Abstract
Background
Mapping T cell epitopes for a pathogen or vaccine requires a complex method for screening hundreds to thousands of peptides with a limited amount of donor sample. We describe an optimized deconvolution process by which peptides are pooled in a matrix format to minimize the number of tests required to identify peptide epitopes.
Methods
Four peptide pool matrices were constructed to deconvolute the HIV-specific T cell response in three HIV-infected individuals. ELISpot assays were used to map peptide antigens.
Results
Many HIV peptides were mapped in all three individuals. However, there were several challenges and limitations associated with the deconvolution process. Peptides that induced low-frequency responses or were masked by peptide competition within a given pool were not identified because they did not meet the threshold criteria for a positive response. Also, amino acid sequence variation limited the ability of this method to map autologous HIV peptides. Alternative analysis strategies and revisions to the original matrix optimizations are presented that address ways to increase peptide identification.
Conclusions
This optimized deconvolution method allows for efficient mapping of T cell peptide epitopes. It is rapid, powerful, efficient, and unrestricted by HLA type.
Keywords: Epitopes, T-Lymphocyte, HIV, Epitope mapping
Introduction
The breadth of an antigen-specific T cell response to a pathogen or vaccine can be measured by mapping T cell epitopes. Mapping is commonly performed using a functional readout following stimulation with overlapping peptides (or pools of such peptides) that span a given protein and presumably encompass all possible T cell epitopes (1-3). However, for whole pathogens, such as HIV, this translates into hundreds of peptides that require large numbers of cells for single testing or a complex strategy for peptide pooling to reduce the amount of sample required to map the epitopes (4-8). We recently optimized a method for construction of peptide pool matrices that allows identification of peptide-specific T cell responses from among a set of several hundred overlapping peptides (9). Here we apply that approach to characterize the method of epitope deconvolution.
Deconvolution of peptide antigens is a two-step process. The first step, in which the sample is tested on all pools of peptides, is used to eliminate those peptides that are not recognized (i.e., a negative response to a pool of 80 peptides eliminates all 80 peptides as possible candidates). After this first round, there is still considerable ambiguity as to which peptides are “real” responses, and which peptides could not be eliminated because they happened to be in pools containing other positive peptides. Thus, during the second round every possible positive peptide is tested individually to determine the total response. The goal in optimized deconvolution is to use the minimum amount of sample over both rounds. Once a response is deconvoluted, further experiments are required to map the minimal epitope sequence, HLA-restriction, and responding T cell subtype of each peptide.
In our previous study, we optimized the deconvolution of a response based on qualitative results (ie “positive” or “negative”). However, T cell responses are generally measured quantitatively (as a frequency of responding cells) by assays such as ELISpot and cytokine flow cytometry. It should be noted that the read-out of these assays impart a limitation for further experiments because only cells responding by secretion of the measured cytokine are identified. Nevertheless, we wanted to test our optimized method experimentally in a situation with multiple peptide-specific responses that ranged in the frequency of responding T cells. We constructed four matrices of pools of peptides (15-mers overlapping by 11) spanning the entire HIV proteome, excluding Vif, Vpr, and Vpu, corresponding to the unique autologous sequences of four acutely infected HIV+ individuals (results from autologous donors to be presented elsewhere). Peptide length and overlap were selected for optimal stimulation of both CD4+ and CD8+ T cells simultaneously and inclusion of all potential epitopes, respectively, in accordance with previous data (3,10). ELISpot assay was chosen as the experimental approach due to its small sample requirement.
The purpose of the current study was to review the design, process, and challenges of the first two rounds of epitope deconvolution using peptide matrices. Therefore, we used these four matrices to identify peptide-specific T cell responses in three HIV+ individuals for whom the peptide sequences within the matrices were non-autologous. This approach is analogous to the common practice of peptide epitope mapping using consensus sequences, as not all consensus sequences will match the HIV sequence of a given HIV+ individual. Here, we present experimental results addressing the utility and reproducibility of our method as well as the effect of peptide sequence variability on identifying T cell peptide epitopes. We also discuss revisions to our original optimizations that may increase our ability to identify T cell epitopes.
Materials and Methods
Patient samples
Three HIV+ individuals were recruited for this study. All three were off antiretroviral therapy and had CD4 counts between 203-685 cells/μL. HIV RNA ranged from 4420-145,000 copies/mL. Study subjects gave informed consent as required by the National Institute of Allergy and Infectious Diseases Institutional Review Board.
Peptides
15-mer peptides (overlapping by 11 amino acids) spanning HIV-1 Gag, Pol, Env, Nef, Rev, and Tat were synthesized in a 96 well format on a scale of 2.5μM/well, peptide purity >75% (New England Peptide, Inc., Gardner, MA). Peptides were dissolved in DMSO at high concentration (typically 100mg/ml, depending on solubility). Peptides were pooled such that the final concentration of each peptide used for stimulation was 2μg/mL. For second round assays, individual peptides were also tested at a final concentration of 2μg/mL. Precise peptide sequences and matrix pool compositions are available upon request.
Deconvolution of HIV peptide-specific T cell response
The software program, Deconvolute This! (Mario Roederer, Vaccine Research Center, NIAID, NIH, readily provided upon request) was used to optimize matrix configuration, instruct pooling of peptides, and determine second round assays based on experimental results. Each matrix included 80 pools of approximately 36 peptides per pool. Second round assays were designed by inputting the number of spot forming cells (SFC)/ 1×105 PBMC measured by ELISpot assay for each peptide pool in the first round of testing. The software outputs the minimum number of spots measured from each of the four pools for each and every peptide in the matrix. Peptides with spot values above the minimum threshold for all four pools where chosen to be tested individually in a second round of assays.
ELISpot assay
Cryopreserved PBMC were rapidly thawed, washed, and resuspended in R10 media (RPMI 1640 supplemented with 10% heat inactivated fetal bovine serum, 100 U/ml penicillin G, 100μg/ml streptomycin sulfate, and 1.7mM sodium glutamate) containing 10U/ml DNAse I (Roche Diagnostics, Indianapolis, IN), and rested for 2-4 h at 37°C. ELISpot plates (Millipore, Bedford, MA) were coated with anti-human IFN-γ (Mabtech, Cincinnati, OH). After extensive washing with PBS, plates were blocked by incubation with R10 media. 1×105 PMC were combined with peptide (2μg/ml/peptide) and plated at 100μl per well. In some experiments cells were plated at 2×105/well. Plates were incubated overnight (18 h) at 37°C.
Following overnight incubation, cells were removed and plates were washed. Spots were developed by addition of colorimetric substrate (NBT/BCIP; Pierce, Rockford, IL) after incubation with a biotinylated, anti-human IFN-γ Ab, followed by streptavidin-conjugated enzyme (both Mabtech) with appropriate washing. After drying, spots were counted using an automated reader (CTL, Cleveland, OH). Negative control wells of cells without peptide were included on each plate and averaged 3.6 SFC per well (range: 0-16) for an input of 1×105 PBMC.
Results
Configuration of peptide pool matrices to T cell epitopes
For future studies, we sought to use peptide matrices based upon distinct HIV sequences. Therefore, we used autologous HIV sequences derived from four acute seroconverters to synthesize overlapping peptides spanning all open reading frames (excluding Vif, Vpr, and Vpu) of HIV from these four subjects. The total number of peptides for each of the four HIV sequences was approximately 720. By running simulations of peptide pool configurations as described in our previous theoretical paper (9), we found that the peptide configuration requiring the least amount of blood sample depends on the predicted number of positive peptides in the set. Unfortunately, this cannot be known in advance. Using our software program, we optimized peptide pool configurations for 720 total peptides based on a number of positive peptides ranging from 1 to 25 (Tables I and II). We chose to construct our matrices based on an estimate of 12 positive peptides, which is close to the median number of peptide responses found previously (8). The optimized configuration for this estimate was a matrix consisting of 80 pools of 36 peptides per pool, such that each peptide was present in four pools (Table I). Peptides were pooled according to this configuration for each HIV sequence; thus, four separate matrices of 80 peptide pools were made.
Table I.
Optimal configurations of peptide pools to deconvolute responses from 720 peptides
| Positive Peptides |
Pool sizea | Coverageb | Number of Pools |
Pipettingsc |
|---|---|---|---|---|
| 1 | 144 | 4 | 20 | 2880 |
| 2 | 144 | 5 | 25 | 3600 |
| 3 | 120 | 5 | 30 | 3600 |
| 4 | 90 | 5 | 40 | 3600 |
| 5 | 80 | 5 | 45 | 3600 |
| 6 | 72 | 5 | 50 | 3600 |
| 8 | 48 | 4 | 60 | 2880 |
| 10 | 48 | 5 | 75 | 3600 |
| 12d | 36 | 4 | 80 | 2880 |
| 15 | 30 | 4 | 96 | 2880 |
| 20 | 24 | 4 | 120 | 2880 |
| 25 | 20 | 4 | 144 | 2880 |
| 30 | 15 | 3 | 144 | 2160 |
Number of peptides per pool
Number of pools to which each peptide belongs
Total number of additions of individual peptides to pools
Configuration chosen for the four matrices in the current study
Each row represents the best configuration of peptide pools to deconvolute a given number of responses (“positive peptides”) with the minimum amount of sample. The number of tests required to deconvolute the response is shown in Table II.
Table II.
Number of tests required to deconvolute peptide responses from 720 possible peptides.
| Configuration | Actual number of positive responses | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 6 | 7 | 10 | 12 | 15 | 20 | 25 | 30 | |
| 1 | 25 | 38 | 66 | 112 | 236 | 369 | 481 | 564 | 648 | 710 | 731 | 737 |
| 2 | 27 | 33 | 49 | 80 | 185 | 315 | 440 | 533 | 631 | 707 | 733 | 742 |
| 3 | 32 | 35 | 43 | 60 | 127 | 226 | 335 | 432 | 548 | 666 | 716 | 737 |
| 4 | 42 | 43 | 46 | 52 | 80 | 131 | 200 | 279 | 395 | 550 | 649 | 702 |
| 5 | 47 | 48 | 50 | 54 | 72 | 109 | 162 | 228 | 333 | 489 | 603 | 673 |
| 6 | 50 | 53 | 54 | 57 | 70 | 97 | 138 | 191 | 285 | 435 | 556 | 639 |
| 7 | 62 | 63 | 64 | 65 | 73 | 88 | 113 | 148 | 214 | 343 | 464 | 563 |
| 8 | 60 | 63 | 64 | 66 | 74 | 88 | 109 | 138 | 190 | 294 | 398 | 492 |
| 9 | 75 | 78 | 79 | 80 | 84 | 91 | 104 | 122 | 162 | 253 | 356 | 455 |
| 10 | 75 | 78 | 79 | 80 | 84 | 91 | 103 | 122 | 162 | 253 | 357 | 454 |
| 12 | 82 | 83 | 84 | 85 | 89 | 96 | 106 | 120 | 148 | 212 | 289 | 367 |
| 15 | 98 | 99 | 100 | 101 | 104 | 108 | 114 | 123 | 141 | 184 | 240 | 304 |
| 20 | 122 | 123 | 124 | 125 | 127 | 130 | 134 | 139 | 150 | 175 | 211 | 255 |
| 25 | 146 | 147 | 148 | 149 | 151 | 153 | 156 | 160 | 167 | 184 | 208 | 240 |
| 30 | 146 | 147 | 148 | 149 | 151 | 155 | 158 | 163 | 171 | 188 | 211 | 237 |
The total number of tests (in both rounds of testing) required to deconvolute a response is shown by the configuration used (each row, as defined by Table I), given a sample with an actual number of positive responses (defined in each column).
Deconvolution of the HIV-specific T cell response of three HIV+ donors
To determine whether our matrix approach would identify peptide-specific responses accurately and efficiently, ELISpot assays were performed using each of the four matrices on three HIV+ individuals who were not those from whom the viral sequences were derived. For each donor, many peptides that elicited T cell responses were identified (Supplemental Table I). Peptides were identified in all HIV gene products tested. The response was broad in two donors (128, 99) and focused mostly on Pol in one donor (113). In many cases two consecutive peptides were identified, strongly suggesting that the epitope is contained within the region of amino acid overlap (Supplemental Figure 1). Many of the peptides have been previously identified (http://www.hiv.lanl.gov/content/immunology/maps/maps.html) and shown to be restricted to defined HLA alleles. Most of the peptides we identified were contained within regions previously restricted to the HLA type of the donor (Supplemental Figure 1 and data not shown).
Threshold determination
Background noise must be discriminated from actual peptide-specific T cell responses when analyzing results from an ELISpot assay. Responses to some epitopes are much higher in frequency than responses to others. It is important to set a threshold that includes low frequency responses without requiring excessive second round testing. Our deconvolution program identifies the SFC value of all four pools for each and every peptide in the matrix. For a peptide to be identified for second round testing, all four pools must exceed the threshold.
Figure 1 shows the number of individual peptides to be tested based on a given threshold setting from a representative first round deconvolution of the response of each donor. In each case, as the threshold increases, there is an exponential decrease in the number of peptides requiring second round testing, until a point at which the curve levels (approximately 15-30 SFC/1×105 PBMC). Figure 1 also displays the percentage of positive peptides that would have been identified using a given threshold. At thresholds in which the number of peptides requiring testing does not vary exponentially (ie the curve becomes level), positive peptides will be missed for second round testing as the threshold is increased.
Figure 1.
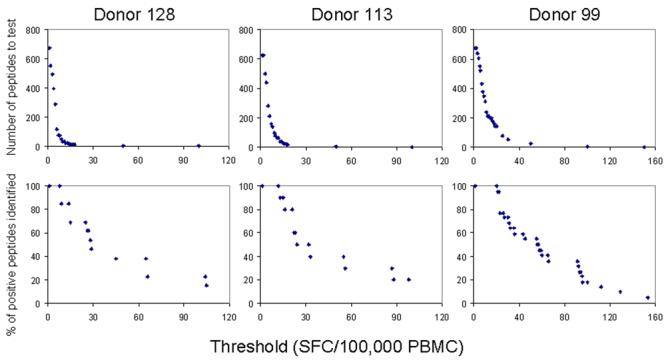
Setting an appropriate threshold involves balancing the number of tests with sensitivity. For three donors, the top panel of graphs shows the number of peptides to be tested in the second round vs. threshold values. The number of peptides requiring testing decreases exponentially as threshold increases. The corresponding graphs below indicate the percent of positive peptides identified vs. threshold. As threshold increases, positive peptides are missed.
The average threshold needed to identify all responses for each donor, expressed as SFC/1×105 PBMC, was: 7 (donor 113, range 2-15), 8.4 (donor 128, range 1-15), and 14 (donor 99, range 8-20). This translates into an average number of second round tests of: 151, 93, and 234, respectively. Accordingly, the average frequency of responding T cells was highest in donor 99 (102 SFC/1×105 PBMC).
Sequence variability affects peptide identification
We were able to assess the effect of sequence variability on peptide identification (or the ability of sequence variants to elicit a T cell response) by using our four unique peptide matrices. Failure to identify a peptide-specific response using a given matrix was most often associated with sequence variability between matrices. For example, in Supplemental Figure 2A, peptide AATNADCAWLEAQEE (Nef53-67) from matrix 3 elicited a positive response in donor 128. However, the similar peptides from the other matrices did not elicit a response. These peptides contained amino acids differences both within the epitope and in the flanking region, which likely reduced their binding to MHC and/or recognition by specific T cells. Some peptide sequence variability was tolerated, as shown in Supplemental Figure 2C, where the corresponding peptides from three matrices were identified despite several amino acid differences that occurred both within and outside of the epitope.
Peptide pool matrices reproducibly identify peptide-specific T cell responses
Mapping experiments were performed three times with each of the four matrices on two donors (128 and 113) using cells from the same cryopreservation lot to determine the reproducibility of peptide identification. In donor 128, we found that the peptides that were most reproducibly identified in our mapping experiments elicited the highest frequencies of T cells when tested singly, (Figure 2). Peptides were less reproducibly identified in donor 113, whose peptide-specific T cell frequencies were generally lower than donor 128 (Figure 2). We speculated that peptide competition within the pools might reduce the ability of a peptide in a given pool to induce a T cell response. Therefore, we performed a more detailed analysis of the results of peptides not reproducibly identified. For example, peptide FRKQNPDMVIYQYMD (Pol329-343, matrix 3) was tested 12 times (four pools, three repetitive experiments). The threshold for first round identification was set at 10 SFC/1×105 PBMC, based on the criteria discussed above. Therefore, for each experiment, all four pools must have scored at least 10 SFC/1×105 PBMC in order to be identified for second round testing. However, this peptide was only identified in two of the three experiments because one of the pools in one experiment fell below this threshold (Supplemental Table II). This pool consistently had a lower T cell response frequency when compared to the other pools, suggesting the composition of this pool may have negatively affected the response to this peptide. The frequency of T cells responding to this peptide in the pools (8-56 SFC/1×105) was much lower than when tested singly (130 SFC/1×105). Table III summarizes the frequency with which a given matrix could identify all of the positive peptides within that matrix. With the exception of matrices 1 and 2 for donor 113, the matrices were highly reproducible for identifying peptides, with an average of identifying 88% of all peptides.
Figure 2.
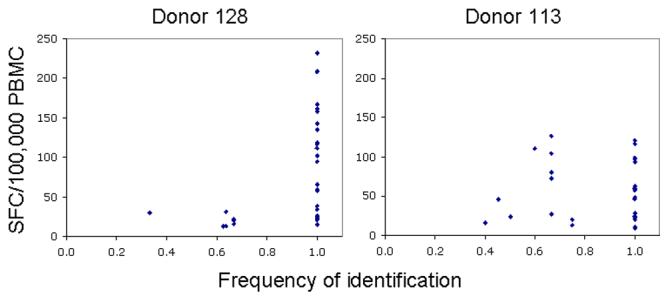
Reproducibility of peptide identification. Mapping experiments were performed multiple times using PBMC from donor 128 and 113. The total number of times a given positive peptide was tested in matrix format depended on how many matrices contained the peptide and how many times the matrices were tested. For example, the same peptide sequence contained in two separate matrices, each tested three times, will have been tested six times (three times for each of the two matrices). On the X-axis is plotted the number of times a given positive peptide was identified divided by the total number of times it was tested (# matrices + # replicates). The Y-axis shows the frequency of the peptide-specific T cell response when the peptide was tested individually.
Table III.
Sensitivity and specificity for peptide matrices
| Donor | Matrix 1 | Matrix 2 | Matrix 3 | Matrix 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity of peptide identification | ||||||||
| n | % | n | % | n | % | n | % | |
| 128 | 13a | 77b | 13 | 100 | 15 | 89 | 12 | 86 |
| 113 | 7 | 67 | 11 | 55 | 19 | 89 | 10 | 97 |
| 99 | 23 | 96 | 22 | 100 | 25 | 100 | 18 | 100 |
| Overall peptide identification capacity | ||||||||
| 128 | 56%c | 72% | 74% | 57% | ||||
| 113 | 21% | 28% | 77% | 44% | ||||
| 99 | 65% | 65% | 74% | 53% | ||||
Number of positive peptides in the matrix
Percent of peptides in the matrix that scored positive (average of 1-3 replicates)
Percent of total peptides (unique peptides identified by any of the four matrices) that scored positive (average of 1-3 replicates). Number of total peptides: Donor 128: 18; Donor 113: 22; Donor 99: 34.
In most cases, identical positive peptides shared by matrices were identified using each matrix in which they were present. However, in rare cases, an identical peptide sequence was identified in one matrix but not another (Supplemental Figure 2B). Such peptides often elicited a low frequency T cell response when tested individually and thus did not fall above the threshold set in the first round of testing, as described above. Accordingly, we found that low frequency peptide-specific responses were identified in mapping experiments where the input cell number was doubled (2×105/well, data not shown). Nevertheless, this result indicates that in some cases, a particular mixture of peptides may inhibit the ability to accurately detect a positive response to one peptide in that mixture. The odds of this occurring increase both for low-level responses and for matrices where each peptide is contained in a larger number of pools (i.e., where the “coverage” increases). Since a single negative response to a pool can eliminate a peptide from second round consideration, this can negatively impact the performance of matrices with greater coverage.
To increase the sensitivity of detection of positive peptides, the deconvolution algorithm can be modified to consider peptides for second round analysis even if they are contained in one pool that is negative (in this example, to use peptides which were scored positive in three of the four pools to which they belong). As expected, proceeding in this way dramatically increased the number of peptides to be tested in the second round of assays (data not shown).
An alternative would be to optimize the original configuration of peptide pools taking into account this modified algorithm, which we term the “n-1 rule.” We modified the program Deconvolute This! to accommodate this variation, and generated the optimal pool configurations shown in Supplemental Table III. As might be predicted, the optimal configurations for any predicted number of positive responses are more complex than for the standard configuration (Table I), in that the coverage is higher. Construction of these pools is more laborious, requiring 50% more pipetting. Similarly, the number of tests to deconvolute a response is about 50% greater using the n-1 rule than the standard approach (Table IV vs. Table II). The improvement in sensitivity remains to be empirically determined. However, for optimal detection of responses under conditions where sample is not as limited, this modified algorithm is suggested.
Table IV.
Number of tests required to deconvolute peptide responses from 720 possible peptides using the “n-1” rule.
| Configuration | Actual number of positive responses | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 6 | 7 | 10 | 12 | 15 | 20 | 25 | 30 | |
| 1 | 32 | 65 | 176 | 345 | 616 | 715 | 741 | 746 | 747 | 747 | 747 | 747 |
| 2 | 37 | 44 | 76 | 142 | 334 | 517 | 637 | 701 | 740 | 754 | 755 | 755 |
| 3 | 50 | 51 | 58 | 80 | 188 | 346 | 499 | 612 | 706 | 757 | 767 | 768 |
| 4 | 56 | 59 | 63 | 74 | 131 | 233 | 356 | 471 | 607 | 723 | 762 | 773 |
| 5 | 72 | 75 | 76 | 79 | 102 | 165 | 268 | 385 | 548 | 707 | 768 | 786 |
| 6 | 80 | 83 | 84 | 86 | 99 | 137 | 204 | 291 | 434 | 624 | 728 | 773 |
| 7 | 90 | 93 | 94 | 95 | 103 | 128 | 179 | 256 | 396 | 601 | 721 | 777 |
| 8 | 96 | 99 | 100 | 101 | 107 | 124 | 160 | 216 | 326 | 518 | 660 | 742 |
| 9 | 105 | 105 | 109 | 110 | 115 | 127 | 151 | 188 | 268 | 426 | 571 | 677 |
| 10 | 120 | 120 | 124 | 125 | 127 | 134 | 149 | 175 | 238 | 385 | 537 | 657 |
| 12 | 120 | 120 | 124 | 125 | 127 | 134 | 149 | 175 | 238 | 385 | 538 | 658 |
| 15 | 126 | 126 | 130 | 131 | 134 | 140 | 152 | 172 | 221 | 337 | 471 | 591 |
| 20 | 160 | 163 | 164 | 165 | 167 | 169 | 174 | 182 | 205 | 277 | 383 | 500 |
| 25 | 192 | 192 | 192 | 197 | 199 | 201 | 203 | 208 | 218 | 256 | 324 | 413 |
| 30 | 210 | 213 | 214 | 215 | 217 | 219 | 221 | 225 | 233 | 260 | 306 | 371 |
Overall performance of peptide pool matrices
By using four matrices, each with a unique set of HIV peptide sequences, on non-autologous HIV+ donors, responses were defined to more peptides than were present in any one single matrix. Combining all peptide-specific responses and replicate testing, the matrix “performance”, or percentage of peptides a given matrix could identify, was determined for each donor (Table III). On average any given matrix could identify 57% of peptide-specific T cell responses.
Discussion
The ability to map T cell epitopes efficiently is important to determine the breadth of the response to a natural infection or vaccine. Our optimized method for configuration of peptide pool matrices enables construction of a single matrix encompassing a set of overlapping peptides spanning a large antigen (several hundred peptides). Peptide matrices have been used to map T cell responses to CMV, a large DNA virus with 213 open reading frames (11-14). The use of one large matrix reduces the amount of sample required to deconvolute the entire T cell response (9). However, as the method is based on peptides, sequence variability between the antigen and the synthetic peptides in the matrix can hamper identification of peptide-specific responses. The sequence variability of HIV both within an individual and between individuals exemplifies this problem for mapping of HIV-specific T cell responses. It has been shown previously that the HIV-specific T cell response is underestimated when using consensus versus autologous sequences for peptide epitope mapping (15). The experimental evaluation of this method in this study yielded several observations: 1) many HIV-derived peptide antigens from several different gene products were identified in three HIV+ individuals using a single matrix of 720 peptides; 2) sequence variability affected peptide identification; 3) the method was reproducible; and 4) setting a frequency threshold for responses is an essential and influential step in the deconvolution process.
The method was efficient and reproducible. Positive peptides were missed in several instances for several reasons. Low frequency peptide-specific T cell responses sometimes fell below the limit of detection. Such peptides either elicited a low frequency T cell response individually or when combined with other peptides in a given peptide pool. Indeed, we found a peptide (FRKQNPDMVIYQYMD, Pol329-343) for which the frequency of responding T cells was much lower when stimulation was performed in the presence of other peptides than when this peptide was tested individually, suggesting peptide competition for binding to HLA can occur and reduce the ability to elicit a peptide-specific response. Despite these limitations, we believe this method holds advantages over other mapping strategies, such as using previously identified or bioinformatically predicted epitopes (6,16,17) , which requires HLA identification and may not be comprehensive, and combinatorial peptide library approaches (18), which require outgrowth of T cell clones, a process that can be highly selective and eliminate marginally-growing clonotypes.
Our initial optimization of matrix configuration assumed a qualitative assessment of positivity. However, analysis of the experimental results required translating a quantitative value into the qualitative determination of “positive” or “negative.” We accomplished this through use of a threshold, below which values were considered negative. Determining the threshold is a tradeoff between the number of second round tests and sensitivity. Lowering the threshold increases the number of peptides requiring testing in the second round. On the other hand, raising the threshold reduces the number of tests but can also eliminate identification of low frequency responses. Thus, the cost of higher sensitivity (and identification of more responses) is the use of a much greater amount of patient sample as well as labor and materials.
Not surprisingly, certain matrices identified responses in some individuals better than others, most likely reflecting the amount of sequence identity shared between the matrix and the HIV species of the donor. With each matrix, some peptides were missed; on average, a given matrix could identify only 57% of responses. Thus, for HIV, this would seem to suggest that epitope mapping should be performed using autologous sequences. While this would be the best approach to identify the complete breadth of the response, it is a very impractical approach. Whole genome sequencing and synthesis of such numbers of peptides are expensive and time-consuming; moreover, matrix construction is extremely labor-intensive. Each of our matrices in this study required >3,000 pipettings, excluding initial reconstitution of peptides and subsequent dilution of individual peptides to be retested in second round assays. Fortunately, mapping of HIV-specific T cell responses in vaccinated individuals, in whom the sequence is pre-determined, enables construction of one matrix that can be used for all vaccinees.
Taken together, the results from these experiments provide the basis for a number of recommendations for epitope deconvolution. (1) Matrices should be optimized for a higher number of epitopes than is predicted in sample material. The number of tests required to deconvolute a response increases exponentially with the number of positive peptides; thus, the cost of having an under-configured matrix is substantial. (2) The number of peptides that are positive is usually about 50% greater than the number of epitopes, due to overlap of epitopes across peptides. The optimization should thus be for the predicted number of peptides (not epitopes). (3) Sample permitting, the “n-1 rule” should be used when optimizing configurations and analyzing first-round results to provide a greater flexibility that accounts for possible peptide interference in pools and allows for lower responses to be detected. (4) Selecting a threshold for positivity in the first round must be done by balancing the number of second round tests required and the sensitivity desired.
When deconvoluting the response to a variable antigen, such as HIV, using multiple matrices that differ in peptide sequence may increase the number of epitopes identified. However, the only potential estimate for how well any given matrix will identify all positive epitopes from an individual is the sequence identity between the matrix and autologous sequence, which is often not known and varies over time. Constructing autologous sequence matrices is very tedious, time-consuming, and expensive. Recently, Frahm and colleagues have reported that using “toggled” peptides, where alternative amino acids are incorporated at variable positions, can increase detection of HIV-specific T cell responses (19). For studies that draw conclusions based on the number of epitopes recognized between individuals, the use of multiple or autologous matrices, or toggled peptide sets, would strengthen the data. However, for routine deconvolution, one matrix may be sufficient.
In conclusion, use of a single peptide pool matrix encompassing hundreds of peptides allows for efficient deconvolution of T cell peptides. Using the ELISpot assay as a functional readout, this method requires a relatively low number of cells (10-20 million for mapping of HIV-specific responses) and is a highly powerful tool for determining the breadth of a pathogen-specific T cell response.
Supplementary Material
Acknowledgments
We thank David Ambrozak, William Adams, and Ribka Ayana for technical assistance. We also thank the VRC Clinical Trials Core for obtaining patient samples.
Footnotes
This work was supported by grants AI 043638, AI 047745 and the UCSD Center for AIDS Research (AI 36214) from the National Institutes of Health.
This work was presented at the Measurement of Antigen-Specific Immune Responses (MASIR) conference in La Plagne, France 2008
References
- 1.Anthony DD, Lehmann PV. T-cell epitope mapping using the ELISPOT approach. Methods. 2003;29(3):260–9. doi: 10.1016/s1046-2023(02)00348-1. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmeister B, Kiecker F, Tesfa L, Volk HD, Picker LJ, Kern F. Mapping T cell epitopes by flow cytometry. Methods. 2003;29(3):270–81. doi: 10.1016/s1046-2023(02)00349-3. [DOI] [PubMed] [Google Scholar]
- 3.Maecker HT, Dunn HS, Suni MA, Khatamzas E, Pitcher CJ, Bunde T, Persaud N, Trigona W, Fu TM, Sinclair E. and others. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry J Immunol Methods 20012551-227–40. [DOI] [PubMed] [Google Scholar]
- 4.Addo MM, Altfeld M, Rosenberg ES, Eldridge RL, Philips MN, Habeeb K, Khatri A, Brander C, Robbins GK, Mazzara GP. and others. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals Proc Natl Acad Sci USA 20019841781–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altfeld M, Addo MM, Eldridge RL, Yu XG, Thomas S, Khatri A, Strick D, Phillips MN, Cohen GB, Islam SA. and others. Vpr is preferentially targeted by CTL during HIV-1 infection J Immunol 200116752743–52. [DOI] [PubMed] [Google Scholar]
- 6.Betts MR, Casazza JP, Patterson BA, Waldrop S, Trigona W, Fu TM, Kern F, Picker LJ, Koup RA. Putative immunodominant human immunodeficiency virus-specific CD8(+) T- cell responses cannot be predicted by major histocompatibility complex class I haplotype. J Virol. 2000;74(19):9144–51. doi: 10.1128/jvi.74.19.9144-9151.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu XG, Addo MM, Rosenberg ES, Rodriguez WR, Lee PK, Fitzpatrick CA, Johnston MN, Strick D, Goulder PJ, Walker BD. and others. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection J Virol 200276178690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA. and others. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load J Virol 20037732081–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roederer M, Koup RA. Optimized determination of T cell epitope responses. J Immunol Methods. 2003;274(1-2):221–8. doi: 10.1016/s0022-1759(02)00423-4. [DOI] [PubMed] [Google Scholar]
- 10.Betts MR, Ambrozak DR, Douek DC, Bonhoeffer S, Brenchley JM, Casazza JP, Koup RA, Picker LJ. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J Virol. 2001;75(24):11983–91. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F. and others. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects J Exp Med 20052025673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kern F, Surel IP, Faulhaber N, Frommel C, Schneider-Mergener J, Schonemann C, Reinke P, Volk HD. Target structures of the CD8(+)-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J Virol. 1999;73(10):8179–84. doi: 10.1128/jvi.73.10.8179-8184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kern F, Bunde T, Faulhaber N, Kiecker F, Khatamzas E, Rudawski IM, Pruss A, Gratama JW, Volkmer-Engert R, Ewert R. and others. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals J Infect Dis 2002185121709–16. [DOI] [PubMed] [Google Scholar]
- 14.Gibson L, Dooley S, Trzmielina S, Somasundaran M, Fisher D, Revello MG, Luzuriaga K. Cytomegalovirus (CMV) IE1- and pp65-specific CD8+ T cell responses broaden over time after primary CMV infection in infants. J Infect Dis. 2007;195(12):1789–98. doi: 10.1086/518042. [DOI] [PubMed] [Google Scholar]
- 15.Altfeld M, Addo MM, Shankarappa R, Lee PK, Allen TM, Yu XG, Rathod A, Harlow J, O’Sullivan K, Johnston MN. and others. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences J Virol 200377137330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalod M, Dupuis M, Deschemin JC, Goujard C, Deveau C, Meyer L, Ngo N, Rouzioux C, Guillet JG, Delfraissy JF. and others. Weak anti-HIV CD8(+) T-cell effector activity in HIV primary infection J Clin Invest 1999104101431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin W, Sbai H, De Groot AS. Bioinformatics tools for identifying class I-restricted epitopes. Methods. 2003;29(3):289–98. doi: 10.1016/s1046-2023(02)00351-1. [DOI] [PubMed] [Google Scholar]
- 18.Sospedra M, Pinilla C, Martin R. Use of combinatorial peptide libraries for T-cell epitope mapping. Methods. 2003;29(3):236–47. doi: 10.1016/s1046-2023(02)00346-8. [DOI] [PubMed] [Google Scholar]
- 19.Frahm N, Kaufmann DE, Yusim K, Muldoon M, Kesmir C, Linde CH, Fischer W, Allen TM, Li B, McMahon BH. and others. Increased sequence diversity coverage improves detection of HIV-specific T cell responses J Immunol 2007179106638–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


