radiotracer approaches in order to detect low levels of translated protein. Structural genomics and synthetic biology efforts have stimulated a new interest in cell-free translation [36–42], and the yield per unit volume of more recently developed systems has reached mg of protein per mL of translation mixture [22, 43], which is comparable to the most highly optimized E. coli expression [44]. Advantages of cell-free translation include automation, scale and speed of operations, opportunities for labeling with non-natural amino acids, and incorporation of simple and more complicated cofactors [45, 46].
In this study, we have shown that cell-free translation circumvented complications of producing an integral membrane enzyme complex whose unregulated function might be deleterious to the cell, such as modification of the lipid composition of the cellular membrane. Thus cell-free translation allowed the facile preparation of an integral membrane enzyme complex that has been otherwise difficult to produce in living cells.
Recent research has identified that some detergents are compatible with cell-free translation [47]. However, these detergents are not necessarily those most desirable for subsequent steps in handling and assaying membrane proteins. Cell-free translation in the presence of liposomes has been successfully applied to the preparation of single polypeptide transporters from Arabidopsis, and a function was demonstrated for both transporters tested [27]. In agreement with the Arabidopsis transporter study, soy lecithin was chosen because it is cheap and available in a highly pure form. However, it is possible to prepare other liposomes or vesicle preparations that might better match the specific tissues where membrane enzymes are located, such as has been done with the incorporation of membrane proteins into E. coli inner membrane preparations [48].
hSCD1 and cytb5 were individually well expressed and captured into liposomes from the wheat germ translation reaction. For reconstitution of the active complex, it was possible to add the mRNA for both hSCD1 and cytb5 to the same translation reaction and to co-translate the proteins. In this case, proteoliposomes containing both proteins were formed based on denaturing electrophoresis and on the detected catalytic activity. In practice, co-translation in the presence of liposomes represents a potentially combinatorial approach to the assembly of functional complexes of membrane proteins. Furthermore, co-translation and reconstitution of cytb5 with the hSCD1 H124A mutant (not shown, a mutation that inactivates SCD1 presumably by removing an essential metal ligand [2]) gave the conclusive result of no activity after an ~3 day work cycle as compared to ~2 weeks required to complete the comparable experiment in a yeast Δole1 auxotroph strain.
Mixing proteoliposomes prepared separately to contain either one or the other of hSCD1 and cytb5 also gave a catalytically active complex, corresponding to merger or transfer of proteins between liposomes to generate a population of proteoliposomes that contained both [34]. Thus separate translation and then recombination of well-characterized individual liposomes represents an additional approach to study protein-protein interactions with membrane proteins that may be facilitated by cell-free translation.
Reconstitution Reaction
Reconstitution of the metal center in rat SCD was suggested by previous in vivo translation studies, but not extensively characterized [49]. Likewise, incorporation of heme into apo-cytb5 was known [24]. Our analysis suggested that the wheat germ extract did not contain enough Fe (~25 μM) or heme (<1 μM) to support stoichiometric incorporation into the translated apo-proteins, given the level of translation observed in our cell-free reactions (50–100 μM of translated polypeptide). In this work, we found that simple addition of ascorbate-stabilized Fe2+ to the extract after the translation was sufficient to reconstitute the active sites of the hSCD1 and give catalytic activity. In addition to the added ascorbate, the reducing environment of the extract may help to solubilize Fe2+, which is generally recognized to be the redox state used for cellular iron trafficking. It is also possible, but not demonstrated, that ferritin, transferrin, and other proteins are present in the wheat germ extract and participate in metal trafficking. The wheat germ extract also contained sufficient heme to yield an active form of cytb5, but the activity could be increased by further supplementation with heme. However, addition of excess heme was inhibitory to catalysis by an unknown mechanism.
In combination, the Fe2+- and heme-activated hSCD1 and cytb5 were assembled in the liposome to produce a catalytically active complex. In these minimally optimized reactions, the specific activity of the cell-free translated and easily purified human SCD1 for oleate production (~10 U/mg) compared quite well with the more difficult to obtain natural rat liver microsomes (~2 U/mg) in a 15 min stopped time assay. It is not possible to compare the specific activity for oleoyl-CoA production of the rat enzyme with hSCD1 obtained from wheat germ cell-free translation, as the previously published purification of rat SCD used an indirect NADPH consumption assay to monitor purification [3].
CONCLUSION
We have shown that a functional human integral membrane desaturase complex can be assembled using cell-free translation. The proteoliposomes containing the enzyme complex were easily purified using a simple density gradient centrifugation. Definitive assays for the conversion of stearoyl-CoA to oleoyl-CoA showed that the purified complex had activity comparable to rat liver microsomes containing a naturally assembled stearoyl-CoA desaturase complex. We have shown that individual proteoliposomes can be mixed to give a functional complex and, furthermore, that individual mRNAs can be co-translated to also give a functional complex. This versatility expands the potential for application of cell-free translation to many other problems in membrane biochemistry. It is thus reasonable to consider that the approaches described herein may be applicable to the study of many other integral membrane proteins, including those from humans with great medical relevance.
Acknowledgments
NIGMS GM-50853 (B.G.F., P.I.) and Protein Structure Initiative U54 GM074901-01 (John L. Markley, P.I., George N. Phillips, Jr. and B.G.F., Co-Investigators) funded this work. M.A.G. acknowledges support from the NSF East Asia and Pacific Summer Institutes Fellowship program. Rat liver microsomes were the generous gift of Dr. M. Flowers and Dr. J.M. Ntambi (University of Wisconsin). The authors warmly thank Dr. Yaeta Endo and Dr. Yuzuru Tozawa (Cell-Free Science and Technology Research Center, Ehime University, Japan) for many helpful discussions.
Abbreviations
- SCD
stearoyl-CoA desaturase
- cytb5
full-length cytochrome b5
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wagner S, Bader ML, Drew D, de Gier JW. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Shanklin J, Whittle E, Fox BG. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 3.Strittmatter P, Spatz L, Corcoran D, Rogers MJ, Setlow B, Redline R. Purification and properties of rat liver microsomal stearyl coenzyme A desaturase. Proc Natl Acad Sci USA. 1974;71:4565–4569. doi: 10.1073/pnas.71.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enoch HG, Catala A, Strittmatter P. Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J Biol Chem. 1976;251:5095–5103. [PubMed] [Google Scholar]
- 5.Dailey HA, Strittmatter P. Structural and functional properties of the membrane binding segment of cytochrome b5. J Biol Chem. 1978;253:8203–8209. [PubMed] [Google Scholar]
- 6.Dailey HA, Strittmatter P. Characterization of the interaction of amphipathic cytochrome b5 with stearyl coenzyme A desaturase and NADPH:cytochrome P-450 reductase. J Biol Chem. 1980;255:5184–5189. [PubMed] [Google Scholar]
- 7.Ntambi JM, Miyazaki M. Regulation of stearoyl-CoA desaturases and role in metabolism. Prog Lipid Res. 2004;43:91–104. doi: 10.1016/s0163-7827(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 8.Dobrzyn A, Ntambi JM. Stearoyl-CoA desaturase as a new drug target for obesity treatment. Obes Rev. 2005;6:169–174. doi: 10.1111/j.1467-789X.2005.00177.x. [DOI] [PubMed] [Google Scholar]
- 9.Holloway PW. A requirement for three protein components in microsomal stearyl coenzyme A desaturation. Biochemistry. 1971;10:1556–1560. doi: 10.1021/bi00785a008. [DOI] [PubMed] [Google Scholar]
- 10.Gomez FE, Miyazaki M, Marwah P, Lardy HA, Ntambi JM, Fox BG. Molecular differences caused by differentiation of 3T3-L1 preadipocytes in the presence of either dehydroepiandosterone (DHEA) or 7-oxo-DHEA. Biochemistry. 2002;41:5473–5482. doi: 10.1021/bi012177r. [DOI] [PubMed] [Google Scholar]
- 11.Heinemann FS, Korza G, Ozols J. A plasminogen-like protein selectively degrades stearoyl-CoA desaturase in liver microsomes. J Biol Chem. 2003;278:42966–42975. doi: 10.1074/jbc.M306240200. [DOI] [PubMed] [Google Scholar]
- 12.Miller CW, Ntambi JM. Peroxisome proliferators induce mouse liver stearoyl-CoA desaturase 1 gene expression. Proc Natl Acad Sci USA. 1996;93:9443–9448. doi: 10.1073/pnas.93.18.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin CE, Oh CS, Jiang Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim Biophys Acta. 2007;1771:271–285. doi: 10.1016/j.bbalip.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Ren J, Knorr C, Huang L, Brenig B. Isolation and molecular characterization of the porcine stearoyl-CoA desaturase gene. Gene. 2004;340:19–30. doi: 10.1016/j.gene.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Kato H, Sakaki K, Mihara K. Ubiquitin-proteasome-dependent degradation of mammalian ER stearoyl-CoA desaturase. J Cell Sci. 2006;119:2342–2353. doi: 10.1242/jcs.02951. [DOI] [PubMed] [Google Scholar]
- 16.Phetsuksiri B, Jackson M, Scherman H, McNeil M, Besra GS, Baulard AR, Slayden RA, DeBarber AE, Barry CE, 3rd, Baird MS, Crick DC, Brennan PJ. Unique mechanism of action of the thiourea drug isoxyl on Mycobacterium tuberculosis. J Biol Chem. 2003;278:53123–53130. doi: 10.1074/jbc.M311209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Y, Fox BG. Identification of Rv3230c as the NADPH oxidoreductase of a two-protein DesA3 acyl-CoA desaturase in Mycobacterium tuberculosis H37Rv. Biochemistry. 2006;45:13476–13486. doi: 10.1021/bi0615285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang Y, Wesenberg G, Bingman CA, Fox BG. In vivo inactivation of mycobacterial integral membrane stearoyl-CoA desaturase DesA3 by a C-terminal specific degradation process. J Bacteriol. 2008 doi: 10.1128/JB.00585-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renesto P, Raoult D. From genes to proteins: in vitro expression of rickettsial proteins. Ann N Y Acad Sci. 2003;990:642–652. doi: 10.1111/j.1749-6632.2003.tb07439.x. [DOI] [PubMed] [Google Scholar]
- 20.Klammt C, Schwarz D, Lohr F, Schneider B, Dotsch V, Bernhard F. Cell-free expression as an emerging technique for the large scale production of integral membrane protein. FEBS J. 2006;273:4141–4153. doi: 10.1111/j.1742-4658.2006.05432.x. [DOI] [PubMed] [Google Scholar]
- 21.Frederick RO, Bergeman L, Blommel PG, Bailey LJ, McCoy JG, Song J, Meske L, Bingman CA, Riters M, Dillon NA, Kunert J, Yoon JW, Lim A, Cassidy M, Bunge J, Aceti DJ, Primm JG, Markley JL, Phillips GN, Jr, Fox BG. Small-scale, semi-automated purification of eukaryotic proteins for structure determination. J Struct Funct Genomics. 2007;8:153–156. doi: 10.1007/s10969-007-9032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawasaki T, Hasegawa Y, Tsuchimochi M, Kasahara Y, Endo Y. Construction of an efficient expression vector for coupled transcription/translation in a wheat germ cell-free system. Nucleic Acids Symp Ser. 2000:9–10. doi: 10.1093/nass/44.1.9. [DOI] [PubMed] [Google Scholar]
- 23.Blommel PG, Martin PA, Wrobel RL, Steffen E, Fox BG. High efficiency single step production of expression plasmids from cDNA clones using the Flexi Vector cloning system. Protein Expr Purif. 2006;47:562–570. doi: 10.1016/j.pep.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Mulrooney SB, Waskell L. High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b(5) Protein Expr Purif. 2000;19:173–178. doi: 10.1006/prep.2000.1228. [DOI] [PubMed] [Google Scholar]
- 25.Sawasaki T, Ogasawara T, Morishita R, Endo Y. A cell-free protein synthesis system for high-throughput proteomics. Proc Natl Acad Sci USA. 2002;99:14652–14657. doi: 10.1073/pnas.232580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klammt C, Lohr F, Schafer B, Haase W, Dotsch V, Ruterjans H, Glaubitz C, Bernhard F. High level cell-free expression and specific labeling of integral membrane proteins. Eur J Biochem. 2004;271:568–580. doi: 10.1111/j.1432-1033.2003.03959.x. [DOI] [PubMed] [Google Scholar]
- 27.Nozawa A, Nanamiya H, Miyata T, Linka N, Endo Y, Weber AP, Tozawa Y. A cell-free translation and proteoliposome reconstitution system for functional analysis of plant solute transporters. Plant Cell Physiol. 2007;48:1815–1820. doi: 10.1093/pcp/pcm150. [DOI] [PubMed] [Google Scholar]
- 28.Gourdon P, Alfredsson A, Pedersen A, Malmerberg E, Nyblom M, Widell M, Berntsson R, Pinhassi J, Braiman M, Hansson O, Bonander N, Karlsson G, Neutze R. Optimized in vitro and in vivo expression of proteorhodopsin: a seven-transmembrane proton pump. Protein Expr Purif. 2008;58:103–113. doi: 10.1016/j.pep.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Kalmbach R, Chizhov I, Schumacher MC, Friedrich T, Bamberg E, Engelhard M. Functional cell-free synthesis of a seven helix membrane protein: in situ insertion of bacteriorhodopsin into liposomes. J Mol Biol. 2007;371:639–648. doi: 10.1016/j.jmb.2007.05.087. [DOI] [PubMed] [Google Scholar]
- 30.le Maire M, Champeil P, Moller JV. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim Biophys Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 31.Sobrado P, Goren MA, James D, Amundson CK, Fox BG. A Protein Structure Initiative approach to expression, purification, and in situ delivery of human cytochrome b5 to membrane vesicles. Protein Expr Purif. 2008;58:229–241. doi: 10.1016/j.pep.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carvalho MD, Carvalho JF, Merrick WC. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Arch Biochem Biophys. 1984;234:603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- 33.Yeh CH, Chang PF, Yeh KW, Lin WC, Chen YM, Lin CY. Expression of a gene encoding a 16.9-kDa heat-shock protein, Oshsp16.9, in Escherichia coli enhances thermotolerance. Proc Natl Acad Sci USA. 1997;94:10967–10972. doi: 10.1073/pnas.94.20.10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enoch HG, Fleming PJ, Strittmatter P. Cytochrome b5 and cytochrome b5 reductase-phospholipid vesicles. Intervesicle protein transfer and oreintation factors in protein-protein interactions. J Biol Chem. 1977;252:5656–5660. [PubMed] [Google Scholar]
- 35.Roberts BE, Paterson BM. Efficient translation of tobacco mosaic virus RNA and rabbit globin 9S RNA in a cell-free system from commercial wheat germ. Proc Natl Acad Sci U S A. 1973;70:2330–2334. doi: 10.1073/pnas.70.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokoyama S. Protein expression systems for structural genomics and proteomics. Curr Opin Chem Biol. 2003;7:39–43. doi: 10.1016/s1367-5931(02)00019-4. [DOI] [PubMed] [Google Scholar]
- 37.Ishihara G, Goto M, Saeki M, Ito K, Hori T, Kigawa T, Shirouzu M, Yokoyama S. Expression of G protein coupled receptors in a cell-free translational system using detergents and thioredoxin-fusion vectors. Protein Expr Purif. 2005;41:27–37. doi: 10.1016/j.pep.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Vinarov DA, Loushin Newman CL, Markley JL. Wheat germ cell-free platform for eukaryotic protein production. FEBS J. 2006;273:4160–4169. doi: 10.1111/j.1742-4658.2006.05434.x. [DOI] [PubMed] [Google Scholar]
- 39.Endo Y, Sawasaki T. Cell-free expression systems for eukaryotic protein production. Curr Opin Biotechnol. 2006;17:373–380. doi: 10.1016/j.copbio.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Calhoun KA, Swartz JR. Total amino acid stabilization during cell-free protein synthesis reactions. J Biotechnol. 2006;123:193–203. doi: 10.1016/j.jbiotec.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Boyer ME, Wang CW, Swartz JR. Simultaneous expression and maturation of the iron-sulfur protein ferredoxin in a cell-free system. Biotechnol Bioeng. 2006;94:128–138. doi: 10.1002/bit.20830. [DOI] [PubMed] [Google Scholar]
- 42.Abe M, Ohno S, Yokogawa T, Nakanishi T, Arisaka F, Hosoya T, Hiramatsu T, Suzuki M, Ogasawara T, Sawasaki T, Nishikawa K, Kitamura M, Hori H, Endo Y. Detection of structural changes in a cofactor binding protein by using a wheat germ cell-free protein synthesis system coupled with unnatural amino acid probing. Proteins. 2007;67:643–652. doi: 10.1002/prot.21341. [DOI] [PubMed] [Google Scholar]
- 43.Jewett MC, Swartz JR. Substrate replenishment extends protein synthesis with an in vitro translation system designed to mimic the cytoplasm. Biotechnol Bioeng. 2004;87:465–472. doi: 10.1002/bit.20139. [DOI] [PubMed] [Google Scholar]
- 44.Blommel PG, Becker KJ, Duvnjak P, Fox BG. Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnol Prog. 2007;23:585–598. doi: 10.1021/bp070011x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abe M, Hori H, Nakanishi T, Arisaka F, Ogasawara T, Sawasaki T, Kitamura M, Endo Y. Application of cell-free translation systems to studies of cofactor binding proteins. Nucleic Acids Symp Ser (Oxf) 2004:143–144. doi: 10.1093/nass/48.1.143. [DOI] [PubMed] [Google Scholar]
- 46.Boyer ME, Stapleton JA, Kuchenreuther JM, Wang CW, Swartz JR. Cell-free synthesis and maturation of [FeFe] hydrogenases. Biotechnol Bioeng. 2008;99:59–67. doi: 10.1002/bit.21511. [DOI] [PubMed] [Google Scholar]
- 47.Klammt C, Schwarz D, Fendler K, Haase W, Dotsch V, Bernhard F. Evaluation of detergents for the soluble expression of alpha-helical and beta-barrel-type integral membrane proteins by a preparative scale individual cell-free expression system. FEBS J. 2005;272:6024–6038. doi: 10.1111/j.1742-4658.2005.05002.x. [DOI] [PubMed] [Google Scholar]
- 48.Wuu JJ, Swartz JR. High yield cell-free production of integral membrane proteins without refolding or detergents. Biochim Biophys Acta. 2008;1778:1237–1250. doi: 10.1016/j.bbamem.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 49.Strittmatter P, Thiede MA, Hackett CS, Ozols J. Bacterial synthesis of active rat stearyl-CoA desaturase lacking the 26-residue amino-terminal amino acid sequence. J Biol Chem. 1988;263:2532–2535. [PubMed] [Google Scholar]