Abstract
Myostatin is a member of the TGF-β superfamily and a potent negative regulator of muscle growth and development in mammals. Its expression is limited primarily to skeletal muscle in mammals, but occurs in many different fish tissues, although quantitative measurements of the embryonic and tissue-specific expression profiles are lacking. A recent phylogenetic analysis of all known myostatin genes identified a novel paralogue in zebrafish, zfMSTN-2, and prompted the reclassification of the entire subfamily to include MSTN-1 and -2 sister clades in the bony fishes. The differential expression profiles of both genes were therefore determined using custom RNA panels generated from pooled (100–150/sampling) embryos at different stages of development and from individual adult tissues. High levels of both transcripts were transiently present at the blastula stage, but were undetectable throughout gastrulation (7 hpf). Levels of zfMSTN-2 peaked during early somitogenesis (11 hpf), returned to basal levels during late somitogenesis and did not begin to rise again until hatching (72 hpf). By contrast, zfMSTN-1 mRNA levels peaked during late somitogenesis (15.5–19 hpf), returned to baseline at 21.5 hpf and eventually rose 25-fold by 72 hpf. In adults, both transcripts were present in a wide variety of tissues, including some not previously known to express myostatin. Expression of zfMSTN-1 was highest in brain, muscle, heart and testes and was 1–3 log orders above that in other tissues. It was also greater than zfMSTN-2 expression in most tissues, nevertheless, levels of both transcripts increased almost 600-fold in spleens of fish subjected to stocking stress. Myostatin expression was also detected in mouse spleens, suggesting that myostatin may influence immune cell development in mammals as well as fish. These studies indicate that zfMSTN-1 and -2 gene expression is differentially regulated in developing fish embryos and in adult tissues. The increased expression of both genes in spleens from stressed fish is further supportive of an immunomodulatory role and may explain increased disease susceptibility associated with stocking stress.
Keywords: Myostatin, GDF-8, Zebrafish, Gene expression, Stress
1. Introduction
Myostatin is a member of the transforming growth factor (TGF)-β superfamily of growth and differentiation factors and is known mostly for its potent abilities to negatively regulate mammalian skeletal muscle growth (Lee, 2004). In fish, however, its function may be more diverse and may influence many other tissues as it is widely expressed (Table 1). The tissue-specific expression pattern previously described for each species is not necessarily the same and is likely due to difficulties in detecting low levels of expression in some tissues. To date, most analyses of myostatin expression in developing and adult fish have been qualitative, which highlights the need for more quantitative analyses. Myostatin protein also appears to be widely distributed as myostatin immunoreactivity was identified in several juvenile and larval tissues of zebrafish, sole and seabream (Radaelli et al., 2003).
Table 1.
Myostatin gene expression in different adult fish tissues
| Tissue | 1 Zebrafish | 2 Catfish | 3 SeabreamMSTN1 | 4 Seabream MSTN-2 | 5 At salmon MSTN-1a/b | 6 rb trout MSTN-1a/b | 7 bk trout MSTN-1a/b | 8 Tilapia |
|---|---|---|---|---|---|---|---|---|
| Brain | + | + | + | + | + | + | + | + |
| Sk muscle | + | + | + | − | + | + | + | + |
| Gill | + | + | NS | NS | + | + | NS | + |
| Eye | + | + | + | + | + | NS | NS | + |
| Skin | NS | NS | NS | NS | NS | NS | NS | NS |
| GI tract | + | + | + | − | + | + | NS | + |
| Heart | + | + | + | − | + | + | NS | − |
| Spleen | NS | + | − | − | + | NS | NS | NS |
| Liver | + | + | − | − | − | + | NS | − |
| Swim bladder | NS | + | NS | NS | NS | NS | NS | NS |
| Kidney | + | + | + | − | − | + | NS | − |
| Ovaries | + | NS | − | + | + | + | + | + |
| Testis | + | NS | − | + | + | + | NS | + |
| Adipose | NS | NS | + | − | NS | NS | NS | NS |
bk, brook; rb, rainbow; At, Atlantic; Sk, skeletal; GI, gastrointestinal; +, detected; −, not detected; NS, not sampled;
Attempts to determine expression patterns with in situ hybridization have failed or have suggested ubiquitous expression, likely because the level of myostatin expression is too low to be detected with high specificity using this method (Amali et al., 2004; Vianello et al., 2003; Xu et al., 2003). Most other analyses have therefore used RT-PCR to evaluate expression of myostatin. In tilapia, myostatin expression was first detected in post-hatching larvae, however it was not detected in fertilized oocytes or in prehatching larvae with eye spots (Rodgers et al., 2001). It was first detected in catfish embyros beginning at day 1 post-fertilization, but this required 35 cycles of amplification and was relatively weak (Kocabas et al., 2002). Similar qualitative analyses detected zebrafish myostatin mRNA in fertilized embryos, which is consistent with maternally deposited transcripts, and throughout the early and late stages of development (Amali et al., 2004; Vianello et al., 2003; Xu et al., 2003). While these studies were able to detect myostatin in fish embryos, only Amali et al. specifically addressed expression during somitogenesis, when myogenesis begins, and none attempted to quantify expression during this significant stage of muscle development. Furthermore, most fish possess two distinct myostatin genes, MSTN-1 and MSTN-2 (Biga et al., 2005; Kerr et al., 2005), that resulted from the duplication of the bony fish genome (Amores et al., 1998; Postlethwait et al., 1998), while salmonids potentially possess four distinct genes (1a, 1b, 2a and 2b) due to the recent tetraploidization of this family (Phillips and Rab, 2001). Previous expression studies were conducted before the discovery of the MSTN-2 subfamily. Some of these studies may therefore be partly unreliable due to complications associated with primer/probe cross-hybridization to the different homologues.
Given that previous studies of myostatin gene expression during the development of fish did not differentiate between MSTN-1 and -2 genes, and that nearly all of the studies were only qualitative assessments, we sought to determine the temporal expression patterns of both zebrafish genes, zfMSTN-1 and -2, and to quantify the relative amounts of each transcript in developing zebrafish embryos and in different adult tissues. Our results indicate that expression of zfMSTN-1 and -2 is indeed differentially regulated and suggest a novel function for myostatin in regulating the immune systems of fish and possibly mammals.
2. Materials and methods
2.1. Animals
The zebrafish Danio rerio used in this study were of a strain originally purchased from Scientific Hatcheries (Huntington Beach, CA) and were housed in a centralized, monitored and recirculating aquatic system at the University of Idaho. The facility was kept at 28.5 °C on a light/dark cycle of 14:10 h. All experiments were performed in accordance with the Animal Care and Use Committees of the University of Idaho and Washington State University under preapproved protocols. Embryos were collected according to developmental stages outlined in The Zebrafish Book (Westerfield, 2000) including mid-blastula (4 h post-fertilization, hpf), mid-gastrula (7 hpf) and several stages of somitogenesis (4 somites—11.3 hpf, 8—13, 13—15.5, 17—17.5, 20—19, 25—21.5, 24, 48 and 72 hpf). Samples were also obtained from adult zebrafish after euthanizing with a lethal dose of buffered tricane methansulfonate (MS222, Argent, argentlabs.com). Individual tissues including brain, skeletal muscle, gill, eye, skin, gastrointestinal tract, heart, spleen, liver, swim bladder and gonads of both sexes were dissected and collected, snap frozen in liquid nitrogen and stored at −80 °C before RNA isolation.
2.2. Analysis of zfMSTN-1 and -2 gene expression
Total RNA was extracted from Danio rerio embryos (100–150 per development stage) and adult tissues with TRIzol (Invitrogen, invitrogen.com) according to the manufacturer’s instructions and cDNA was synthesized using 1 μg total RNA from each sample and the First Strand cDNA synthesis kit (Invitrogen). A qualitative analysis of zfMSTN-1 and -2 mRNA levels was first performed by reverse transcriptase polymerase chain reaction (RT-PCR). The same gene specific primers were used in this analysis and in quantitative real time RT-PCR (see below). For all analyses of gene expression in this study, the identity of each amplicon was verified by sequencing transcripts from different tissues including spleen. For each primer pair, the forward primer targeted the coding region in exon 1 and the reverse primer in exon 2 (Kerr et al., 2005; Xu et al., 2003). Thus, amplicons from both primer sets spanned the first intron of each respective gene. The sequence for the zfMSTN-1 forward primer was 5′-ACA TGC CAC CAC AGA AAC CA-3′ and the reverse was 5′-CAA CAC TTC GGT TTC CGA TCT AC-3′. The forward zfMSTN-2 primer was 5′-GCA AGC AGC GAG ACC ATC AT-3′ and the reverse was 5′-CAT GCA ACA CTT CGG CAT TC-3′. Expression of elongation factor 1-α (EF1-α) was used as a reference control. Its forward primer sequence was 5′-GCA TAC ATC AAG AAG ATC GGC-3′ and its reverse primer was 5′-GCA GCC TTC TGT GCA GAC TT G-3′. After an initial denaturation at 94 °C for 4 min, samples were amplified for 40 cycles (30 s at 94 °C, 15 s at 58 °C and 1 min at 72 °C) with a final elongation step of 5 min at 72°C.
2.3. Primer validation
The high conservation of the zfMSTN-1 and -2 genes complicate PCR-based analyses of individual transcript levels. Therefore gene-specific primers were designed and validated using a primer mismatch assay that paired zfMSTN-1 primers with zfMSTN-2 cDNA plasmids (1 ng) and vice versa. A temperature titration assay was also performed for each primer, using skeletal muscle cDNA, to determine the maximum range of annealing temperatures that would avoid primer cross-hybridization (Fig. 1a). All primers successfully amplified appropriate template even at the highest temperatures tested, which were 3–8 °C above the respective calculated Tm. Each primer set specifically amplified only the target gene and did not cross-hybridize to the other paralogue (Fig. 1b). Primer specificity was also verified by sequencing cDNA amplicons.
Fig. 1.
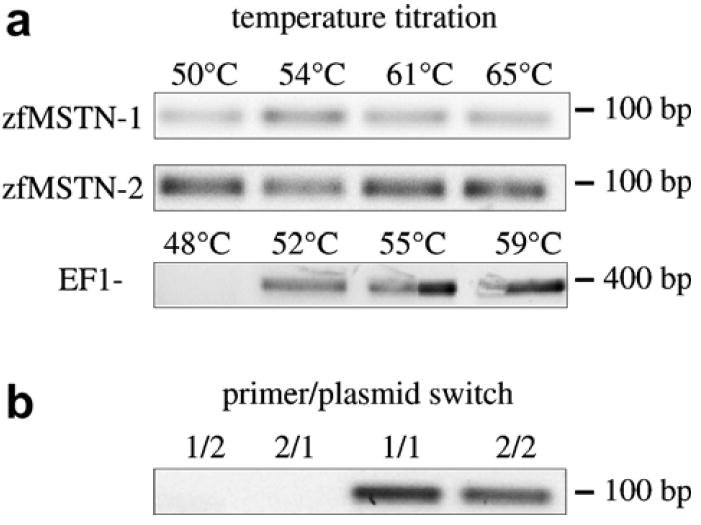
Primer validation. (a) Primers for zfMSTN-1, -2 and EF1-α were used to amplify adult zebrafish skeletal muscle cDNA using the annealing temperatures indicated. (b) Assessment of potential primer cross-reactivity. Plasmids containing cDNA for each myostatin were amplified with specific or non-specific primers (1/2 = zfMSTN-1 plasmid + zfMSTN-2 primers, etc.).
2.4. Real-time PCR analysis of myostatin gene expression
Real-time RT-PCR was performed using the same primers as described for RT-PCR and the iCyler IQ real time PCR detection system (Bio-Rad, bio-rad.com). Samples were amplified for 45 cycles (95 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s) using primer concentrations that were emperically determined (zfMSTN-1 forward = 900 nM, reverse = 600 nM; zfMSTN-2 forward = 900 nM, reverse = 50 nM). Melt curve analysis was also performed to once again assure primer specificity. For each assay, a master mix was first created using IQ SYBR Green Mastermix (Bio-Rad), which was then aliquoted before the addition of cDNA for each timepoint or tissue pool. Each individual aliquot was divided into three equal volumes and gene-specific primers were then added. Samples were run in triplicate on each plate and each analysis was repeated. Three control wells were also included on each plate that consisted of a no template control (NTC) and a RT- (RNA only) control. The cycle threshold (CT) was calculated for each amplicon using Q-Gene software (Muller et al., 2002) and CT values for both myostatin transcripts were normalized to those of EF1-α.
2.5. Stocking density stress and myostatin expression
The effects of stocking density stress on zfMSTN-1 and -2 mRNA levels in brain, skeletal muscle and spleen were investigated using two densities. High density (HD) or “stressed” fish were housed at 40 fish/L (approximately 32 g/L) and low density (LD) fish were housed at 5 fish/L (4 g/L). Three tanks housed fish at either density for a total of 6 tanks. Fish were watched when fed to ensure satiety was reached and there were no deaths or illnesses observed during these experiments. Tissues were dissected from euthanized fish and processed as described above.
2.6. Analysis of murine myostatin expression
A skeletal muscle biopsy (gastrocnemius) and spleen from a FVB/NJ mouse were provided by Dr. Derek McLean (Dept. of Animal Sciences, WSU). Both tissues were snap frozen in liquid nitrogen and total RNA was isolated and cDNA was synthesized as described above. The expression of myostatin and β-actin was determined by RT-PCR using intron-spanning primers specific for the mouse genes (MSTN forward, 5′-GCT GAT TGC TGC TGG CCC AGT GG-3′; MSTN reverse, 5′-GAG CAC CCA CAG CGG TCT ACT ACC-3′; actin forward, 5′-CGC TGC GCT GGT CGT CGA CAA CG-3′; actin reverse, 5′-ATC GTA CTC CTG CTT GCT GAT CCA C-3′). cDNA from both samples was initially amplified for 30 cycles (94 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min) and then a 5 μl aliquot was re-amplified using the same protocol for an additional 30 cycles.
2.7. Statistical analysis
The CT was calculated for each gene product using Q-Gene software (Muller et al., 2002). Normalized expression values were first analyzed by an analysis of variance to determine the presence of tank effect, which was determined not to exist. Differences between HD and LD means (±SEM; n = 15) were therefore determined using a t-test (p ≤ 0.05).
3. Results
3.1. Developmental expression of zfMSTN-1 and -2
Levels of zfMSTN-1 and -2 transcripts were assessed qualitatively by normal RT-PCR and quantitatively by real-time RT-PCR. The expression patterns and levels of both transcripts were determined using pooled samples of 100–150 embryos per stage of development, which thereby diluted individual variation. Both transcripts were weakly detected throughout development using the qualitative assay, although zfMSTN-1 levels were more readily detected at each timepoint (Fig. 2). Nevertheless, differences between different stages could not be resolved as expression levels were at the limit of detection. This was not due to poor sample quality as RNA fidelity was previously verified by gel electrophoresis and because EF1-α was easily amplified in all samples. Rather than optimizing this assay for semi-quantitative results only, we assayed the same samples using real-time RT-PCR.
Fig. 2.

Qualitative assessement of zfMSTN-1, -2 and EF1-α expression throughout embryogenesis. Levels of each transcript were qualitatively assessed by RT-PCR using embryos collected at the indicated times (hpf, hours post-fertilization). Adult RNA was extracted from skeletal muscle. (NTC, no template control; 11.3 hpf, 4 somite; 13, 8 somite; 15.5, 13 somite; 17.5, 17 somite; 19, 20 somite; 21.5, 25 somite).
High levels of both zfMSTN-1 and -2 transcripts were present at the blastula stage (4 hpf), but quickly dropped to almost undetectable levels throughout gastrulation (7 hpf). The presence of both transcripts during the blastula stage is likely attributed to the transfer of maternal components. Both transcript levels increased steadily following somitogenesis, although the rate and absolute levels of zfMSTN-1 were substantially greater than those of zfMSTN-2 (Fig. 3a). Indeed, zfMSTN-1 levels were almost always greater than those of zfMSTN-2. A closer evaluation of the gene expression patterns during somitogensis (Fig. 3b) reveals a zfMSTN-2 peak with an initial 5-fold increase during early somitogenesis (11.3 hpf). Expression returned to basal levels during late somitogenesis and did not begin to rise again until hatching (72 hpf). By contrast, zfMSTN-1 mRNA levels were low during early somitogenesis (11.3–13 hpf), but increased 30-fold during late somitogenesis (15.5–19 hpf) before returning to basal levels at the end of this developmental period (21.5 hpf). These data indicate that expression of zfMSTN-1 and -2 is differentially regulated in developing embryos, specifically during early and late somitogenesis, and during the events that precede hatching.
Fig. 3.
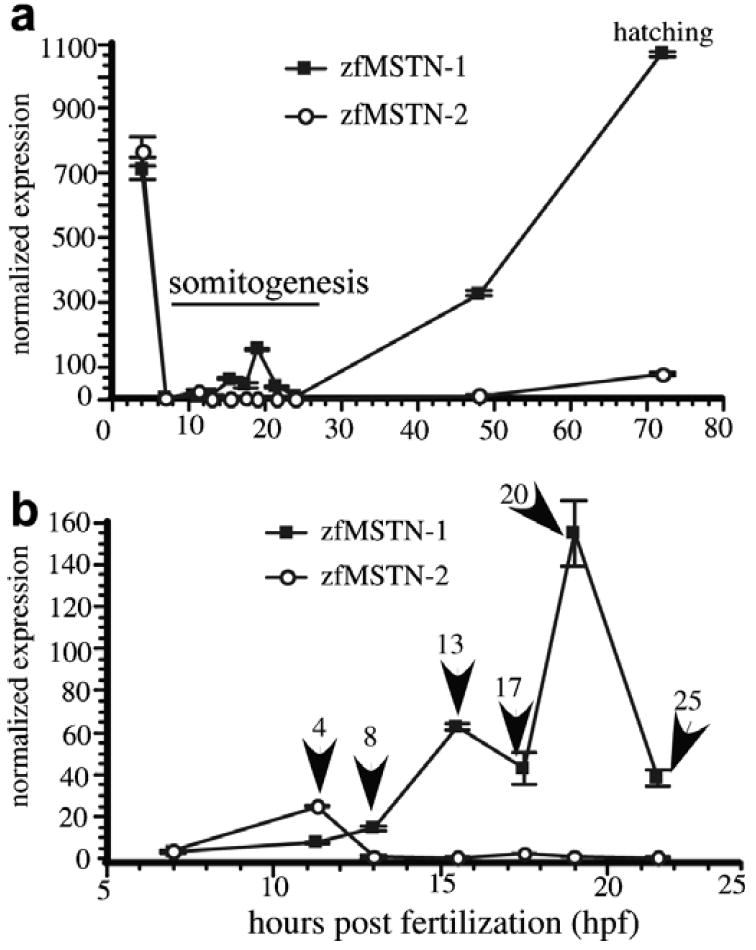
Quantitative analysis of zfMSTN-1 and -2 mRNA levels in developing embryos. A comprehensive RNA panel was constructed by staging and individually sampling 100–150 embryos at the time periods indicated (4, 7, 11.3, 15.5, 17.5, 19, 21.5, 24, 48 and 72 hpf). (a) Normalized expression (mean ± SEM) of both zfMSTN-1 and -2 using real-time RT-PCR. Values for each myostatin transcript were normalized to those of EF1-α. (b) Expression during early development only; 7–21.5 hpf. Arrows indicate the number of somites present in each embryo. Assays were performed on pooled samples, therefore, error bars represent assay variance only.
3.2. Adult tissue-specific expression of zfMSTN-1 and -2
Previous studies identified myostatin expression in many different fish tissues (Table 1). With one exception (Maccatrozzo et al., 2001a), however, these analyses only studied MSTN-1 expression patterns using primers that may have potentially cross-hybridized to either or both MSTN transcripts. Total RNA was therefore isolated from twelve tissues of zebrafish adults and was analyzed by real-time RT-PCR to determine individual tissue expression patterns. Both zfMSTN transcripts were present in brain, muscle, gill, eyes, skin, gastrointestinal tract, heart, spleen, liver, swim bladder and gonads of both sexes (Fig. 4). This distribution includes tissues not previously known to express myostatin, including liver, heart and spleen. Normalized expression of zfMSTN-1 was highest in brain, muscle, heart and testes, 1–3 log orders above that in other tissues and was greater than zfMSTN-2 expression in all tissues except eyes and swim bladder. Normalized expression of zfMSTN-2 was greatest in brain, eyes, spleen and testes and was 1–4 log orders above that of other tissues. These studies suggest distinct functional roles for zfMSTN-1 and -2 during embryogenesis and in adult tissues.
Fig. 4.
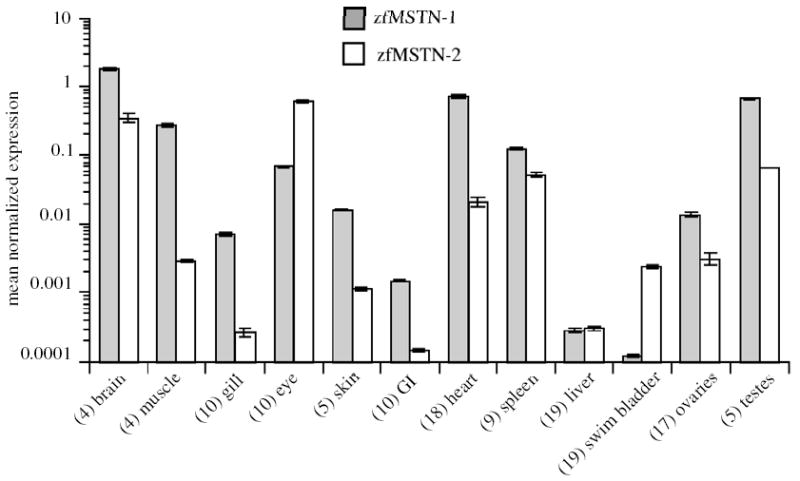
Adult tissue distribution of zfMSTN-1 and -2 mRNA. Total RNA was isolated from tissues of adult zebrafish and analyzed using qRT-PCR. Transcript levels were normalized to EF1-α. Assays were repeated in triplicate and tissues were sampled twice (representative assay shown). Mean values are shown for each tissue, which represents a pooled sampling from several animals (numbers indicated). Error bars (±SEM) therefore represent assay variation only.
3.3. Stocking stress
Previous studies with fish and mammals indicate that myostatin expression is influenced by different stressors (Carlson et al., 1999; Gonzalez-Cadavid and Bhasin, 2004; Vianello et al., 2003; Wehling et al., 2000). Adult zebrafish were therefore subjected to 3 days of acute stocking density stress to determine changes in zfMSTN-1 and -2 gene expression in muscle, brain and spleen. Due to the dramatic difference in absolute levels and the degree of changes, transcript levels of both genes are shown as normalized expression on a logarithmic scale and as percent of control (Fig. 5). There was no significant change in the expression of either gene in muscle and brain. By contrast, levels of both zfMSTN-1 and -2 mRNA in spleens of HD fish were almost 3 log orders of magnitude greater than those in LD spleens. This dramatic increase in expression of both genes suggests that myostatin may play an important role in modulating immune system function during conditions of stress. The changes in mRNA do not appear to be systemic responses, rather tissue-specific, as only spleen levels were affected (Fig. 6).
Fig. 5.
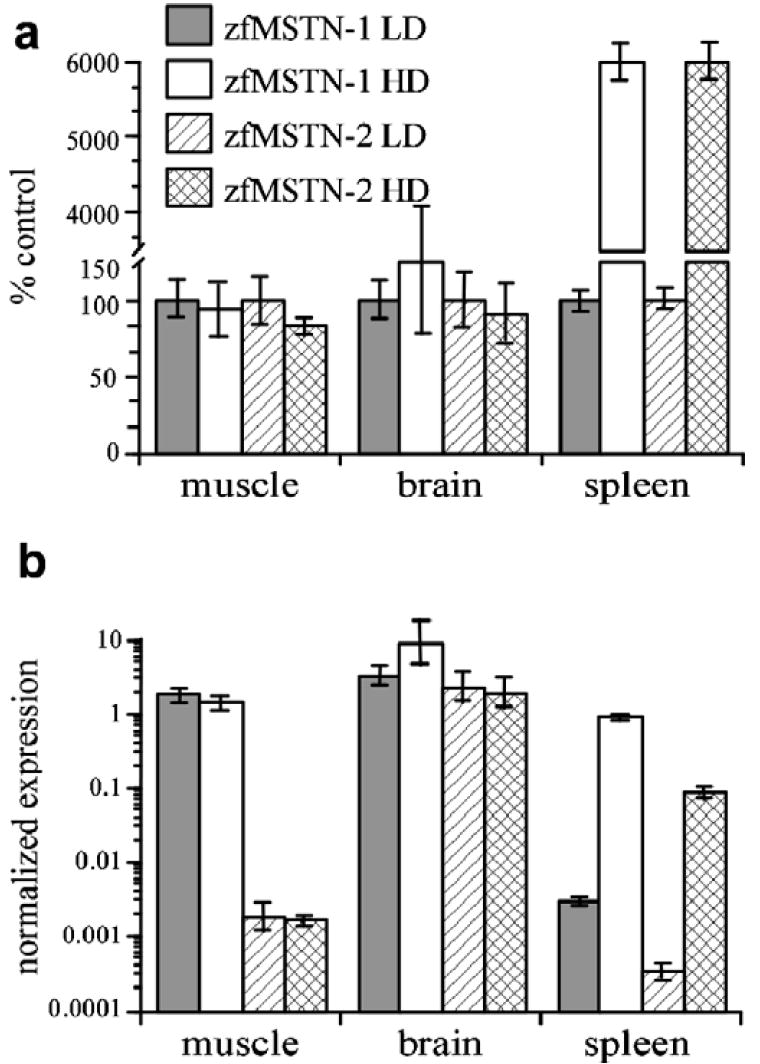
Stocking density stress and zebrafish myostatin expression. The effect of acute stocking density stress on the expression of zfMSTN-1 and -2 in muscle, brain and spleen was determined by qRT-PCR. High density (HD) fish were housed at a density of 40 fish/L water. Low density (LD) Fish were housed at a density of 5 fish/L. Fish were housed in 3 tanks per treatment group to control for tank effect. HD and LD treatments were repeated twice. (a) Relative expression levels of zfMSTN-1 and -2 as a percentage of the LD controls. (b) Mean normalized levels (±SEM) of zfMSTN-1 and -2 expression in logarithmic scale.
Fig. 6.
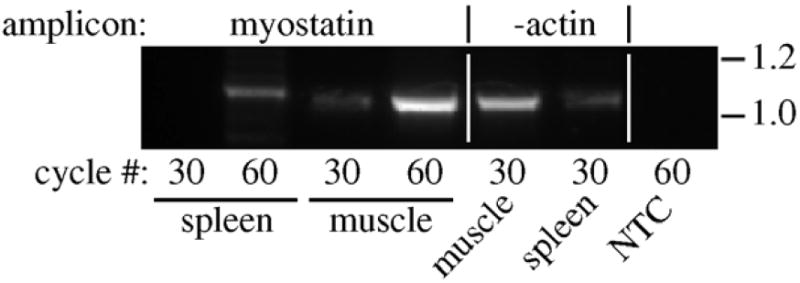
Myostatin expression in mouse spleen. Expression was analyzed by RT-PCR as described in Section 2 using total RNA from spleen and skeletal muscle. Myostatin cDNA was amplified for 30 cycles and then a 5 μl aliquot was re-amplified for an additional 30 cycles for a total of 60. β-actin was also amplified to assure cDNA quality and a no template control (NTC, with myostatin primers) was also included. Amplicon sizes (kb) are indicated on the right.
3.4. Myostatin expression in mouse spleens
Although myostatin expression in mammals occurs mostly in skeletal muscle, we sought to determine whether it could be detected in the mouse spleen as the stress-induced increase in transcript levels of both zfMSTNs suggests that even low levels of basal expression could be physiologically relevant. After 30 cycles of PCR amplification, myostatin expression was only detected in skeletal muscle, although β-actin transcripts were present in both muscle and spleen, confirming RNA integrity. However, expression was readily detected in both tissues after 60 cycles, suggesting that myostatin is at least minimally expressed in the mouse spleen as well.
4. Discussion
Myostatin function has been extensively studied in mammalian systems, although its function may be quite different in fish where its expression occurs in many more adult and developing tissues. A recent phylogenetic analysis (Kerr et al., 2005) has identified two distinct myostatin clades, suggesting that all fish express at least two myostatin genes. Genomic and cDNA clones for MSTN-2 have been cloned from zebrafish (Biga et al., 2005; Kerr et al., 2005) and from a select few other species and relatively little is known about their relative expression patterns. This highlights the need for more extensive studies of MSTN-2 expression during embryonic development and in adult tissues.
In this study, we characterized and quantified the expression of zfMSTN-1 and -2 genes in developing zebrafish embryos and in adult tissues. High expression levels of both genes during blastula stages are indicative of remaining maternal transcripts, which is consistent with previous studies using zebrafish (Biga et al., 2005; Vianello et al., 2003), tilapia (Rodgers et al., 2001) and brook trout (Roberts and Goetz, 2003) as myostatin expression is present in ovaries, unfertilized eggs and even in newly fertilized eggs at the 2 cell stage (Biga et al., 2005). Both transcripts were nearly undetectable during gastrulation. This indicates that neither is necessary at this stage and is consistent with previous assessments of developing embryos from different fish species including tilapia (Rodgers et al., 2001), zebrafish (Biga et al., 2005), rainbow trout (Garikipati et al., 2006), brook trout (Roberts and Goetz, 2003) and even in mammals. Indeed, in situ hybridization studies of myostatin expression in mice determined that it is first detected in the myotome compartment of somites (McPherron et al., 1997). As mouse somitogenesis progresses, levels of myostatin message increase, although expression is limited almost exclusively to developing skeletal muscle. In the present study, temporal expression patterns of either zfMSTN-1 or -2 do not necessarily follow the patterns of muscle development, although the changes in expression of both genes during somitogenesis are consistent with the early stages of myogenesis. Levels of zfMSTN-1 mRNA steadily rose throughout somitogenesis while zfMSTN-2 levels peaked and then dropped during early somitogenesis. The formation of trunk and tail somites is similarly controlled, although some of the factors that influence this process are different. This is best illustrated by comparing somite formation and MRF gene expression in spadetail (spt), one-eyed pinhead (Oep) and no-tail (ntl) mutants or in spt:Oep double mutants (Griffin and Kimelman, 2002; Weinberg et al., 1996). Paraxial mesoderm segregates in the cranial to caudal directions and results in the temporal formation of somites and in the development of the somitic myotome. Therefore, the early expression of zfMSTN-2 takes place in parallel with the onset of myogenesis in the trunk while zfMSTN-1 expression takes place in parallel with the same extensive process that occurs in the tail. A definitive role for each myostatin remains to be determined. Nevertheless, the temporal expression patterns of both genes in zebrafish suggest that they are both likely involved in the early stages of muscle development. Curiously, the selective knockdown of MSTN-1 expression with antisense morpholino oligonucleotides was reported to increase the expression of some muscle-specific genes at 10 hpf and to increase the developmental rate and size of somites (Amali et al., 2004)—a surprising finding given that (1) zfMSTN-1 expression is virtually undetectable at this developmental time and lower than the corresponding level of zfMSTN-2 expression (Fig. 3) and (2) that other studies of zebrafish (Kerr et al., 2005), tilapia (Rodgers et al., 2001), rainbow trout (Garikipati et al., 2006 ), brook trout (Roberts and Goetz, 2003) and even mice (McPherron et al., 1997) all suggest that myostatin is not expressed during gastrulation. Xu et al. (2003) also generated null fish by overexpressing a dominant-negative zfMSTN-1 (i.e. pro-domain a.k.a. LAP), but did not reproduce the results described by Amali et al. Thus, a comparative analysis of loss-of-function phenotypes following MSTN-1 and/or MSTN-2 knockdown is needed to resolve this issue.
In addition to skeletal muscle, weak myostatin expression has been detected in mammalian Purkinje fibers and cardiomyocytes (Morissette et al., 2006; Sharma et al., 1999; Shyu et al., 2005) as well as in mammary glands (Ji et al., 1998). Although recent studies indicate that myostatin may influence cardiac muscle growth (Morissette et al., 2006; Shyu et al., 2005) and possibly adipocyte differentiation (Kim et al., 2001; Rebbapragada et al., 2003; Zimmers et al., 2002), its primary function is to negatively regulate skeletal muscle growth. This has been demonstrated in numerous studies using skeletal muscle cell lines and in myostatin null mice, domesticated cattle and in a child (Grobet et al., 1997; Kambadur et al., 1997; Lee and McPherron, 1999; McPherron et al., 1997; McPherron and Lee, 1997; Schuelke et al., 2004). Its more ubiquitous expression in fish tissues (Table 1 and Fig. 4) suggests that its functions may be even more diverse in the fishes. Previous expression studies, however, were performed before the discovery of the two distinct myostatin gene families in fish. Thus, they either assessed only zfMSTN-1 expression or may have inadvertently sampled both transcripts simultaneously. Furthermore, none of these studies were quantitative and thus, the relative contribution of each gene product within a specific tissue was unknown. We determined that both zfMSTN-1 and -2 genes are expressed in a wide variety of zebrafish tissues. Both transcripts were detected in every tissue sampled, although levels of expression were quite different among tissues and between genes. The tissues with the highest expression of both genes are brain, muscle, eyes, heart, testes and surprisingly spleen, although levels in the latter were an order of magnitude lower. Expression of zfMSTN-1 was greater than that of zfMSTN-2 in all tissues except eyes and swim bladder (Fig. 4). In fact, zfMSTN-2 message was almost 10-fold higher in eyes while zfMSTN-1 message was barely detected in swim bladder. These data suggest that zfMSTN-1, as in development, is predominantly expressed despite zfMSTN-2’s potentially more specific role in eyes and swim bladder.
Levels of both zfMSTN transcripts were comparatively low in spleens of non-stressed fish, but rose to levels similar to those in skeletal muscle as a consequence of stocking stress. Thus, even very low levels of myostatin expression could be indicative of potential function in some tissues. This could also be true for mammals as weak myostatin expression was also detected in mouse spleen, but could approach physiologically relevant levels with the appropriate stimuli. This tissue is composed primarily of immune cells, both B and T lymphocytes, and to a lesser extent of aged and defective erythrocytes. It is also supported by fibrous connective tissue and a thin layer of smooth muscle. The exact cellular source of myostatin expression in spleens cannot be definitively determined at this time. However, we recently identified myostatin expression (MSTN-1a and -1b) in peripheral blood lymphocytes as well as spleens of rainbow trout (Garikipati et al., 2006), which is indicative of immune cell origins of expression rather than non-parenchymal cell origins. These data together suggest that myostatin may influence immune cell development in both fish and mammals and that it may additionally participate in stress-mediated changes in immune function.
The possibility that myostatin may have different functional roles in fish versus mammals is only just beginning to be studied. In mammals, myostatin expression increases in muscles undergoing atrophy (Carlson et al., 1999; Lalani et al., 2000; Ma et al., 2003; Wehling et al., 2000) and in response to some stressors including thermal injury (Lang et al., 2001), microgravity (Lalani et al., 2000) and chronic fasting (Jeanplong et al., 2003). In contrast, chronic crowding stress decreases myostatin expression in zebrafish skeletal muscle (Vianello et al., 2003). A similar result was observed in tilapia subjected to chronic nutritional stress (Rodgers et al., 2003) and during sexual maturation in rainbow trout, a time when muscle undergoes atrophy (Rescan et al., 2001). These studies indicate differential responses to stress in mammals versus fish. Although any particular stressor can impact tissues and organisms differently, hypercortisolemia commonly develops regardless of the stressor. Glucorticoids increase myostatin gene expression in mammals (Artaza et al., 2002; Lang et al., 2001; Ma et al., 2001; Ma et al., 2003), but decrease MSTN-1 message in tilapia larvae (Rodgers et al., 2003), which could explain the differential response to stress in mammals and fish. Nevertheless, glucocorticoids are not likely responsible for the stressed-induced increase in splenic zfMSTN-1 and -2 expression (Fig. 5) as brain and skeletal muscle expression was unaffected. Overcrowding is a common problem encountered in the aquaculture industry and often results in increased incidence of disease. Myostatin’s precise immune function is unknown, however, if its inhibitory role in muscle is conserved in the spleen and possibly other immune tissues, the upregulation of both MSTN-1 and -2 genes could contribute to stress induced immunocompromization.
The expression patterns described herein further support a role for both zfMSTN genes during myogenesis, although the ubiquitous expression pattern in different adult tissues suggests that the cytokines may influence many different tissues and cell types. The presence of multiple fish genes—two in zebrafish and potentially four in salmonids—that are differentially expressed throughout development and in adult tissues also suggests that the precise role of a specific gene may vary between tissues. Low levels of expression in any particular tissue may not necessarily represent retrograde transcription as significant changes in expression, as seen in spleens of stressed zebrafish, could have substantial effects on tissue development and organismal physiology. A better understanding of physiological factors that influence the expression of each gene and of the transcriptional machinery involved will therefore help distinguish the potential divergent actions of myostatin in fish and mammals.
Acknowledgments
This research was funded by a grant from the United States Department of Agriculture (2004-34468-15199) to Buel D. Rodgers.
References
- Amali AA, Lin CJ, Chen YH, Wang WL, Gong HY, Lee CY, Ko YL, Lu JK, Her GM, Chen TT, Wu JL. Up-regulation of muscle-specific transcription factors during embryonic somitogenesis of zebrafish (Danio rerio) by knock-down of myostatin-1. Dev Dyn. 2004;229:847–856. doi: 10.1002/dvdy.10454. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Artaza JN, Bhasin S, Mallidis C, Taylor W, Ma K, Gonzalez-Cadavid NF. Endogenous expression and localization of myostatin and its relation to myosin heavy chain distribution in C2C12 skeletal muscle cells. J Cell Physiol. 2002;190:170–179. doi: 10.1002/jcp.10044. [DOI] [PubMed] [Google Scholar]
- Biga PR, Roberts SB, Iliev DB, McCauley LA, Moon JS, Collodi P, Goetz FW. The isolation, characterization, and expression of a novel GDF11 gene and a second myostatin form in zebrafish, Danio rerio. Comp Biochem Physiol B Biochem Mol Biol. 2005;141:218–230. doi: 10.1016/j.cbpc.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Carlson CJ, Booth FW, Gordon SE. Skeletal muscle myostatin mRNA expression is fiber-type specific and increases during hindlimb unloading. Am J Physiol. 1999;277:R601–R606. doi: 10.1152/ajpregu.1999.277.2.r601. [DOI] [PubMed] [Google Scholar]
- Garikipati D, Gahr SA, Rodgers BD. Identification, characterization and quantitative expression analysis of rainbow trout myostatin-1a and -1b genes. J Endocrinol. 2006;190(3):879–888. doi: 10.1677/joe.1.06866. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cadavid NF, Bhasin S. Role of myostatin in metabolism. Curr Opin Clin Nutr Metab Care. 2004;7:451–457. doi: 10.1097/01.mco.0000134365.99523.7f. [DOI] [PubMed] [Google Scholar]
- Griffin KJ, Kimelman D. One-eyed pinhead and spadetail are essential for heart and somite formation. Nat Cell Biol. 2002;4:821–825. doi: 10.1038/ncb862. [DOI] [PubMed] [Google Scholar]
- Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, Fries R, Hanset R, Georges M. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [see comments]. [DOI] [PubMed] [Google Scholar]
- Jeanplong F, Bass JJ, Smith HK, Kirk SP, Kambadur R, Sharma M, Oldham JM. Prolonged underfeeding of sheep increases myostatin and myogenic regulatory factor Myf-5 in skeletal muscle while IGF-I and myogenin are repressed. J Endocrinol. 2003;176:425–437. doi: 10.1677/joe.0.1760425. [DOI] [PubMed] [Google Scholar]
- Ji S, Losinski RL, Cornelius SG, Frank GR, Willis GM, Gerrard DE, Depreux FF, Spurlock ME. Myostatin expression in porcine tissues: tissue specificity and developmental and postnatal regulation. Am J Physiol. 1998;275:R1265–R1273. doi: 10.1152/ajpregu.1998.275.4.R1265. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7:910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- Kerr T, Roalson EH, Rodgers BD. Phylogenetic analysis of the myostatin gene sub-family and the differential expression of a novel member in zebrafish. Evol Dev. 2005;7:390–400. doi: 10.1111/j.1525-142X.2005.05044.x. [DOI] [PubMed] [Google Scholar]
- Kim HS, Liang L, Dean RG, Hausman DB, Hartzell DL, Baile CA. Inhibition of preadipocyte differentiation by myostatin treatment in 3T3-L1 cultures. Biochem Biophys Res Commun. 2001;281:902–906. doi: 10.1006/bbrc.2001.4435. [DOI] [PubMed] [Google Scholar]
- Kocabas AM, Kucuktas H, Dunham RA, Liu Z. Molecular characterization and differential expression of the myostatin gene in channel catfish (Ictalurus punctatus) Biochim Biophys Acta. 2002;1575:99–107. doi: 10.1016/s0167-4781(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Lalani R, Bhasin S, Byhower F, Tarnuzzer R, Grant M, Shen R, Asa S, Ezzat S, Gonzalez-Cadavid NF. Myostatin and insulin-like growth factor-I and -II expression in the muscle of rats exposed to the microgravity environment of the NeuroLab space shuttle flight. J Endocrinol. 2000;167:417–428. doi: 10.1677/joe.0.1670417. [DOI] [PubMed] [Google Scholar]
- Lang CH, Silvis C, Nystrom G, Frost RA. Regulation of myostatin by glucocorticoids after thermal injury. FASEB J. 2001;15:1807–1809. doi: 10.1096/fj.00-0849fje. [DOI] [PubMed] [Google Scholar]
- Lee SJ. Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- Lee SJ, McPherron AC. Myostatin and the control of skeletal muscle mass. Curr Opin Genet Dev. 1999;9:604–607. doi: 10.1016/s0959-437x(99)00004-0. [DOI] [PubMed] [Google Scholar]
- Ma K, Mallidis C, Artaza J, Taylor W, Gonzalez-Cadavid N, Bhasin S. Characterization of 5′-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. Am J Physiol Endocrinol Metab. 2001;281:E1128–E1136. doi: 10.1152/ajpendo.2001.281.6.E1128. [DOI] [PubMed] [Google Scholar]
- Ma K, Mallidis C, Bhasin S, Mahabadi V, Artaza J, Gonzalez-Cadavid N, Arias J, Salehian B. Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. Am J Physiol Endocrinol Metab. 2003;285:E363–E371. doi: 10.1152/ajpendo.00487.2002. [DOI] [PubMed] [Google Scholar]
- Maccatrozzo L, Bargelloni L, Cardazzo B, Rizzo G, Patarnello T. A novel second myostatin gene is present in teleost fish. FEBS Lett. 2001a;509:36–40. doi: 10.1016/s0014-5793(01)03124-6. [DOI] [PubMed] [Google Scholar]
- Maccatrozzo L, Bargelloni L, Radaelli G, Mascarello F, Patarnello T. Characterization of the myostatin gene in the gilthead seabream (Sparus aurata): sequence, genomic structure, and expression pattern. Mar Biotechnol (NY) 2001b;3:224–330. doi: 10.1007/s101260000064. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morissette MR, Cook SA, Foo S, McKoy G, Ashida N, Novikov M, Scherrer-Crosbie M, Li L, Matsui T, Brooks G, Rosenzweig A. Myostatin regulates cardiomyocyte growth through modulation of Akt signaling. Circ Res. 2006;99:15–24. doi: 10.1161/01.RES.0000231290.45676.d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374. 1376, 1378–9. [PubMed] [Google Scholar]
- Ostbye TK, Galloway TF, Nielsen C, Gabestad I, Bardal T, Andersen O. The two myostatin genes of Atlantic salmon (Salmo salar) are expressed in a variety of tissues. Eur J Biochem. 2001;268:5249–5257. doi: 10.1046/j.0014-2956.2001.02456.x. [DOI] [PubMed] [Google Scholar]
- Phillips R, Rab P. Chromosome evolution in the Salmonidae (Pisces): an update. Biol Rev Camb Philos Soc. 2001;76:1–25. doi: 10.1017/s1464793100005613. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Radaelli G, Rowlerson A, Mascarello F, Patruno M, Funkenstein B. Myostatin precursor is present in several tissues in teleost fish: a comparative immunolocalization study. Cell Tissue Res. 2003;311:239–250. doi: 10.1007/s00441-002-0668-y. [DOI] [PubMed] [Google Scholar]
- Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol Cell Biol. 2003;23:7230–7242. doi: 10.1128/MCB.23.20.7230-7242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescan PY, Jutel I, Ralliere C. Two myostatin genes are differentially expressed in myotomal muscles of the trout (Oncorhynchus mykiss) J Exp Biol. 2001;204:3523–3529. doi: 10.1242/jeb.204.20.3523. [DOI] [PubMed] [Google Scholar]
- Roberts SB, Goetz FW. Myostatin protein and RNA transcript levels in adult and developing brook trout. Mol Cell Endocrinol. 2003;210:9–20. doi: 10.1016/j.mce.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Rodgers BD, Weber GM, Kelley KM, Levine MA. Prolonged fasting and cortisol reduce myostatin mRNA levels in tilapia larvae; short-term fasting elevates. Am J Physiol Regul Integr Comp Physiol. 2003;284:R1277–R1286. doi: 10.1152/ajpregu.00644.2002. [DOI] [PubMed] [Google Scholar]
- Rodgers BD, Weber GM, Sullivan CV, Levine MA. Isolation and characterization of myostatin complementary deoxyribonucleic acid clones from two commercially important fish: Oreochromis mossambicus and Morone chrysops. Endocrinology. 2001;142:1412–1418. doi: 10.1210/endo.142.4.8097. [DOI] [PubMed] [Google Scholar]
- Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, Braun T, Tobin JF, Lee SJ. Myostatin mutation associated with gross muscle hypertrophy in a child. N EnglJ Med. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- Sharma M, Kambadur R, Matthews KG, Somers WG, Devlin GP, Conaglen JV, Fowke PJ, Bass JJ. Myostatin, a transforming growth factor-beta superfamily member, is expressed in heart muscle and is upregulated in cardiomyocytes after infarct. J Cell Physiol. 1999;180:1–9. doi: 10.1002/(SICI)1097-4652(199907)180:1<1::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Shyu KG, Ko WH, Yang WS, Wang BW, Kuan P. Insulin-like growth factor-1 mediates stretch-induced upregulation of myostatin expression in neonatal rat cardiomyocytes. Cardiovasc Res. 2005;68:405–414. doi: 10.1016/j.cardiores.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Vianello S, Brazzoduro L, Dalla Valle L, Belvedere P, Colombo L. Myostatin expression during development and chronic stress in zebrafish (Danio rerio) J Endocrinol. 2003;176:47–59. doi: 10.1677/joe.0.1760047. [DOI] [PubMed] [Google Scholar]
- Wehling M, Cai B, Tidball JG. Modulation of myostatin expression during modified muscle use. FASEB J. 2000;14:103–110. doi: 10.1096/fasebj.14.1.103. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene: 2000. [Google Scholar]
- Xu C, Wu G, Zohar Y, Du SJ. Analysis of myostatin gene structure, expression and function in zebrafish. J Exp Biol. 2003;206:4067–4079. doi: 10.1242/jeb.00635. [DOI] [PubMed] [Google Scholar]
- Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, McPherron AC, Wolfman NM, Lee SJ. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]


