Abstract
DNA polymerases play a crucial role in the cell cycle due to their involvement in genome replication and repair. Understanding the reaction mechanism by which these polymerases carry out their function can provide insights into these processes. Recently, the crystal structures of human DNA polymerase λ (Polλ) have been reported both for pre- and post- catalytic complexes (García-Díaz et al., DNA Repair, 3, 1333, 2007). Here we employ the pre-catalytic complex as a starting structure for the determination of the catalytic mechanism of Polλ using ab initio quantum mechanical/molecular mechanical methods. The reaction path has been calculated using Mg2+ and Mn2+ as the catalytic metals. In both cases the reaction proceeds through a two step mechanism where the 3′-OH of the primer sugar ring is deprotonated by one of the conserved Asp residues (D490) in the active site before the incorporation of the nucleotide to the nascent DNA chain. A significant charge transfer is observed between both metals and some residues in the active site as the reaction proceeds. The optimized reactant and product structures agree with the reported crystal structures. In addition, the calculated reaction barriers for both metals are close to experimentally estimated barriers. Energy decomposition analysis to explain individual residue contributions suggests that several amino acids surrounding the active site are important for catalysis. Some of these residues, including R420, R488 and E529, have been implicated in catalysis by previous mutagenesis experiments on the homologous residues on Polβ. Furthermore, Polλ residues R420 and E529 found to be important from the energy decomposition analysis, are homologous to residues R183 and E295 in Polβ, both of which are linked to cancer. In addition, residues R386, E391, K422 and K472 appear to have an important role in catalysis and could be a potential target for mutagenesis experiments. There is partial conservation of these residues across the Pol X family of DNA polymerases.
Keywords: DNA polymerase lambda, QM/MM, molecular dynamics, simulations, polymerase catalysis
1 Introduction
DNA based organisms must replicate their genomes accurately and also remove and replace damaged nucleotides to maintain genome stability. These are critical processes since errors in replication and/or repair can result in mutations, some of which can lead to disease or even death [1, 2, 3]. DNA polymerases are the proteins in charge of the processes involved in DNA replication and repair. They are classified into several distinct families based on differences in the primary structure of their catalytic subunits [4].
All transactions associated with the processes involving DNA polymerases depend on the reaction catalyzed by these enzymes. This reaction is based on a nucleophylic attack of the O3′ of the primer-terminal nucleotide on the Pα of the incoming nucleotide to form an O-P bond, with subsequent release of pyrophosphate (PPi). The mechanism involved in this transfer has been hypothesized to occur with the help of two metal ions [5, 6]. Experimental and theoretical studies on the reaction mechanism of several polymerases support this hypothesis [7, 8, 9, 10, 11, 12, 13].
Theoretical studies on the reaction mechanism of DNA polymerase β (Polβ) have suggested that the reaction is largely associative. One study using QM calculations on model systems [14] has proposed a direct proton transfer from the O3′ to an O atom on the Pα. In another study, a full QM/MM treatment on the same system suggests the proton transfers from the primer O3′ to an aspartate residue in the active site [15]. This latter mechanism is similar to the one proposed for T7 DNA polymerase [16, 17]. Conversely, theoretical calculations on the mechanism of DNA polymerase IV from Sulfolobus solfataricus have proposed the proton transfer to happen through an ordered water [18]. This pathway has also been proposed previously as a likely mechanism for Polβ [9, 19]. In all cases the phosphate-breaking step has been proposed to be associative-like, although the general base may differ.
In addition to the chemistry involved in the catalytic step, the question arises as to the nature of the metal. Recent studies on DNA polymerase λ (Polλ) have suggested a slight preference for Mn2+ over Mg2+ as the activating metal based on lower activation energy for the former metal [20]. However, for the homologous Polβ such preference is not observed [21].
Polλ is a family X polymerase that fills short gaps during DNA repair. It has been implicated in both non-homologous end-joining (NHEJ) of double stranded DNA breaks and in base excision repair of damaged bases [22, 23, 24, 25, 26]. Polλ shares 35% amino acid identity with Polβ, another family X polymerase [27, 28, 29, 30]. Experimentally, it has been shown that Polβ under-goes a large conformational change involving subdomain motion needed for catalysis [12]. On the other hand this large subdomain motion is not observed in Polλ [31, 32]. However, this conformational change appears not to be rate limiting and it is possible that the rate limiting step is the same (or similar) for both Polλ and Polβ.
Recently, several X-ray structures of human Polλ have been obtained [33]. These structures include a pre-catalytic ternary complex (pdb id 2PFO) with the primer terminus nucleotide poised for an in-line attack of the ribose O3′ to the Pα of the incoming non-hydrolyzable dUPNPP, with a Mn2+ in the catalytic metal site and a Mg2+ in the “dNTP-binding” position. In addition, a post-catalytic structure (pdb id 2PFQ) was obtained where a dCTP has been covalently added to the primer terminus with both metal sites occupied by Mn2+.
In the present contribution, we take the reported pre-catalytic X-ray structure (2PFO) as the starting point for quantum mechanical/molecular mechanical (QM/MM) calculations [34, 35, 36]. Two sets of calculations are performed with either Mg2+ or Mn2+ as the divalent metal ions in the active site to investigate whether there is a preference and/or different mechanism in the activation process. This is the first such theoretical study that compares the catalysis of a DNA polymerase with two different metals (Mg2+ and Mn2+) in the active site. Our calculations provide further support for the hypothesized two metal mechanism and show that both metals experience a significant charge transfer to some residues in the active site during the reaction. In addition, an energy decomposition analysis provides insight into the catalytic role of amino acid residues surrounding the active site, several of which are conserved among family X polymerases [37].
2 Methods
Molecular dynamics (MD) [38, 39, 40] and QM/MM [41, 42, 43, 44, 45, 46] calculations have been performed to investigate the reaction mechanism of human Polλ. MD simulations were carried out with the AMBER9 suite of programs [47]. All QM/MM calculations were performed with modified versions of Gaussian03 and TINKER for the QM/MM reaction path calculations [48, 49]. The QM/MM optimizations were performed with an iterative method as described in [35] and [36] using the pseudobond model for all boundary atoms [34, 50]. Reaction paths were obtained using a combination of the quadratic string method (QSM) [51], reaction coordinate driving (RCD) [52] and quadratic synchronous transit (QST3) [53, 54].
The initial structure of Polλ was taken from the pre-catalytic X-ray structure (pdb id 2PFO) [33]. This pre-catalytic structure contains a non-hydrolyzable nucleotide analog (dUPNPP), a Mg2+ as the dNTP-binding metal and a Mn2+ in the catalytic site. This structure was modified by replacing the dUPNPP by dUTP and the Mn2+ by Mg2+. Hydrogens were added using the XLEaP program in AMBER9 [47]. This program was also used to solvate the structure in a box of 5856 water molecules. The system was minimized and subjected to molecular dynamics (MD) simulations using the PMEMD module of AMBER9 [47]. A total of 2 ns of MD was carried out to equilibrate the system and the last snapshot was selected as the starting point for the QM/MM calculations. The final configuration was modified by retaining all atoms in the protein and all water molecules within 30 Å of the catalytic metal for the subsequent QM/MM calculations (see Supplementary Information for details).
Following equilibration, the QM subsystem was chosen to include both active site metals, side chains of D427, D429, D490, the primer dC nucleotide (excluding C5′ and the phosphate group), a part of the incoming dUTP nucleotide (triphosphate and C5′) and two water molecules that complete the coordination spheres of the metals, for a total of 72 QM atoms including 5 boundary atoms (Cα of aspartates, C5′ of dC and C4′ of dUTP). The total charge of the dUTP was set to −3 with one protonated oxygen on the γP as done in previous theoretical studies on Pol β and HIV reverse transcriptase [11, 15]. Note that we have not changed the incoming nucleotide to the canonical Watson-Crick pair (dTTP). This was done in order to avoid the docking of dTTP in the place of dUTP, and to remain as close as possible to the initial X-ray structure.
Two sets of calculations were performed with either Mg2+ or Mn2+ as active site metals. In the case of Mn2+ as the catalytic metal, all calculations were performed assuming a high spin state with all d electrons on both Mn atoms unpaired (see Supplementary Information) [55, 56, 57]. The QM subsystem was treated at the B3LYP level [58, 59] with a combined basis set where the 6–31G* basis was used for all reactive atoms (OD1 from D490, OD1 from D279, O in the ordered water, O3′, O3α and Pα), except for the Pα where an additional diffuse function was included. All other (non-reactive) QM atoms were represented with 3–21G. The Mn2+ atoms were treated with the LANL2DZ pseudopotential. The remaining atoms were included in the MM subsystem and treated with the parm99 force field [47].
Although there is a post-catalytic structure for Polλ [33], it contains a different nucleotide (dTTP) as the newly added base than the reactant structures. Because of this, the product structures for both systems (Mg2+ and Mn2+) were produced in silico from the reactant structure and subjected to QM/MM optimization. The optimized end-points were employed for the reaction path calculation with the quadratic string method (QSM) [51]. This is a “chain-of-replica” method where the path is represented by a discrete number of structures [44, 60, 61, 62, 63]. In QSM the intermediate structures are obtained by a linear interpolation between the end points. Subsequently, all structures are optimized to the minimum energy path simultaneously. This affords an unbiased optimization of the path, which is highly efficient because all images on the path are calculated in parallel. The initial QSM paths consist of 10 points including end-points and were iteratively optimized with constrained MM-optimization [64, 65, 63]. After the initial paths were converged, the highest energy points were optimized to the closest transition state (TS) with the quadratic synchronous transit (QST3) method [53, 54]. Following the TS optimization the paths between the critical points were calculated with either QSM or RCD, see supplementary materials for details.
To verify the applicability of the basis set, an initial QSM path with 10 total points was calculated for the Mg2+ with all atoms being treated at the B3LYP/6-31G* level (6-31+G* for the Pα). This path was found to be the same as the ones calculated with the combined basis set and the barrier was ≈2.5 kcal/mol higher. In this case the highest energy point has not been optimized to the TS and the energy could drop around 1–1.5 kcal/mol. Therefore, based on this result we decided to perform the remaining calculations with the combined basis set for computational efficiency.
3 Results and Discussion
3.1 Reaction Paths
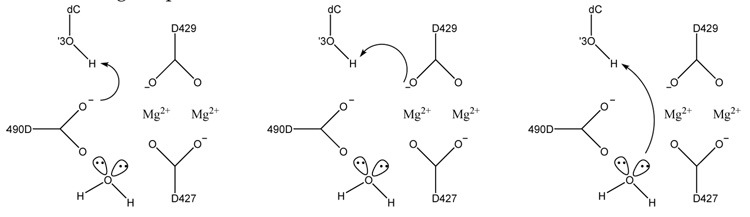
In this subsection we provide the results for the reaction mechanisms. In this case, we have decided to explore different reaction pathways based on previous studies [15, 18]. The different schemes representing the possible pathways differ in the general base used to deprotonate the O3′ of the primer nucleotide. These possible bases include D490, D429 or an ordered water that completes the coordination sphere of the catalytic metal (see Fig. 1).
Figure 1.
Tested reaction schemes for the Polλ catalyzed reaction. Scheme 1 (left): proton transfer to D490, scheme 2 (middle): proton transfer to D429, scheme 3 (right): proton transfer to ordered water.
The first scheme corresponds to the proton transfer from O3′ to the closest OD of D490. This scheme is equivalent to the one proposed by Lin et al. for human DNA polymerase β [15]. In scheme 2, the proton is transferred to the closest OD in D429. Scheme 3 is based on a recently proposed mechanism for DNA polymerase IV from Sulfobolus solfataricus where the proton is transferred to an ordered water molecule [18]. In this case only the structure with Mg2+ was taken into account to determine the proton acceptor.
The calculated potential energy barrier associated with the reaction path for scheme 1 is 17.6 kcal/mol (see Fig. S1 in supplementary materials ). The calculated reaction barrier for scheme 2 is very close to that of scheme 1, however, the optimized path (Fig. S2 in supplementary materials) shows that the reaction proceeds by an initial proton transfer to D490. This is followed by the breaking of the Pα-Oβ bond and ending with the direct proton transfer from D490 to D429. Based on this, we have ruled out scheme 2 since the initial step for this mechanism is equivalent to scheme 1.
In the case of the proton transfer to the ordered water, we obtained a reaction barrier of 35.4 kcal/mol. Another related mechanism involves a proton transfer through this water molecule to an O on the Pα [18]. The reaction energy for this mechanism was found to be only 2 kcal/mol lower than the one for scheme 3. Note that in the present study, one oxygen is protonated in the Pγ, whereas Wang et al. used a deprotonated triphosphate for their calculations [18]. In order to test this difference in protonation states, the reactant and product for scheme 3 were re-optimized with the deprotonated triphosphate. In our hands, the calculated reaction energy for scheme 3 in Polλ with the deprotonated triphosphate is 41.7 kcal/mol. In addition, scheme 1 was also tested with the deprotonated phosphate structures, with a resulting reaction energy of 25.2 kcal/mol. Experimentally, two proton transfers have been observed for this reaction [66], one corresponds to the deprotonation of the O3′ and the other is hypothesized to be the protonation of the phosphate. This protonation could be done by a protonated side chain or from a proton in the solvent. In this case, present and previous results suggest that the O3′ deprotonation is rate limiting, and could suggest that the protonation of the phosphate may happen after binding in the active site (as assumed in the present case), or after PPi formation as in [9, 14] with a lower barrier than for the first proton transfer. Also, experimentally it is not known if the PPi is deprotonated based on its pKa values which range between 7.0–9.0 [66].
Based on the above observations for the three different reaction mechanisms, our results point to scheme 1 with the protonated phosphate as the preferred mechanism for Polλ. This scheme is similar to the previously proposed mechanism by Lin et al. [15] for human Polβ where the proton is transfered to the homologous D256 residue. Conversely, in our hands the water-mediated mechanism proposed by Wang et al. [18] appears to not be viable for Polλ because of the high reaction energies associated with this scheme. From these results as well as those previously published it is difficult to generalize these reaction mechanisms to all the members of the X and Y family polymerases at this point [16, 17, 19, 14, 15, 9]. However, although difficult to generalize based on the general base, it appears that the mechanism is associative-like [66].
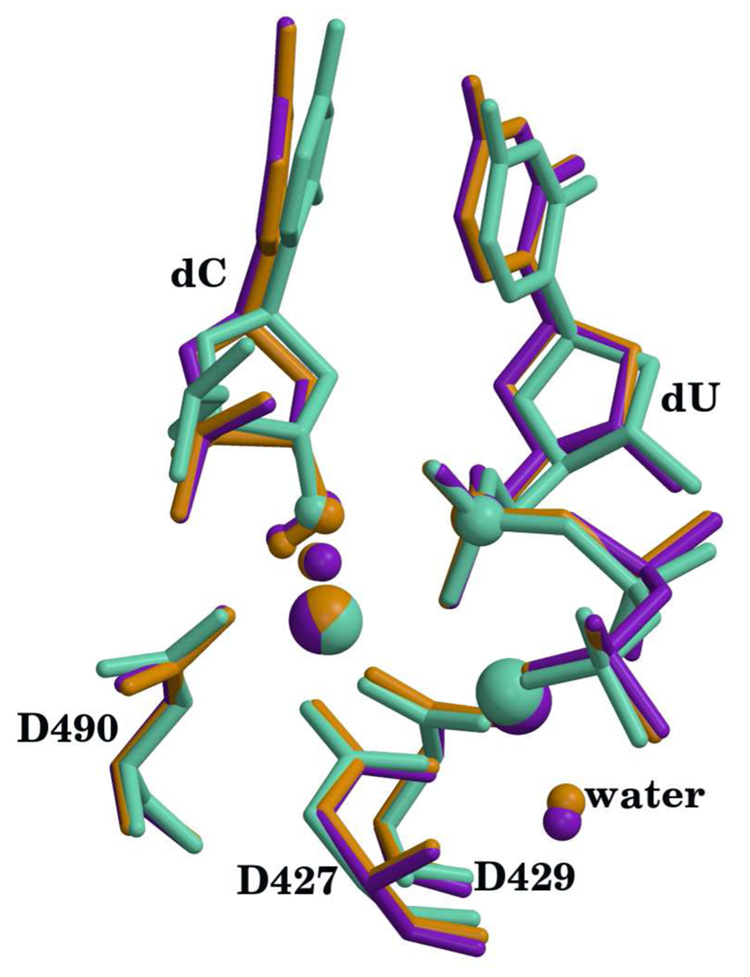
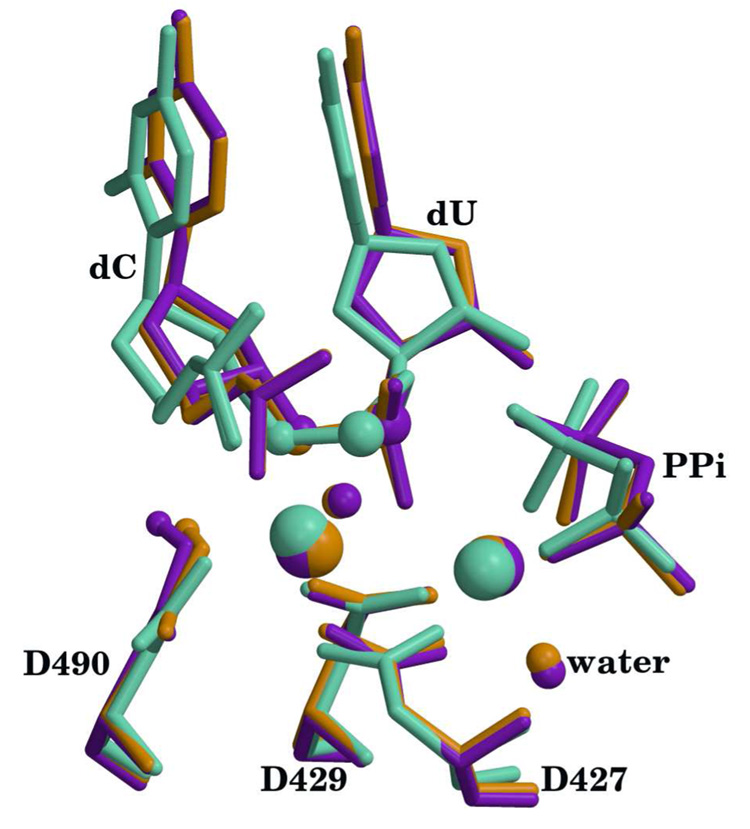
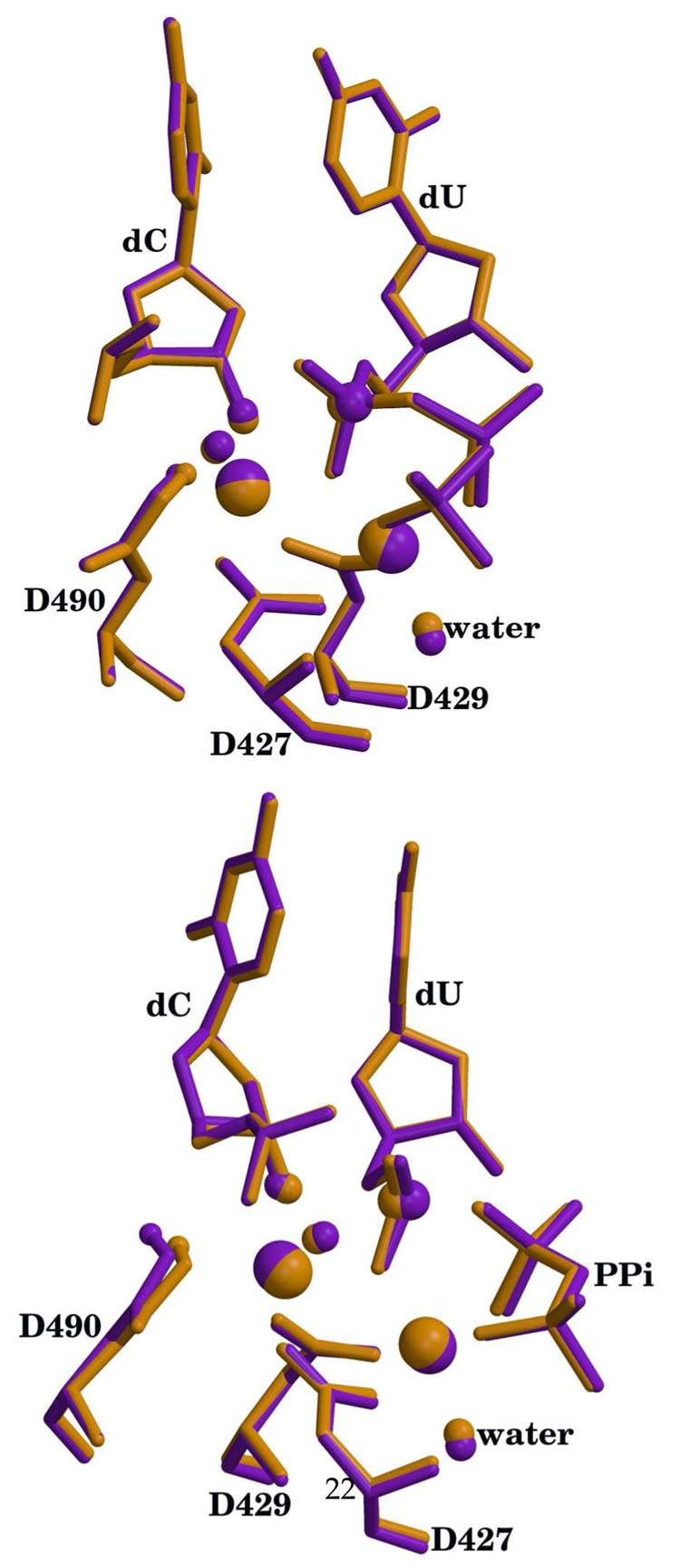
The relaxed and optimized reactant systems with Mg2+ and Mn2+ both show an RMSD of 1.7 Å for backbone atoms with respect to the 2PFO X-ray reference. This difference comes from the MD relaxation of the entire enzyme. However, the superposition of the active site residues (D427, D429, D490, dC, dUTP and metals) yields an RMSD below 0.3 Å for all heavy atoms in both Mg2+ and Mn2+ systems (see Figure 2). In the case of the product systems, the RMSD between the relaxed and optimized structures to the X-ray reference (2PFQ) is 1.9 Åfor all heavy atoms. Again, as in the reactant case, the RMSD for all heavy atoms in the active site residues remains around 0.2 Å (see Fig. 3). Note that this is a very good agreement, especially since the product structure was directly generated from the reactant via an optimization with the QM/MM method.
Figure 2.
Superposition of active sites for the calculated reactants with the X-ray crystal structure (pdb id 2PFO). X-ray structure is shown in green, Mn2+ structure is shown in purple and Mg2+ structure is shown in orange. Hydrogen atoms except for the O3′-H have been omitted for clarity. The metals, O3′, Pα, waters that complete the metal coordination and the H atom on O3′ are highlighted as spheres. Catalytic metal is on the left and nucleotide binding metal is on the right. Metal ligand distances are reported in table S1 (supplementary materials).
Figure 3.
Superposition of active sites for the calculated products with the X-ray crystal structure (pdb id 2PFQ). X-ray structure is shown in green, Mn2+ structure is shown in purple and Mg2+ structure is shown in orange. Hydrogen atoms except for the transferred proton have been omitted for clarity. The metals, O3′, Pα, waters that complete the metal coordination and the transferred proton are highlighted as spheres. Catalytic metal is on the left and nucleotide binding metal is on the right. Metal ligand distances are reported in table S1 (supplementary materials).
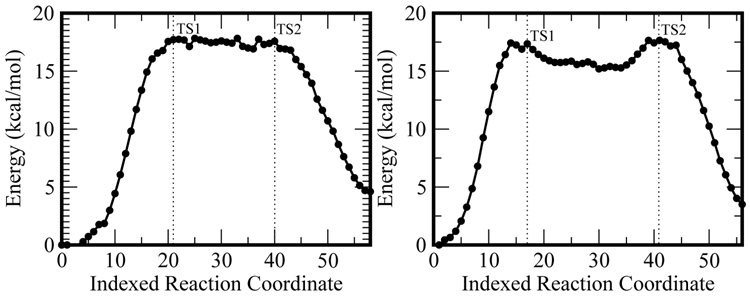
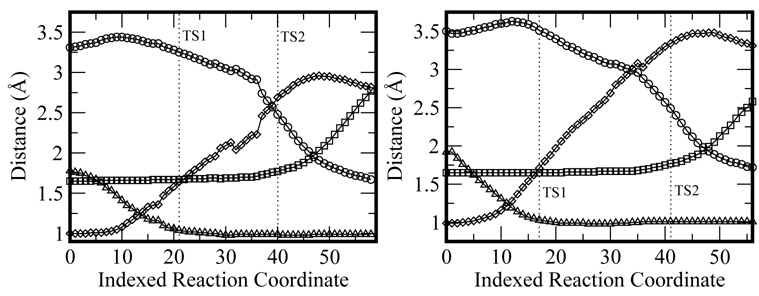
As seen in Figure 4 both the Mg2+ and Mn2+ paths show a large initial energy increase, which corresponds to the proton transfer. This is followed by a high energy, rugged valley characterized by the breaking of the Pα-Oβ bond. Finally the nucleotide is transferred and pyrophosphate is formed in the product structure. Note that in both cases, the product structures are around 4–5 kcal/mol higher in energy than the reactants. Overall, this mechanism is similar to the one proposed for Polβ, with the exception that the reaction energy for Polβ is negative and we do not observe a stable low energy intermediate after the proton transfer [15].
Figure 4.
Calculated reaction paths for the Mg2+(left) and Mn2+ (right) catalyzed reactions.
Two TSs are observed along the paths for Mg2+ and Mn2+ In the case of the Mg2+ the optimized transition states (TSs) give a total energy barrier of 17.6 and 16.4 kcal/mol with respect to the reactant and a frequency calculation revealed a single imaginary (negative) frequency for each maximum. For the Mn2+ the TSs are located at 16.8 and 17.6 kcal/mol respectively. A single imaginary frequency was obtained for each of these structures as well. Additionally in both cases the first TS was observed to correspond to the proton transfer and the second to the Pα-Oβ bond break (see Fig. 5).
Figure 5.
Superposition of active sites for the calculated transition states (TS1 top, TS2 bottom). Mg2+ structures are shown in orange and Mn2+ structures are shown in purple. Hydrogen atoms except for the transferred proton have been omitted for clarity. The metals, O3′, Pα, waters that complete the metal coordination and transferred proton are highlighted as spheres. Catalytic metal is on the left and nucleotide binding metal is on the right. Metal ligand distances are reported in table S1 (supplementary materials).
The calculated barriers are close to the experimental counterpart, where a kcat of 6.0 s−1 has been reported for the correct insertion of dT opposite dA by human Polλ in the presence of Mg2+. This corresponds to a barrier of 16.6 kcal/mol [8], calculated from pre-steady state kinetic studies for the incorporation of a single nucleotide via the transition state formula (TST) [67]. The barriers estimated using TST assume that the catalytic step is rate limiting and thus that the activation free energy ΔG‡ corresponds to the energy barrier. Note that our calculated barriers correspond to potential energy and the estimated experimental barriers correspond to free energy. Usually, the calculated potential energy barriers provide an upper bound to the corresponding free energy barriers in QM/MM calculations [36, 18]. Therefore, it can be expected that a free energy calculation would lower the value of the barriers closer to the experimental estimate.
Furthermore, it has been estimated experimentally that the energy barrier for the Mn2+ catalyzed reaction in human Polλ is between 0.4–0.9 kcal/mol lower than the corresponding Mg2+ one [20]. In the present work, the energy difference in the barriers is not large enough to determine an ion preference or whether either TS is rate limiting. In addition, based on the approximations inherent in the methodology, the estimated experimental difference is too small and falls within the error of the method.
Figure 5 shows the superposition of the optimized transition states for both metals. As can be seen, the first TS of the Mg2+ system is very similar to the first TS of the Mn2+ system, where the proton has been transferred to D490. In the case of the second TS both structures show that the Pα-Oβ bond is broken. The only other major difference between the second TS of the Mg2+ and that of the Mn2+ systems is a slightly larger distance between the transfered proton and O3′ in the Mn2+ system. These trends are best observed in Figure 6, where selected distance changes along the reaction coordinate are presented. Note that as the reaction progresses from the first to the second TS, the O3′-Pα decreases considerably while the Pα-Oβ increases. Additionally, as can be seen in Figure 5, the Pα in the second TS shows a distorted trigonal bipyramidal phosphorane, resulting in an associative-like mechanism.
Figure 6.
Selected distance changes along the reaction coordinate for the Mg2+(left) and Mn2+ (right) catalyzed reactions. Circles: O3′-Pα; squares: Pα-O; rhombus: O3′-H; triangles: H-O(D490).
It is important to note that in our calculations the distance between the metals, either Mg2+ or Mn2+, remains almost constant along the path. The observed fluctuations for the inter-metal distances are less than 0.1 Å over the range of the reaction, with an average distance of 3.5 Å for Mg2+ and 3.7 Å for Mn2+, metal-ligand distances for reactants, TSs and products are given in the supplementary materials.
The analysis of the charge distribution in the QM subsystem presents an interesting picture of the reaction mechanism. Changes in charge for selected groups in the active site at the stationary points on the path (TSs and end-points) are shown in Table 1. As can be seen the overall charge transfer to D490, O3′, αP and the PPi are similar for the Mg2+ and Mn2+ catalyzed reactions, with a significant reduction in the negative charge on O3′ and a similar reduction in the positive charge on the PPi. Note however, that in the case of the Mg2+ both metals experience a large charge transfer to D427, D490 and the water molecule coordinated to the dNTP-binding Mg2+. On the other hand, for Mn2+ there is an observed charge transfer from the metals in the TSs, however, there is no overall change between the reactant and the product. The difference in charge transfer from the metals to the ligands may be due to the use of different basis sets/methodology for Mg2+ and Mn2+.
Table 1.
Changes in ESP fitted charges in the QM subsystem with respect to the Reactant (in electron units). Only changes larger than |0.1| units in a given step are reported.
| Mg2+ | Mn2+ | |||||
|---|---|---|---|---|---|---|
| TS1 | TS2 | Product | TS1 | TS2 | Product | |
| D427 | .26 | .28 | .15 | 0. | 0. | 0. |
| D429 | .11 | 0. | −.12 | −.1 | 0. | −.1 |
| D490 | .10 | .35 | .28 | 0. | .16 | .28 |
| dNTP-binding Mea | −.31 | −.19 | −.16 | 0. | .13 | 0. |
| catalytic Meb | −.24 | −.52 | −.14 | .16 | −.12 | 0. |
| dC sugar | 0. | 0. | −.10 | 0. | 0. | 0. |
| O3′ | .10 | .28 | .48 | 0. | .2 | .46 |
| PPi (including O) | 0. | −.10 | −.39 | 0. | 0. | −.38 |
| Pα (including O) | 0. | .14 | .10 | 0. | .12 | .10 |
| H2O (on struct. Me)c | .12 | .11 | .1 | 0. | 0. | 0. |
Metal (Me) that coordinates the PPi.
Metal that coordinates the O3′and Pα.
Water molecule coordinated to the dNTP-binding metal.
3.2 Residue Analysis
Further insights into the catalytic mechanism of Polλ may be obtained by understanding the role of individual residues in catalysis. To that end, the non-bonded interaction energy between the MM environment and the QM subsystem can be decomposed into contributions per residue [68, 69]. Note that this “breakdown” is only obtained in an approximate manner and is only qualitative. In this analysis the differences in interaction energies are calculated between individual residues and the QM subsystem when the system goes from reactant to TS(s). A negative contribution denotes a stabilizing contribution to the TS and a positive one denotes a destabilizing contribution. We consider a residue to provide a significant contribution to (de)stabilization if the interaction is greater than 1 kcal/mol, see supplementary materials for details.
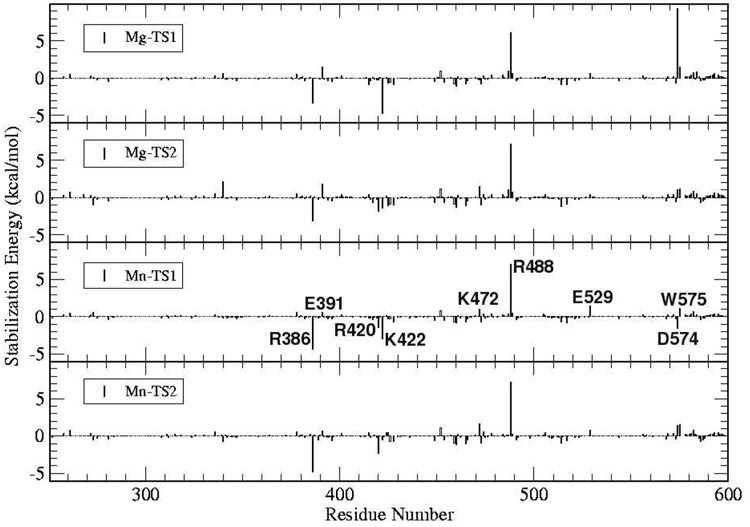
Several residues are observed to have a significant effect on the TS barrier (see Fig. 7). Coulomb and Van der Waals analysis show that a significant part of the stabilization provided by the MM environment comes from the solvent (see Supplementary Material). Overall most of the interaction differences observed are electrostatic. The only Van der Waals (VdW) interactions that were observed were for the TSs of Mg2+ where an MM water has a contribution of approximately 3 kcal/mol, and for the second TS of the Mn2+ reaction, where R488 and a water molecule have an interaction of around 1 kcal/mol (see Supplementary Material).
Figure 7.
Electrostatic stabilization energy per residue for the Mg2+ and Mn2+ catalyzed reactions (protein only).
From Fig. 7 it can be seen that there are a number of residues that have a significant contribution to the stabilization of the TSs. In particular R386, E391, R420, K422, K472, R488, E529, D574 and W575 are observed for both Mg2+ and Mn2+. None of these residues have been mutated experimentally on Polλ to our knowledge. However, the homologous residues to R420, R488 and E529 in Polβ have been experimentally studied. Note that the comparison between our calculations and the experimental mutagenesis results is only qualitative since the determined energy change is only a first order approximation to the TS (de)stabilization and does not include structural or dielectric screening contributions.
R420
This residue shows an important stabilizing contribution to the TSs for both catalytic metals of around −1 kcal/mol in Mg2+ and −2 kcal/mol for Mn2+. Mutation of the equivalent Arg-183 to Ala in Polβ produces a reduction greater than 99% in enzyme activity [70]. Moreover, this mutation has been observed in some esophageal cancers [2]. Castro et al. have suggested the possibility of homologous residues in other polymerases to act as a general acid that protonates the pyrophosphate in their hypothesized two proton transfer mechanism [66]. Incidentally, R420 happens to be the corresponding residue for this proposed mechanism. The protonation of the triphosphate by this residue is currently under investigation.
R488
Experimental studies have shown that the mutation of the homologous R254 in rat Polβ to Ala decreases the kcat from 51 min−1 to 1 min−1 for an experimental ΔΔG of 2.33 kcal/mol [71]. From Figure 8 it can be seen that R488 forms a salt bridge with D490. In our analysis, this residue provides a significant destabilizing contribution of around 6 kcal/mol. The protonation of D490 reduces its acid strength and thereby reduces the strength of the R488 interaction. The resulting destabilization of the TS by R488 can be attributed to the tight coupling between these two residues. This destabilization does not take into account any structural changes that result from the experimental mutation of this residue. The experimentally observed reduction in kcat could be due to the fact that the role of R488 may also be structural, by orienting D490 for a proper geometry of the coordination sphere of the catalytic metal and its proper positioning to deprotonate the O3′. Indeed, salt bridges between R488 and D490 are observed in the X-ray structures with Na, Mg and Mn (pdbid 2PFN 2PFO and 2PFQ respectively) with average distance of 2.8 Å. Therefore, by mutating the homologous R254 to A in Polβ, the salt bridge is removed and the structural integrity of the catalytic aspartate could be compromised. However, a minor alteration in the D256-R254 salt bridge distance is noticed (2.95 Å in the 1BPY X-ray structure).
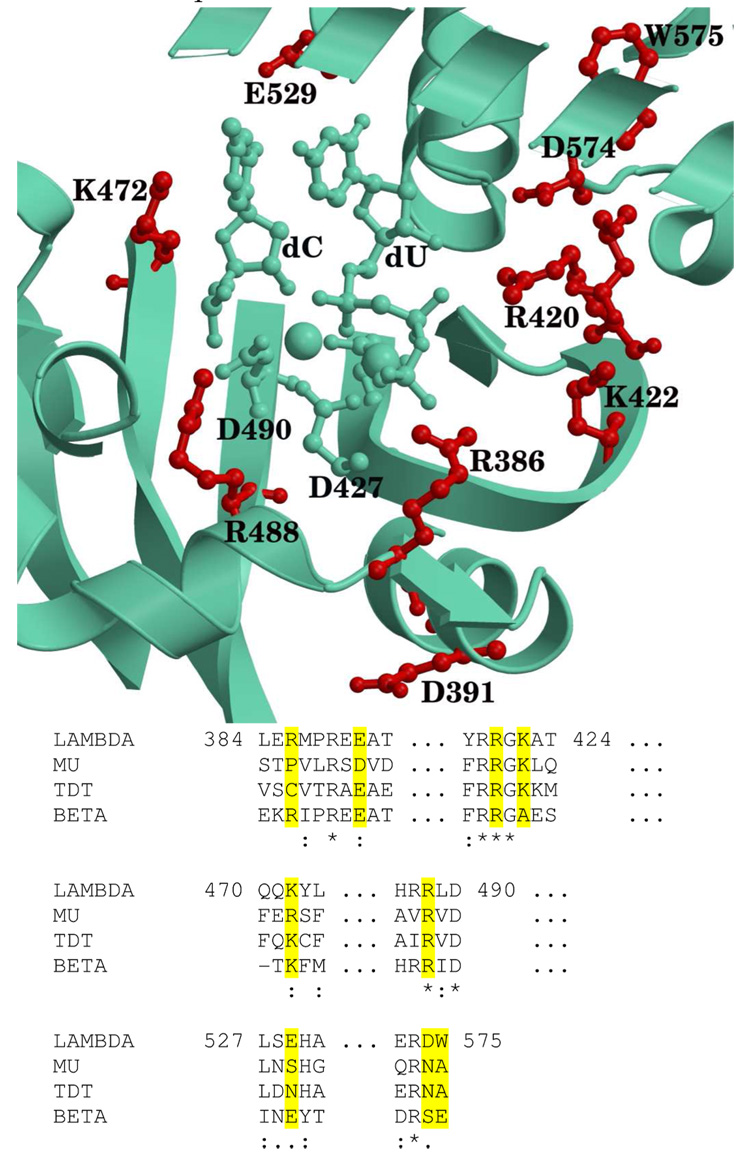
Figure 8.
Active site positions of selected residues in the Mg2+ system (top) that contribute to the (de)stabilization of the TS during catalysis. The residues shown in red correspond to those which are common to all TSs or have a large contribution (see text). Bottom: Sequence alignment of the X family polymerases for the selected residues (highlighted in yellow). Key: “*”indicates residues conserved in all sequences,“:” indicates residues conserved in at least three of the four sequences, “.” indicates homologous residues present in at least three of the four sequences.
E529
Our results show that this residue also results in a destabilizing contribution of around 1 kcal/mol for both metals. This may be the result of the interaction with the dC base. Experimentally, this residue has been linked to gastric cancer in Pol β [1, 2]. Mutation of the homologous residue in Polβ to a lysine inhibits the base excision repair mechanism [1].
To our knowledge these are the only residues that have been studied experimentally and correspond to any of the residues suggested to have a significant contribution to catalysis in our residue analysis. The remaining residues could provide good targets for experimental mutagenesis studies. In particular K472 is located at the end of a loop region that is five residues longer in Polλ than in Polβ. The role of this loop and the residues involved in this loop are currently under experimental investigation. Although the residues obtained from this analysis are catalytically important, they have not been included in the QM subsystem due to computational limitations and due to the fact that it has been previously shown that residues that show significant non-bonded interactions with the QM subsystem can be adequately modeled with the MM force field [69].
To further understand the catalytic role of these residues, we have performed a sequence alignment of Polλ with the sequences of other X family polymerases as shown in Figure 8 bottom (see Fig. S11 in supplementary materials for full alignment), as in a previous study [37]. This alignment, performed with T-Coffee [72], shows that the equivalent residues for R420 and R488 in Polλ are totally conserved among X family polymerases. K472 is highly conserved except in Polµ where there is an arginine instead. The remaining residues are conserved to some degree, either by identity in a couple of polymerases, or by residue function, e.g., charge of the residue. These results suggest that the conserved residues could have a role in the enzyme function and non-conserved residues could be involved in the specificity and/or selectivity of the different polymerases.
4 Conclusions
The reaction mechanism for human Polλ has been determined by means of QM/MM calculations for two catalytic metals. Three reaction schemes were tested to determine the general base that deprotonates the O3′. Our results point to D490 as the general base, the other two schemes (D429 or H2O) appear to not be viable in Polλ. In the case of D429 as the proton acceptor, the proton is transfered first to D490. For the H2O mediated scheme, the calculated reaction energies were too high. After determining the scheme, the reaction path was calculated for two systems, Mg2+ and Mn2+. In both cases, the reaction mechanism proceeds in two steps. The first step corresponds to the abstraction of the proton on O3′ by D490, followed by the breaking of the Pα-Oβ bond. This mechanism agrees with the previously proposed mechanism for human Polβ. The calculated energy barriers are around 17 kcal/mol for the Mg2+ and Mn2+ catalyzed reactions. These barriers are relatively close to the experimental determination of 16.6 kcal/mol for the incorporation of dT opposite dA with Mg2+. For both Mg2+ and Mn2+ a significant charge transfer is observed from the catalytic and dNTP-binding metals to some residues in the active site.
Energy decomposition analysis to explain individual residue contributions to catalysis was carried out. This analysis suggests that there are several residues that have a significant effect on the TS barrier. Sequence alignment studies show that some of these residues are strictly conserved among different enzymes of the Polymerase X family, pointing to a functional role for these amino acids. The other residues that are not conserved may be involved in specific enzyme requirements due to their unique roles. Among the calculated residues from the decomposition, three have been studied experimentally, R420, R488 and E529, on the corresponding residues on Polβ and two of these have been observed in certain cancers. The remaining residues have not been studied experimentally and could be an interesting target for mutagenesis studies.
Supplementary Material
Detailed methods for initial structure, QM/MM structure optimization and path optimization are given, as well as QSM paths for selected reaction schemes, metal-ligand distances, residue analysis for all residues and waters, and sequence alignment of Polλ with other Family X polymerases.
Acknowledgments
This research was supported by the intramural research program of the NIH and NIEHS. Computing time from the NIEHS and UNC computing facilities are gratefully acknowledged. GAC would like to thank Drs. S.K. Burger, Z. Lu and Prof. W. Yang at Duke University for the QSM code and Prof. Y. Zhang at NYU for enlightening discussions. We would like to thank the reviewers as well as Drs. T. Darden, B. Beard, G. Muller at NIEHS for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Iwanaga A, Ouchida M, Miyazaki K, Hori K, Mukai T. Functional mutation of DNA polymerase β found in human gastric cancer - inability of the base excision repair in vivo. Mutation Res. 1999;vol. 435:121–128. doi: 10.1016/s0921-8777(99)00036-1. [DOI] [PubMed] [Google Scholar]
- 2.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase β and cancer? Cell Cycle. 2004;vol. 3:98–1001. [PubMed] [Google Scholar]
- 3.Kunkel TA. Considering the cancer consequences of altered DNA polymerase function. Cancer Cell. 2003;vol. 3:105–110. doi: 10.1016/s1535-6108(03)00027-8. [DOI] [PubMed] [Google Scholar]
- 4.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. In Prot. Chem. 2004;vol. 69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 5.Cowan J. Structural and catalytic chemistry of magnesium-dependent enzymes. BioMetals. 2002;vol. 15:225–235. doi: 10.1023/a:1016022730880. [DOI] [PubMed] [Google Scholar]
- 6.Steitz TA. DNA- and RNA- dependent DNA polymerases. Curr. Opin. Struct. Bio. 2003;vol. 3:31–38. [Google Scholar]
- 7.Abashkin YG, Erickson JW, Burt SK. Quantum chemical investigation of enzymatic activity in DNA polymerase β. a mechanistic study. J. Phys. Chem. B. 2001;vol. 105:287–292. [Google Scholar]
- 8.Fiala KA, Abdel-Gawad W, Suo Z. Pre-steady state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase λ. Biochemistry. 2004;vol. 43:6751–6762. doi: 10.1021/bi049975c. [DOI] [PubMed] [Google Scholar]
- 9.Radhakrishnan R, Schlick T. Correct and incorrect nucleotide incorporation pathways in DNA polymerase β. Biochem. Biophys. Res. Comm. 2006;vol. 350:521–529. doi: 10.1016/j.bbrc.2006.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Showalter AK, Lamarche BJ, Bakhtina M, Su M-I, Tang K-H, Tsai M-D. Mechanistic comparison of high-fidelity and error-prone DNA polymerases and ligases involved in DNA repair. Chem. Rev. 2006;vol. 106:340–360. doi: 10.1021/cr040487k. [DOI] [PubMed] [Google Scholar]
- 11.Rungrotmongkol T, Mulholland AJ, Hannongbua S. Active site dynamics and combined quantum mechanics/molecular mechanics (QM/MM) modelling of a hiv-1 reverse transcriptase/DNA/dttp complex. J. Mol. Graph. Model. 2007;vol. 26:1–13. doi: 10.1016/j.jmgm.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Bakhtina M, Roettger MP, Kumar S, Tsai M-D. A unified kinetic mechanism applicable to multiple DNA polymerases. Biochemistry. 2007;vol. 46:5463–5472. doi: 10.1021/bi700084w. [DOI] [PubMed] [Google Scholar]
- 13.Xiang Y, Warshel A. Quantifying free energy profiles of proton transfer reactions in solution and proteins by using a diabatic FDFT mapping. J. Phys. Chem. B. 2008;vol. 112:1007–1015. doi: 10.1021/jp076931f. [DOI] [PubMed] [Google Scholar]
- 14.Bojin MD, Schlick T. A quantum mechanical investigation of possible mechanisms for the nucleotidyl transfer reaction catalyzed by DNA polymerase β. J. Phys. Chem. B. 2007;vol. 111:11244–11252. doi: 10.1021/jp071838c. [DOI] [PubMed] [Google Scholar]
- 15.Lin P, Pedersen LC, Batra VK, Beard WA, Wilson SH, Pedersen LG. Energy analysis of chemistry for correct insertion by DNA polymeraseβ. Proc. Natl. Acad. Sci. 2006;vol. 36:13294–13299. doi: 10.1073/pnas.0606006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Florián J, Goodman MF, Warshel A. Computer simulation of the chemical catalysis of DNA polymerases: Discriminating between alternative nucleotide insertion mechanisms for T7 DNA polymerase. J. Am. Chem. Soc. 2003;vol. 125:8163–8177. doi: 10.1021/ja028997o. [DOI] [PubMed] [Google Scholar]
- 17.Florián J, Goodman MF, Warshel A. Computer simulations of protein functions: Searching for the molecular origin of the replication fidelity of DNA polymerases. Proc. Natl. Acad. Sci. 2005;vol. 102:6819–6824. doi: 10.1073/pnas.0408173102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Yu X, Hu P, Broyde S, Zhang Y. A water-mediated and substrate-assisted catalytic mechanism for sulfobolus solfataricus DNA polymerase IV. J. Am. Chem. Soc. 2007;vol. 129:4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberts IL, Wang Y, Schlick T. DNA polymerase β catalysis: Are different mechanisms possible? J. Am. Chem. Soc. 2007;vol. 129:11100–11110. doi: 10.1021/ja071533b. [DOI] [PubMed] [Google Scholar]
- 20.Blanca G, Shelev I, Ramadan K, Villani G, Spadari S, Hübscher U, Maga G. Human DNA polymerase λ diverged in evolution from DNA polymerase β toward specific Mn++ dependence: a kinetic and thermodynamic study. Biochemistry. 2003;vol. 42:7467–7476. doi: 10.1021/bi034198m. [DOI] [PubMed] [Google Scholar]
- 21.Battra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Structures of DNA polymerase β with active site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell. 2008 doi: 10.1016/j.molcel.2008.02.025. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bebenek K, García-Díaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda: Implications for function. J. Biol. Chem. 2003;vol. 278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 23.Shevelev I, Blanca G, Villani G, Ramadan K, Spadari S, Húbscher U, Maga G. Mutagenesis on human DNA polymerase λ: essential roles of tyr505 and phe506 for both DNA polymerase and terminal transferase activities. Nucl. Ac. Res. 2003;vol. 31:6919–6925. doi: 10.1093/nar/gkg896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García-Díaz M, Bebenek K, Gao G, Pedersen LC, London RE, Kunkel TA. Structure-function studies of DNA polymerase λ. DNA Rep. 2005;vol. 4:1358–1367. doi: 10.1016/j.dnarep.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 25.García-Díaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;vol. 124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 26.Foley MC, Schlick T. Simulations of DNA Pol λ R517 mutants indicate 517's crucial role in ternary complex stability and suggest DNA slippage origin. J. Am. Chem. Soc. 2008;vol. 130:3967–3977. doi: 10.1021/ja077982t. [DOI] [PubMed] [Google Scholar]
- 27.Braithwaite EK, Prasad R, Shock DD, How EW, Beard WA, Wilson SH. DNA polymerase λ mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J. Biol. Chem. 2005;vol. 280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 28.Braithwaite EK, Kedar PS, Lan L, Polosina YY, Asagoshi K, Poltoratsky VP, Horton JK, Miller H, Teebor GW, Yasui A, Wilson SH. DNA polymerase λ protects mouse fibroblasts against oxidative DNA damage and is regruited to sites of DNA damage/repair. J. Biol. Chem. 2005;vol. 280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 29.Tano K, Nakamura J, Asagoshi K, Arakawa H, Sonoda E, Braithwaite EK, Prasad R, Buerstedde J-M, Takeda S, Watanabe M, Wilson SH. Interplay between DNA polymerase β and λ in repair of oxidation DNA damage in chicken dt40 cells. DNA Rep. 2007;vol. 6:869–875. doi: 10.1016/j.dnarep.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon AF, García-Díaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pedersen LC. The X family portrait: Structural insights into biological functions of X family polymerases. DNA Rep. 2007;vol. 6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Díaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed confromation for the pol λ catalytic cycle. Nat. Struct. Mol. Bio. 2005;vol. 12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 32.Foley MC, Arora K, Schlick T. Sequential side-chain residue motions transform the binary into the ternary state of DNA polymerase λ. Biophys. J. 2006;vol. 91:3182–3195. doi: 10.1529/biophysj.106.092080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.García-Díaz M, Bebenek K, Pedersen LC, Kunkel TA. Role of the catalytic metals during polymerization by DNA polymerase lambda. DNA Rep. 2007;vol. 6:1333–1340. doi: 10.1016/j.dnarep.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Lee T, Yang W. A pseudo-bond approach to combining quantum mechanical and molecular mechanical methods. J. Chem. Phys. 1999;vol. 110:46–54. [Google Scholar]
- 35.Zhang Y, Liu H, Yang W. Free energy calculation on enzyme reactions with an efficient iterative procedure to determine minimum energy paths on a combined ab initio QM/MM potential energy surface. J. Chem. Phys. 2000;vol. 112:3483–3491. [Google Scholar]
- 36.Zhang Y, Liu H, Yang W. Computational Methods for Macromolecules-Challenges and Applications. Heidelberg, Germany: Springer Verlag; 2002. Ab Initio QM/MM and Free Energy Calculations of Enzyme Reactions. [Google Scholar]
- 37.Frouin I, Toueille M, Ferrari E, Shevelev I, Hübscher U. Phosphorilation of human DNA polymerase λ by the cyclin-dependent kinase cdk2/cyclin a complex is modulated by its association with proliferating cell nuclear antigen. Nucl. Ac. Res. 2005;vol. 33:5354–5361. doi: 10.1093/nar/gki845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCammon JA, Gelin BR, Karplus M. Dynamics of folded proteins. Nature. 1977;vol. 267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 39.MacKerrell AD, Jr, Brooks B, Brooks CL, III, Roux NB, Won Y, Karplus M. Encyclopedia of computational Chemistry. New York, NY: John Wiley & Sons Ltd; 1998. CHARMM: The energy function and its parametrization with an overview of the program. [Google Scholar]
- 40.Leach AR. Molecular modelling; principles and applications. 2nd Ed. Harlow, UK: Prentice Hall; 2001. [Google Scholar]
- 41.Warshel A, Levitt M. Theoretical studies of enzymatic reactions: dielectric electrostatic and steric stabilization of the carbonium ion in the reaction of lyzozyme. J. Mol. Biol. 1977;vol. 103:227. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 42.Kollman P, Kuhn B, Donini O, Perakyla M, Stanton R, D B. Elucidating the nature of enzyme catalysis utilizing a new twist on an old methodology: Quantum mechanical-free energy calculaitons on chemical reactions in enzymes and in aqueous solution. Acc. Chem. Res. 2001;vol. 34:72–79. doi: 10.1021/ar000032r. [DOI] [PubMed] [Google Scholar]
- 43.Náray-Szabó G, Berente I. Computer modelling of enzyme reactions. J. Mol. Struct. 2003;vol. 666–667:637–644. [Google Scholar]
- 44.Woodcock H, Hodošček M, Sherwood P, Lee Y, Schaefer HI, BR B. Exploring the quantum mechanical/molecular mechanical replica path method: a pathway optimization of the chorismate to prephenate claisen rearrangement catalyzed by chorismate mutase. Theo. Chem. Acc. 2003;vol. 109:140–148. [Google Scholar]
- 45.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MHM. Electrostatic basis for enzyme catalysis. Chem. Rev. 2006;vol. 106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 46.Senn H, Thiel W. Atomistic approachs in modern biology. Berlin, Germany: Springer Berlin/Heidelberg; 2007. QM/MM methods for biological systems. Topics in Current Chemistry. [Google Scholar]
- 47.Case DA, Cheatham TE, III, Darden TA, Gohlke H, Luo R, Merz KM, Jr, Onufirev A, Simmerling C, Wang B, Woods RJ. The amber biomolecular simulation programs. J. Comp. Chem. 2005;vol. 26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision D.02. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 49.Ponder J. TINKER, Software Tools for Molecular Design, Version 3.6: the most updated version for the TINKER program can be obtained from J.W. Ponder's. St. Louis: Washington University; 1998. WWW site at http://dasher.wustl.edu/tinker. [Google Scholar]
- 50.Zhang Y. Improved pseudobonds for combined ab initio quantum mechanical/ molecular mechanical (QM/MM) methods. J. Chem. Phys. 2005;vol. 122:24224. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 51.Burger SK, Yang W. Quadratic string method for determining the minimum-energy path based on multiobjective optimization. J. Chem. Phys. 2006;vol. 124:054109. doi: 10.1063/1.2163875. [DOI] [PubMed] [Google Scholar]
- 52.Williams I, Maggiora G. Use and abuse of the distinguished-coordinate method for transition state structure searching. J. Mol. Struct. 1982;vol. 89:365–378. [Google Scholar]
- 53.Gonzalez C, Schlegel H. Steepest descent path following. J. Chem. Phys. 1990;vol. 94:5523. [Google Scholar]
- 54.Schlegel H. Exploring potential energy surfaces for chemical reactions: an overview of some practical methods. J. Chem. Phys. 2003;vol. 24:1514–1527. doi: 10.1002/jcc.10231. [DOI] [PubMed] [Google Scholar]
- 55.Siegbahn PEM. A quantum chemical study of the mechanism of manganese catalase. Theo. Chem. Acc. 2001;vol. 105:197–206. [Google Scholar]
- 56.Ivanov I, Klein ML. Dynamical flexibility and proton transfer in the arginase active site probed by ab initio molecular dynamics. J. Am. Chem. Soc. 2005;vol. 127:4010–4020. doi: 10.1021/ja043693i. [DOI] [PubMed] [Google Scholar]
- 57.Liu S, Perera L, Pedersen LG. Binuclear manganese(II) complexes in biological systems. Mol. Phys. 2007;vol. 105:2893–2898. [Google Scholar]
- 58.Becke AD. Density-functional thermochemistry. III, the role of exact exchange. J. Chem. Phys. 1993;vol. 98:5648–5652. [Google Scholar]
- 59.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation energy formula into a functional of the electron density. Phys. Rev. B. 1988;vol. 37:785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 60.Elber R, Karplus M. A method for determining reaction paths in large molecules: Application to myoglobin. Chem. Phys. Lett. 1987;vol. 139:375–380. [Google Scholar]
- 61.Maragakis P, Stefan A, Brumer Y, Reichman D, Kaxiras E. Adaptive nudged elastic band approach for transition state calculation. J. Chem. Phys. 2002;vol. 117:4651–4658. [Google Scholar]
- 62.Liu H, Lu Z, Cisneros GA, Yang W. Parallel iterative reaction path optimization in ab initio quantum mechanical/molecular mechanical modeling of enzyme reactions. J. Chem. Phys. 2004;vol. 121:697–706. doi: 10.1063/1.1759318. [DOI] [PubMed] [Google Scholar]
- 63.Cisneros GA, Liu H, Lu Z, Yang W. Reaction path determination for quantum mechanical/molecular mechanical modeling of enzyme reactions by combining first order and second order “chain-of-replicas” methods. J. Chem. Phys. 2005;vol. 122:114502-1–114502-9. doi: 10.1063/1.1860560. [DOI] [PubMed] [Google Scholar]
- 64.Jhih-Wei C, Trout B, Brooks B. A super-linear minimization scheme for the nudged elastic band method. J. Chem. Phys. 2003;vol. 119:12708–12717. [Google Scholar]
- 65.Xie L, Liu H, Yang W. Adapting the nudged elastic band method for determining minimum energy paths of chemical reactions in enzymes. J. Chem. Phys. 2004;vol. 120:8039–8052. doi: 10.1063/1.1691404. [DOI] [PubMed] [Google Scholar]
- 66.Castro C, Smidansky E, Maksimchuk KR, Arnold JJ, Korneeva VS, Götte M, Konigsberg W, Cameron CE. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by rna- and DNA-dependent rna and DNA polymerases. Proc. Natl. Acad. Sci. 2007;vol. 104:4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu H, Yang W. Free energies of chemical reactions in solution and in enzymes with ab initio quantum mechanics/molecular mechanics methods. Ann. Rev. Phys. Chem. 2008;vol. 59:573–601. doi: 10.1146/annurev.physchem.59.032607.093618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu H, Zhang Y, Yang W. How is the active-site of enolase organized to achieve overall efficiency in catalyzing a two step reaction. J. Am. Chem. Soc. 2000;vol. 122:6560. [Google Scholar]
- 69.Cisneros GA, Liu H, Zhang Y, Yang W. Ab-initio QM/MM study shows there is no general acid in the reaction catalyzed by 4-oxalocrotonate tautomerase. J. Am. Chem. Soc. 2003;vol. 134:10348–10393. doi: 10.1021/ja029672a. [DOI] [PubMed] [Google Scholar]
- 70.Date T, Yamamoto S, Tanihara K, Nishimoto Y, Liu N, Matsukage A. Site-directed mutagenesis of recombinant rat DNA polymerase β: Involvement of arginine-183 in primer recognition. Biochemistry. 1990;vol. 29:5027–5034. doi: 10.1021/bi00473a005. [DOI] [PubMed] [Google Scholar]
- 71.Menge KL, Hostomsky Z, Nodes BR, Hudson GO, Rahmati S, Moomaw EW, Almassy RJ, Hostomska Z. Structure-function analysis of the mamailan DNA polymerase β active site: Role of aspartic acid 256, arginine 254 and arginine 258 in nucleotidyl transfer. Biochemistry. 1995;vol. 34:15934–15942. doi: 10.1021/bi00049a008. [DOI] [PubMed] [Google Scholar]
- 72.Notredame C, Higgins DG, Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;vol. 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed methods for initial structure, QM/MM structure optimization and path optimization are given, as well as QSM paths for selected reaction schemes, metal-ligand distances, residue analysis for all residues and waters, and sequence alignment of Polλ with other Family X polymerases.