Abstract
MicroRNAs (miRNAs) are 20−24 nucleotide RNA molecules that play essential roles in posttranscriptional regulation of target genes. In animals, miRNAs bind to target mRNA through imperfect complementary sequences that are usually located at the 3’ untranslated regions (UTRs), leading to translational repression or transcript degradation. In plants, miRNAs predominately mediate degradation of target mRNAs via perfect or near-perfect complementary sequences. MicroRNA targets include a large number of transcription factors, suggesting a role of miRNAs in the control of regulatory networks and cellular growth and development. Many miRNAs and their targets are conserved among plants or animals, whereas some are specific to a few plant or animal lineages. Conserved miRNAs do not necessarily exhibit the same expression levels or patterns in different species or at different stages within a species. Therefore, sequence and expression divergence in miRNAs between species may affect miRNA accumulation and target regulation in interspecific hybrids and allopolyploids that contain two or more divergent genomes, leading to developmental changes and phenotypic variation in the new species.
Keywords: microRNAs, hybrids, polyploidy, interspecies, gene expression, evolution
Introduction
MicroRNAs (miRNAs) are a class of small RNAs that serve as post-transcriptional negative regulators of gene expression in plants and animals [1-7]. The 20−24 nucleotide single-stranded miRNAs repress the target genes by mRNA degradation or translational repression [8-11]. MIRNA genes are transcribed by RNA polymerase II [12, 13] from intergenic and/or coding sequences that are independent of their target genes, generating primary miRNA (pri-miRNA) that is processed by nuclear RNaseIII-like enzymes [14], such as Dicer and Drosha in animals [14] and DICER-LIKE proteins (e.g., DCL1) in plants [15]. The resulting pre-miRNA contain miRNA:miRNA* intermediate duplex formed by self-complementary foldback structure. HEN1 methylates the 2’ hydorxy of the 3’ terminal nucleotide of plant miRNAs [16], which protects the 3’ end from uridylation and degradation [17]. The pre-miRNAs are transported into cytoplasm by HASTY (HST) [18], a homolog of Exportin-5 that is involved in transport of pre-miRNAs [19, 20] and tRNAs [21] in animals. HYL1, a double-stranded RNA binding protein, is also required for miRNA accumulation [22, 23]. The double-stranded miRNA that is unwound by a helicase-like enzyme, and the miRNA strand whose 5’-end is less tightly paired [9, 24] is usually incorporated into the effector RNA-induced silencing complex (RISC) [2, 25]. One or more ARGONAUTE proteins such as AGO1 in the active miRNA-containing RISC complex help guide the targets by complementary sequences [26, 27]. As a result, most plant miRNAs function as negative regulators to guide miRNAs to mRNA targets for degradation [2, 28]. Furthermore, some miRNAs may play a role in chromatin modifications and gene transcription. The genes encoding two Arabidopsis miRNA targets (PHABULOSA and PHAVOLUTA) are heavily methylated downstream of the miRNA complementary sites, and the methylation is reduced in phb-1d and phv-1d mutants [29]. Although the cause of reduced DNA methylation is unknown, the data suggest a link between miRNAs and transcriptional regulation.
Plants produce many short-interfering RNAs (siRNAs) that are often generated from endogenous loci and repeats or by exogenous agents such as virus [5, 28, 30]. These siRNAs are different from miRNA loci because siRNAs are derived from aberrant double-stranded RNAs and require the activity of RNA-dependent RNA polymerases (RDRs) [31]. Biogenesis of siRNAs requires the functions of DCL2, DCL3, DCL4, RDR2, RDR6, and RNA polymerase IV [32-34]. In some cases, endogenous siRNAs trigger epigenetic effects on target loci and are associated with RNA-directed DNA methylation and chromatin remodeling [30, 35-38]. RDR6 plays a role in silencing of transgenes, some viruses, and specific endogenous mRNAs that are targets of trans-acting siRNAs (ta-siRNAs) [39, 40]. ta-siRNAs are formed by miRNA-guided cleavage within pre-ta-siRNA transcripts (phasing) and through DICER-LIKE processing of dsRNA that is dependent on RDR6/SDS3 activity [39-41]. The phased ta-siRNAs direct the degradation of target mRNAs as do plant miRNAs [28].
After the first miRNA (lin-4) was discovered in Caenorhabditis elegans [42], and since then, thousands of miRNAs have been identified in plants and animals [43]. Estimates indicate that 1−5% of the transcribed genes in animals contain miRNAs, making them one of most abundant and dynamic classes of genetic regulators [44]. The collection of miRNAs is growing in the miRNA Registry (http://microrna.sanger.ac.uk/sequences/). Release 10.1 (December 2007) listed 5,395 miRNA locus entries, including 564 in human, 461 in mouse, 193 in zebra fish, 137 in nematode, 147 in fruit fly, 199 in Arabidopsis, 215 in poplar, 243 in rice, 96 in maize, 263 in moss, and 72 in alga [43]. In plants many of miRNAs have relatively few targets because target recognition requires near-perfect complementarity, whereas target recognition for the animal miRNAs requires a relatively low level of sequence complementarity [2, 3, 45].
Complementarity to the core region (positions 1−10) of miRNA is often sufficient for effective regulation in animals. Therefore, one miRNA can affect transcript and protein levels of hundreds of targets in animals [46]. Consequently, these miRNAs control a wide range of physiological and developmental processes in animals and in plants as well. In plants, miRNAs mediate leaf development [11, 47-49] including radial patterning in shoots [50], organ identity and flowering [8, 51-55], phytohormone signaling [56-59], and responses to biotic and abiotic stresses [59-62] that are also mediated by natural cis-antisense siRNAs (nat-siRNA)[63] and 30−40 nt small RNAs [64]. ta-siRNAs affect temporal regulation of leaf and trichome development [40, 65].
Although the role of miRNAs in animal and plant development has been extensively studied, little is known about how miRNAs are conserved between species, how the conserved miRNAs play similar or different roles in distinct and related species, how spatial and temporal regulations of conserved miRNAs change among the related species, and whether the expression patterns of miRNAs and their targets in the progenitors are also maintained in the interspecific hybrids and new allopolyploid species that are derived from two ore more divergent species [66-69]. There is evidence for sequence variation and expression divergence of miRNAs between species as well as co-evolution of miRNA loci and their targets within species [70-72]. In this brief review, we analyzed and reported computational and experimental data documented in the last decade, and presented some hypotheses on miRNA regulation between related species and in interspecific hybrids and allopolyploids.
Conservation and diversity of miRNAs between species
The sequences of mature miRNAs are generally conserved in animal or plant kingdom [2, 5, 45], and a few are even conserved between animal and plant kingdom [73]. In animals, many miRNAs, including let-7 and mir-1, appear at a branch from basal metazoan to the common ancestors of invertebrates, vertebrates, and mammals [71, 74]. These conserved microRNAs often have similar functions in the different animal lineages. For example, mir-1 displays muscle-specific expression in nematode, fruit fly, and human, suggesting an essential role of mir-1 in muscle development throughout the animal kingdom [75-77].
Major miRNA renovations occurred at the emergence of vertebrates and placental mammals [71]. Many miRNAs, such as miR-126 and miR-206, are expressed in vertebrate-specific organs [78]. Moreover, ∼42% of the 800 primate miRNAs are primate-specific and are absent in other mammals [79]. Using the massively parallel signature sequencing (MPSS) technique, Berezikov et al. (2006) identified 477 new miRNA genes in the brains of humans and chimpanzee [80]. Among all small RNAs cloned, 75% were known human and primate miRNAs, 14% were conserved in vertebrates, 10% were primate-specific, and 1% were human-specific. The data suggest that miRNA evolution is an on-going process. Some miRNAs are species-specific, and others are expanded in a species through duplication events as observed in plants [81, 82].
Like animal miRNAs, the majority of plant miRNAs are conserved. Among the first set of 16 miRNAs identified in Arabidopsis, eight match orthologous loci in rice (Oryza sativa L. ssp indica) [83]. These miRNAs and stem-loop precursor structures are evolutionarily conserved between a monocot and an eudicot. The mature miRNA sequences are almost invariable, and the high level of sequence conservation extends to the stem-loop precursor duplex. However, the sequences outside the mature miRNAs are highly variable. The data suggest an important role of secondary precursor structure in miRNA processing and biogenesis [28, 81].
Since miRNA precursors are transcribed by RNA polymerase II and polyadenylated [12, 13], some miRNAs should be present in a plethora of expressed sequence tags (ESTs). Indeed, by analyzing ∼6 million ESTs, Zhang et al. (2006) identified 481 miRNAs that belong to 37 miRNA families in 79 plant species, and many of them are conserved across major plant lineages from mosses and gymnosperms to monocots and eudicots [72]. Among 41 miRNA families further examined, nine (miR156/157, 172, 170/171, 165/166, 159/319, 396, 168, 160, and 390) are highly conserved and present in 10 or more plant species and families. miR156/157, miR172, and 170/171 are found in 45, 24, and 22 different plant species corresponding to 21,12, and 12 plant families, respectively. Ten (miR394, 164, 169, 167, 162, 398, 414, 393, 397, and 163) are moderately conserved and present in 5−9 plant families, sixteen (miR95, 408, 399, 158, 403, 161, 406, 173, 419, 415, 413, 416, 417, 418, 420, and 426) are lowly conserved and found in 2−4 plant families, and only six (miR400, 401, 402, 404, 405, and 407) are specific to one plant species (Arabidopsis). Note that some non-conserved miRNAs may be present in many other species after additional ESTs and genomic sequences become available. For example, miR163 and 158 are thought to be specific to Arabidopsis [2]. However, recent studies suggest that they are present in five and four different plant species, respectively [72].
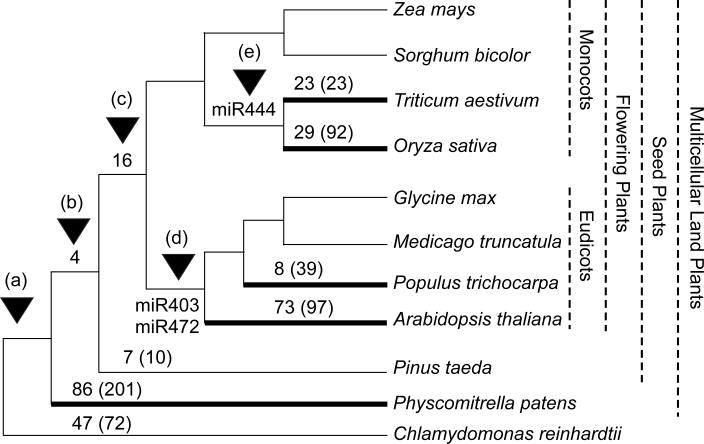
In spite of a high level of sequence conservation of miRNAs in plants [43, 72], many miRNAs are not conserved in different plant species [72, 84], and several major renovations of miRNAs occurred during plant evolution from unicellular alga to vascular and seed plants [43] as shown in the abbreviated phylogeny [85] (Figure 1 and Supplementary Table 1). There are 310 miRNA families and 1271 miRNA loci identified in 1 unicellular and 10 multicellular plants. Chlamydomonas reinhardtii (green alga) contains 47 unique miRNA families in 72 loci [86, 87], and 86 families in 201 miRNA loci are found only in Physcomitrella patens (moss) [88-90], suggesting an explosion of miRNAs in the multicellular mosses relative to the unicellular algae. In contrast, the expansion of miRNAs from gymnosperms (pine, Pinus taeda) [91] to angiosperms (flowering plants) is relatively small (only 16 new miRNA families) probably because of relatively under-sequenced genomes in some species such as pine. Major renovations of miRNAs occur within the eudicots and monocots, respectively. For example, 73 miRNA families (97 loci) are specific to Arabidopsis [61, 83, 92-97], and 8 new miRNA families (39 loci) are found in poplar (Populus trichocarpa) [62, 98]. Among 48 non-conserved miRNA loci in Arabidopsis, many are encoded by single-copy genes [94]. Sequence similarity analyses of miRNA precursor foldback arms revealed evidence for recent evolutionary origin of 16 MIRNA loci through inverted duplication events from protein-coding gene sequences [81, 82, 94]. A small number of microRNAs identified in soybean [99] and Medicago [100] is probably due to a low coverage of genomic sequences in these species. In monocots, rice and wheat have 23 and 29 specific miRNA families, respectively [101-104].
Figure 1.
Evolution of miRNAs in plants. An abbreviated phylogeny is adapted from a previous study [85] using the miRNA Registry (http://microrna.sanger.ac.uk/sequences/) (Release 10.1, December 2007) [43]. Thick lines indicate the species with plant phyla that have undergone major miRNA renovations. A major miRNA renovation (a) gives rise to 47 miRNA families (72 loci) that are uniquely present in the unicellular Chlamydomonas reinhardtii. Four miRNA families (b) are present in all multicellular land plants including mosses that show the second renovation, giving rise to additional 86 miRNA families and a total of 201 loci in land plants. The next renovation of miRNAs (c) is associated with seed plants that have resulted in 7 (10 loci) and 16 miRNA families in gymnosperms and angiosperms, respectively, and is then followed by renovations in eudicots (d) and monocots (e). There are a couple of major miRNA renovations within each lineage, although only a few miRNAs are common in each lineage (miR403 and miR472 in eudicots and miR444 in wheat and rice). The number of miRNA loci is shown in parenthesis next to the number of miRNAs or miRNA families. The numbers of miRNA families and loci identified in unicellular Chlamydomonas, moss, and various plant species [43] and used for constructing the phylogenetic tree are shown in Supplementary Table 1.
These data suggest that miRNA evolution is an on-going process through a “death and birth” pathway [94]. Consistent with this idea, except for a few young miRNA loci in Arabidopsis, miRNA loci in higher plants are more mature than those in mosses and the unicellular Chlamydomonas [81, 86, 88, 89]. Indeed, the miRNA loci in Arabidopsis and higher plants are predominately short hairpin loci. In contrast, the majority of miRNA loci in Chlamydomonas and mosses are long hairpin loci with the characteristics of an early stage in miRNA evolution [86, 88]. Moreover, many miRNA loci in Chlamydomonas and mosses are derived from protein-coding regions [86, 88, 90], while many others are arranged in closely linked clusters of two or more miRNA stem loops as observed occasionally in Arabidopsis [105] and maize [106]. The data suggest that miRNA loci undergo rapid changes in plant evolution.
Expression variation of miRNAs among different species
Although sequence conservation of miRNAs and target genes may suggest conservation of expression patterns and functions, this assumption does not hold true for many conserved miRNAs. By comparing expression patterns of ∼100 miRNAs that are conserved in sequences in fish, chicken, and mouse, Ason et al. (2006) indicated that the timing and location of miRNA expression is not strictly conserved [107]. Several conserved miRNAs such as miR-454a, miR-145, and miR-205 clearly displayed spatial expression differences between two closely related species, medaka and zebrafish. It is conceivable that the spatial and temporal regulation of conserved miRNAs may also play an important role in shaping developmental and physiological changes during animal evolution [71].
In plants, many conserved miRNAs are expressed in diverse species. In a study using miRNA microarrays and RNA blot analysis, Axtell et al. (2005) found that out of 23 miRNAs examined, 19 were expressed in A. thaliana rosette leaves, 13 were expressed in tobacco leaves, 12 were expressed in wheat germ lysate, 13 accumulated in rice seedlings, 13 accumulated in magnolia leaves, 11 accumulated in pine leaves, eight were detected in fern leaves and stems, three were expressed in lycopod leaves and stems, and two were expressed in moss leaf gametophytes [108]. Even the most conserved miRNAs, such as miR160 and miR390, exhibited expression differences between species. Expression of miR390 was not detected in lycopod, pine, or tobacco, but it was expressed in moss. These expression variations may suggest that miR390 expression was lost in some lineages during evolution. Alternatively, expression levels of miR390 could be below the detection level in some species. The expression variation of conserved miRNAs in plants and animals may be underestimated because of several reasons. RNA blot analysis and miRNA microarrays using pooled tissues may not detect real-time changes in cell types, tissues, or organs. Moreover, developmental variation may exist among different species. Alternative techniques such as miRNA in situ hybridization may reveal subtle changes in spatial and temporal expression among different organs and between different species.
Given that the miRNA regulatory systems mediate gene expression important to biological pathways and developmental variation, many target genes may be under evolutionary pressure to maintain or avoid miRNA complementary target sites [109]. In animals, the majority of target sequences of miRNAs are found in their 3’ untranslated regions (UTRs) [110, 111]. Computational analyses have shown that only ∼10% of predicted target sites are conserved across species, whereas the majority (∼90%) is not conserved [112, 113]. Microarray analyses further demonstrated tissue-specific regulation of conserved and non-conserved targets. The genes with non-conserved targeting sites tend to be expressed in tissues and organs in which the miRNAs are not expressed, whereas the genes with conserved targeting sites are often expressed in the same tissues or organs but the miRNAs are expressed prior to or after the expression of the targets [46]. This suggests evolutionary constraints for the genes with conserved targeting sites and neutral fitness for the genes with non-conserved targeting sites, leading to tissue-specific and developmental dependent regulation of miRNAs and their targets during evolution.
Target prediction in plants is relatively simpler than that in animals because plant miRNA targets have near-perfect miRNA binding sites [83, 114, 115]. For example, not only are the mature miR166 and 165 conserved between Arabidopsis and gold club moss (Selaginella kraussiana), but also are the binding sites of their targets that encode for the class-III homeodomain-leucine zipper (HD–Zip) gene family conserved in all lineages of land plants, including mosses, ferns, and seed plants [116]. Outside the miRNA binding site, the sequence conservation is much lower, and only 8% of the third bases in the condon are identical. The high level of sequence conservation between miRNAs and their target binding sites may underestimate sequence variation of target genes across different plant species. In the flowering plants, the miR159/319 family is highly conserved and targets members of MYB and TCP transcription factor gene families [47, 115]. In moss, none of the predicted targets using the published method is homologous to the MYB or TCP genes [88]. One of the predicted targets is a gene with a cyclin domain in moss. The cleavage sites of the predicted target were validated by the RNA ligase-mediated rapid amplification of 5’-cDNA ends (RLM 5’-RACE) method. By reducing the stringency of search for miR319 targets in moss, Axtrell et al. (2007) identified two additional genes that are homologous to MYB transcription factor genes in flowering plants. RLM 5’RACE also confirmed that these two genes are cleaved by miR319 in moss as in flowering plants [88]. Therefore, accurately identifying miRNA targets in distantly related species may require modifying computational parameters and performing additional experiments.
MicroRNAs and interspecific variation of gene expression in allopolyploids
Although the regulatory mechanisms for miRNAs and their target recognition are highly conserved between species, expression variation of miRNAs and their targets exists among different species and even closely related species. This may reflect divergence of miRNA regulatory networks between species. Transcription of miRNA precursors is mediated by RNA polymerase II [12, 13], suggesting a role of cis- and trans-acting factors and chromatin modifications in the transcription of miRNA producing loci. Moreover, processing factors such as Drosha in animals and DCL1 in plants and effector complex such as Argonaute proteins directly affect maturation of miRNAs and efficiency of regulation on target genes [2, 28].
Differences in miRNA accumulation and target recognition between species may be caused by several possible reasons. First, the recently evolved miRNA loci underwent frequent “death and birth” [94], and these genes may have a distinct evolutionary path. Some non-conserved miRNAs may have drifted from parental gene families and have no interactions with the original gene transcripts, whereas some young MIRNA loci clearly originated from one gene family but form miRNAs that target transcripts in another family. Duplicate miRNA loci may lead to an expansion of miRNA families across different species and differential expression of miRNA members in the same family [52, 82, 105]. Only a subset is stably maintained in similar regulatory networks across species. Second, both non-conserved and conserved MIRNA loci may exhibit spatial and temporal expression patterns due to changes in cis-regulatory elements, trans-acting factors, and/or chromatin modifications. This may result in tissue-specific expression patterns of miRNAs and possibly their targets as well [107]. For example, miRNAs from miR56/157 family are upregulated in seedlings and siliques and down-regulated in inflorescences and stems, whereas miR172 has opposite expression patterns. miR163, miR398, and miR396 accumulate at higher levels in leaves and roots than that in stems and inflorescences [108]. Third, non-conserved targeting and targeting avoidance may increase expression variation of the targets across species, even though coevolution of miRNAs and their targets is thought to constrain inter-species expression variation by reducing stochastic noise and buffering expression variation [70]. Indeed, the expression patterns of the majority of miRNA targets are generally negatively correlated with those of their corresponding miRNAs. However, some components of miRNA machinery are part of feedback regulation by miRNA accumulation. DCL1 mRNA is subjected to negative feedback regulation through the activity of miR162 [117], and AGO1 expression is negatively regulated by miR168 in a feedback loop [27].
In comparison with miRNA genes from 12 Drosophila species Stark et al. (2007) suggested constraints on evolution of the foldback structure of miRNA loci [118]. This is probably related to relative homogenous size (∼80-bp) of animal miRNA foldbacks. As a result, over 30 newly discovered miRNAs in 12 Drosophila species appear to target an overall similar gene set of known miRNAs. Plant miRNA foldback sequences are highly variable and may contribute to the specificity of miRNA targeting.
An evolutionary role of miRNA sequence divergence and target selection has been demonstrated in Arabidopsis [119]. The miR159 and miR319 families share sequence identity in 17 of 21nucleotides but affect different developmental processes through distinct targets. MiR159 regulates 8 genes encoding MYB transcription factors, whereas miR319 predominantly target the six TCP transcription factor genes. The effects of miR319 on MYB targets are relatively minor. In contrast, miR159 has no effect on TCP targets. This targeting specificity is determined by sequence variation in miR159 and miR319. Mutations in the miR319 nucleotide sites that differ between miR519 and miR319 alter the specificity of targeting MYB genes but retain the ability of targeting TCP genes, while sequence modifications in miR159 do not show any changes in targeting specificity. Moreover, suppressor screen identified mutants defective in nucleotide positions relevant for miRNA activity, some of which may partially affect miRNA-guided target cleavage. Thus, functional specialization of miRNAs is achieved through changes in sequences and expression.
In a recent study of the MIR319a locus in Arabidopsis and its related species, Warthmann et al. (2008) found that this miRNA locus is relatively conserved among A. thaliana, A. lyrata, A. halleri, and Capsella rubella but is highly divergent between A. thaliana and Brassica oleracea. Interestingly, overexpression of miR319a foldback sequence derived from A. thaliana and A. lyrata, and Sibara virginica in A. thaliana induces cotyledon epinasty, crinkled leaves and siliques, probably because of miR319-guided degradation of mRNAs encoding a series of related TCP transcription factor genes [47, 119], whereas overexpression of B. oleracea miR319, a divergent ortholog, has no effects on leaf and flower development [120]. RNA blot analysis indicated that B. oleracea miR319 was not detectable in the transgenic plants. This suggests that the B. oleracea miR319 transgene is silenced or expressed at low levels. Alternatively, the B. oleracea miR319 primary transcripts and precursors may be poorly processed in A. thaliana.
It is reasonable to speculate that expression variation of miRNAs and their targets must be modulated in the cell nuclei of interspecific hybrids and allopolyploids that contain two or more sets of genomes from divergent species. Polyploidy is a major evolutionary feature of many plants and some animals [121, 122]. Both allopolyploids and autopolyploids (duplication of a single genome) are prevalent in nature [69, 123-126]. Recent work has shown that polyploid genomes undergo rapid changes in genome structure and function via genetic and epigenetic changes [66, 68], including chromosomal rearrangements (e.g., translocation, deletion, and transposition), DNA sequence elimination and mutations, and chromatin and RNA-mediated changes that give rise to gene expression changes that are not associated with changes in DNA sequence.
Genome-wide expression analyses in plant allopolyploids [127-129] and hybrids between inbred lines [130] have indicated that many genes are expressed nonadditively (differently from the mid-parent value), which is consistent with nonadditive variation in phenotypes (e.g., dominance and novel variation) in allopolyploids and hybrids. Interestingly, over 50% miRNA targets are among the nonadditively expressed genes in the allopolyploids (M. Ha and Z. J. Chen, unpublished data), suggesting a link between differential expression of miRNA targets and accumulation of corresponding miRNAs in the interspecific hybrids and allopolyploids. Interspecies differences in growth and development may be mediated by miRNA pathways because many miRNA targets encode transcription factors or proteins that are important to growth and development in animals and plants [2, 11, 47, 83]. In Arabidopsis, 199 miRNA loci have been identified and grouped into 105 different miRNA families [43, 92, 94-96, 115]. As of December 2007, 51 miRNA families derived from 118 loci have known targets [43]. Among them, at least 21 families from 70 loci have been shown to target genes that encode transcription factors, suggesting an important role for miRNAs in the control of regulatory networks.
RNA interference (RNAi) works in allopolyploids as in diploids and is an effective method for down-regulating redundant genes in polyploids [131, 132]. Overexpessing double-stranded RNAs induces dominant negative effects on the target genes in Arabidopsis suecica, a natural allotetraploid derived from A. thaliana and A. arenosa [133, 134]. Chen et al. (2008) reported that RNAi of met1, a gene encoding DNA methyltransferase, reduced DNA methylation and altered the expression of ∼200 genes, many of which encode transposons, predicted proteins, and centromeric and heterochromatic RNAs [135]. Derepression of transposons, heterochromatic repeats, and centromeric small RNAs was primarily derived from A. thaliana genome. A high level of A. thaliana centromeric small RNA accumulation was correlated with hypermethylation of A. thaliana centromeres. The greater effects of reduced DNA methylation on transposons and centromeric repeats in A. thaliana than in A. arenosa are consistent with the repression of many genes that are expressed at higher levels in A. thaliana than in A. arenosa in the resynthesized allotetraploids. The data suggest differential siRNA accumulation and DNA methylation between closely related species, which may trigger homoeologous-specific RNA-mediated DNA methylation in the allopolyploids.
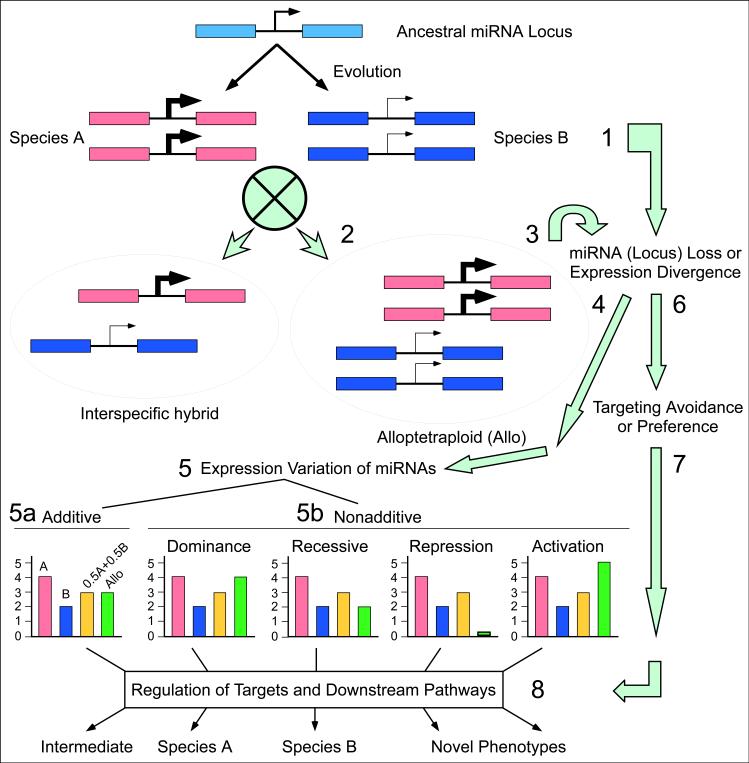
We propose a model to explain nonadditive expression of target genes and phenotypic variation in interspecific hybrids and allopolyploids (Figure 2). MicroRNA loci in different species inherited from ancestral species may diverge in sequence and expression patterns (e.g., tissue-specificity), gain new expression patterns, or undergo gene loss, as a consequence of genetic and epigenetic changes during evolution [66, 68, 136, 137]. Over time, the expression differences are fixed such that the regulatory networks are finely tuned in each species. Combination of two miRNA loci with similar or differential expression patterns will perturb the regulatory balance of miRNAs and their targets in the interspecific hybrids and new allopolyploids. As a result, the accumulation levels of miRNAs may be additive or nonadditive [66, 127]. The latter includes several probable states of gene expression such as dominance, recessive, repression, and activation or overdominance.
Figure 2.
A model for mediating interspecies variation of miRNAs and target gene regulation. The miRNA locus is represented by an arrow between coding sequences (pink, blue, and pale blue boxes). The circled cross represents a genetic cross between the two species, resulting in an interspecific hybrid or allotetraploid (arrow heads). Pale green arrows along with numbers (1−8) indicate possible changes. MicorRNA loci may diverge between species A and B or undergo mutation or loss in one species and/or exhibit expression changes (1). If both loci are present, they may express differently due to genetic and epigenetic mechanisms (2) [66]. Thick and thin arrows in the interspecific hybrid or allotetraploid indicate high and low levels of miRNA accumulation, respectively. Changes in homoeologous chromosomes in the allotetraploid may also induce miRNA locus loss or sequence changes (3). Sequence and expression changes in miRNA loci (4) may lead to expression variations of miRNAs between species and in the interspcific hybrid or allotetraploid. The possible miRNA accumulation levels in an interspecific hybrid or alloteraploid are shown as additive (5a) and nonadditive (5b, dominance, recessive, repression, and activation or overdominance) in histograms. Relative expression levels of miRNAs are shown in red (species A), blue (species B), yellow (mid-parent value, equivalent to an equal mixture of parental RNAs), and green (interspecific hybrid or allotetraploid), respectively. MiRNA sequence divergence or loss may also facilitate target avoidance or preference of one homoeologous locus over another in the interspecific hybrid and allotetraploid (6), which may also induce expression variation of the target genes (7). Expression variation of miRNAs affects regulation of targets and associated regulatory pathways (8) that may result in different outcomes of phenotypic variation (parental types, intermediate to the parents, and novel, see text for discussion).
If the miRNAs originating from related species have similar target binding sites, the miRNA expression variation in the allopolyploids will lead to dosage-dependent negative effects on target genes and their downstream pathways in the allopolyploids. The miRNA-mediated pathways may, on the other hand, give rise to phenotypic outcomes such as intermediate between the progenitors, novel, or similar to one species or another. If the miRNA sequences diverge or target specificities change over time, the expression variation may be caused by one or many factors in combination. But the phenotypic outcomes may still fall in these categories. Some novel phenotypes may depend on intrinsic interactions between miRNAs and divergent and/or new targets, leading to target preference or avoidance [71]. Non-conserved targeting and targeting avoidance may increase expression variation of the targets across species. Changes in target preference or avoidance of homoeologous loci may lead to species-specific expression, morphological, and developmental variation. Furthermore, miRNAs and expression of targets tend to be co-regulated [70], which suggests that miRNA biogenesis proteins such as DCLs, AGOs, and RDRs have concerted regulation within species and diversified modifications between species. There is evidence for rapid evolution of the genes related to antiviral RNAi function (DCR2, R2D2, and AGO2), and they evolve significantly faster than paralogous genes that mediate “housekeeping” functions [138]. Regulatory interactions between divergent miRNAs and their targets, as well as sequence divergence and expression changes in the genes encoding miRNA biogenesis proteins between different species, may reprogram miRNA-mediated biological pathways in the interspecific hybrids and new allopolyploids and thus, shape the growth and development during allopolyploid evolution.
In addition to miRNAs, other small RNAs may also be produced in the polyploids as a general response to the “genomic shock” [139]. Recently, 24- to 30-nt PIWI-interacting RNAs (piRNAs), also known as repeat-associated siRNAs (rasiRNAs), are identified in animal germline cells [140-145]. Biogenesis of piRNAs is Dicer-independent but requires three piwi members (MIWI, MILI/PIWIL2, and MIWI2/PIWIL4) and are crucial to germline development and male fertility [145-147]. The piRNA pathway provides an adaptive defense against transposons and genome invaders [148]. Similarly, 30−40-nt RNAs in Arabidopsis are induced by bacterial pathogens and silence genes encoding protein factors involved in disease resistance [64].
In Drosophila virilis, endogenous siRNAs are derived from the transposon Penelope in both males and females but only maternally loaded in embryos, which may suggest maternal transmission of Penelope siRNA in the repression of transposition [149]. The genome-wide repression of A. thaliana homoeologous loci [127] and accumulation of A. thaliana-centromeric siRNAs associated with changes in DNA methylation [135] may be similar to the maternal repression of transposons because A. thaliana is used as a maternal parent in production of allotetraploids [132, 150]. We failed to produce interspecific hybrids or allotetraploids using A. arenosa as the maternal parent, which is reminiscent of hybrid dysgenesis in Drosophila [151-153] and Peromyscus [154]. It is reasonable to speculate that RNA-mediated pathways involving miRNAs, siRNAs, nat-siRNAs, and piwiRNAs play important roles in hybrid performance such as dysgenesis, sterility, and vigor. In plants, interspecific hybrids and allopolyploids generally display growth vigor. It will be interesting to investigate why and how similar small RNAs in the same allotetraploid cells would cause homoeologous specific changes in gene expression, leading to interspecies variation in growth and development.
Future perspectives
MicroRNAs play an essential role in the evolution of regulatory networks and biological pathways in plants and animals. Although miRNAs are generally conserved and share similar functions in target gene regulation, many miRNAs are not conserved between distantly related species or even closely related species. The notion is supported by the increasing number of miRNAs that have been cloned and sequenced and/or predicted from a rapid expansion of genomic and EST sequences that are available across different species and in many ecological strains within a species. Moreover, the conserved miRNAs are found not to have the same expression patterns among different species or at different developmental stages. Expression variation of miRNAs and sequence divergence between non-conserved miRNA loci may accelerate evolutionary rates of miRNA-mediated pathways. On one hand, miRNAs and their targets coevolve and are co-regulated, leading to the constraints on interspecies expression variation of miRNA targets; on the other hand, non-conserved or duplicate (multiple) miRNA loci diverge under selection, creating new target sites and/or avoiding old ones between species and in different developmental stages within a species. Non-conserved targeting and targeting avoidance may produce a major source of expression variation in miRNA-mediated pathways across species. Finally, cis- and trans-acting factors, chromatin components, Dicer proteins and processing cofactors, and Argonaute proteins, which evolve independently among distantly related species, may also modulate interspecies variation in the transcription of miRNA loci, miRNA processing and maturation, and target regulation. Using the stable allopolyploids as a novel model system, we can monitor how divergent miRNA-mediated pathways originating from different species function in the same cells and how these pathways shape physiological and morphological evolution of the interspecific hybrids and new allopolyploid species.
Supplementary Material
Acknowledgements
We thank Ruth Chang for critical reading of the manuscript. The work was supported by the grants from the National Science Foundation (DBI0501712 and DBI0624077) and the National Institutes of Health (GM067015).
References
- 1.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Bartel B, Bartel DP. MicroRNAs: At the root of plant development? Plant Physiol. 2003;132:709–717. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 5.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 6.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 7.Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 10.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. Rna. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 15.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer Homolog, and HEN1, a Novel Protein, Act in microRNA Metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 20.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calado A, Treichel N, Muller EC, Otto A, Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. Embo J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 23.Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci U S A. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 25.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 26.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 29.Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 31.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 32.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 35.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299 doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 36.Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 37.Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. Embo J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S, Saini HK, Dongen SV, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 45.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 46.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 47.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 48.Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region. Embo J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 51.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 52.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007;134:1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- 53.Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131:4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- 54.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou GK, Kubo M, Zhong R, Demura T, Ye ZH. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007;48:391–404. doi: 10.1093/pcp/pcm008. [DOI] [PubMed] [Google Scholar]
- 56.Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 58.Wu MF, Tian Q, Reed JW. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- 59.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 60.Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell. 2005;17:2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 68.Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, Lee HS, Comai L, Madlung A, Doerge RW, Colot V, Martienssen RA. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003;19:141–147. doi: 10.1016/s0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- 69.Comai L. The advantages and disadvantages of being polyploid. Nature Reviews Genetics. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 70.Cui Q, Yu Z, Purisima EO, Wang E. MicroRNA regulation and interspecific variation of gene expression. Trends Genet. 2007;23:372–375. doi: 10.1016/j.tig.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 71.Niwa R, Slack FJ. The evolution of animal microRNA function. Curr Opin Genet Dev. 2007;17:145–150. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 72.Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 73.Arteaga-Vazquez M, Caballero-Perez J, Vielle-Calzada JP. A family of microRNAs present in plants and animals. Plant Cell. 2006;18:3355–3369. doi: 10.1105/tpc.106.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, Hofacker IL, Stadler PF. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brennecke J, Stark A, Cohen SM. Not miR-ly muscular: microRNAs and muscle development. Genes Dev. 2005;19:2261–2264. doi: 10.1101/gad.1363905. [DOI] [PubMed] [Google Scholar]
- 76.Nguyen HT, Frasch M. MicroRNAs in muscle differentiation: lessons from Drosophila and beyond. Curr Opin Genet Dev. 2006;16:533–539. doi: 10.1016/j.gde.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 77.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 79.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 80.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 81.Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 82.Maher C, Stein L, Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006;16:510–519. doi: 10.1101/gr.4680506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lindow M, Krogh A. Computational evidence for hundreds of non-conserved plant microRNAs. BMC Genomics. 2005;6:119. doi: 10.1186/1471-2164-6-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bowe LM, Coat G, dePamphilis CW. Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales’ closest relatives are conifers. Proc Natl Acad Sci U S A. 2000;97:4092–4097. doi: 10.1073/pnas.97.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 87.Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fattash I, Voss B, Reski R, Hess WR, Frank W. Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol. 2007;7:13. doi: 10.1186/1471-2229-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 91.Lu S, Sun YH, Amerson H, Chiang VL. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 2007;51:1077–1098. doi: 10.1111/j.1365-313X.2007.03208.x. [DOI] [PubMed] [Google Scholar]
- 92.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC. High-Throughput Sequencing of Arabidopsis microRNAs: Evidence for Frequent Birth and Death of MIRNA Genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, Henderson IR, Jacobsen SE, Wang W, Green PJ, Meyers BC. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and Silencing-Associated Small RNAs in Plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Dejardin A, Depamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjarvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leple JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouze P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, Van de Peer Y, Rokhsar D. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 99.Zhang B, Pan X, Anderson TA. Identification of 188 conserved maize microRNAs and their targets. FEBS Lett. 2006;580:3753–3762. doi: 10.1016/j.febslet.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 100.Dezulian T, Remmert M, Palatnik JF, Weigel D, Huson DH. Identification of plant microRNA homologs. Bioinformatics. 2006;22:359–360. doi: 10.1093/bioinformatics/bti802. [DOI] [PubMed] [Google Scholar]
- 101.Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biol. 2007;8:R96. doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang JF, Zhou H, Chen YQ, Luo QJ, Qu LH. Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res. 2004;32:1688–1695. doi: 10.1093/nar/gkh332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V. CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 2007;35:D829–833. doi: 10.1093/nar/gkl991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li A, Mao L. Evolution of plant microRNA gene families. Cell Res. 2007;17:212–218. doi: 10.1038/sj.cr.7310113. [DOI] [PubMed] [Google Scholar]
- 106.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 107.Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc Natl Acad Sci U S A. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Massirer KB, Pasquinelli AE. The evolving role of microRNAs in animal gene expression. Bioessays. 2006;28:449–452. doi: 10.1002/bies.20406. [DOI] [PubMed] [Google Scholar]
- 110.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 111.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 112.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 113.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 114.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 115.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 116.Floyd SK, Bowman JL. Gene regulation: ancient microRNA target sequences in plants. Nature. 2004;428:485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- 117.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 118.Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, Carrington JC, Weigel D. Sequence and expression differences underlie functional specialization of arabidopsis microRNAs miR159 and miR319. Dev Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 120.Warthmann N, Das S, Lanz C, Weigel D. Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Mol Biol Evol. 2008 doi: 10.1093/molbev/msn029. [DOI] [PubMed] [Google Scholar]
- 121.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 122.Grant V. Plant Speciation. Second ed. Columbia University Press; New York: 1981. [Google Scholar]
- 123.Tate JA, Soltis PS, Soltis DE. Polyploidy in Plants, The Evolution of the Genome. Academic Press; New York: 2004. [Google Scholar]
- 124.Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 125.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 126.Levy AA, Feldman M. The impact of polyploidy on grass genome evolution. Plant Physiol. 2002;130:1587–1593. doi: 10.1104/pp.015727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, Chen ZJ. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hegarty MJ, Jones JM, Wilson ID, Barker GL, Coghill JA, Sanchez-Baracaldo P, Liu G, Buggs RJ, Abbott RJ, Edwards KJ, Hiscock SJ. Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Mol Ecol. 2005;14:2493–2510. doi: 10.1111/j.1365-294x.2005.02608.x. [DOI] [PubMed] [Google Scholar]
- 129.Hegarty MJ, Barker GL, Wilson ID, Abbott RJ, Edwards KJ, Hiscock SJ. Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 130.Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc Natl Acad Sci U S A. 2006;103:6805–6810. doi: 10.1073/pnas.0510430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lawrence RJ, Pikaard CS. Transgene-induced RNA interference: a strategy for overcoming gene redundancy in polyploids to generate loss-of-function mutations. Plant J. 2003;36:114–121. doi: 10.1046/j.1365-313x.2003.01857.x. [DOI] [PubMed] [Google Scholar]
- 132.Wang J, Tian L, Madlung A, Lee HS, Chen M, Lee JJ, Watson B, Kagochi T, Comai L, Chen ZJ. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jakobsson M, Hagenblad J, Tavare S, Sall T, Hallden C, Lind-Hallden C, Nordborg M. A unique recent origin of the allotetraploid species Arabidopsis suecica: Evidence from nuclear DNA markers. Mol Biol Evol. 2006;23:1217–1231. doi: 10.1093/molbev/msk006. [DOI] [PubMed] [Google Scholar]
- 134.O'Kane S, Schaal B, Al-Shehbaz I. The origins of Arabidopsis suecica (Brassicaceae), as indicated by nuclear rDNA sequences, and implications for rDNA evolution. Systematic Botany. 1995;21:559–566. [Google Scholar]
- 135.Chen M, Ha M, Lackey E, Wang J, Chen ZJ. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics. 2008 doi: 10.1534/genetics.107.086272. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Matzke MA, Scheid OM, Matzke AJ. Rapid structural and epigenetic changes in polyploid and aneuploid genomes. Bioessays. 1999;21:761–767. doi: 10.1002/(SICI)1521-1878(199909)21:9<761::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 137.Chen ZJ, Ni Z. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays. 2006;28:240–252. doi: 10.1002/bies.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 139.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 140.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 141.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 142.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 143.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 144.Hartig JV, Tomari Y, Forstemann K. piRNAs--the ancient hunters of genome invaders. Genes Dev. 2007;21:1707–1713. doi: 10.1101/gad.1567007. [DOI] [PubMed] [Google Scholar]
- 145.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 146.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 147.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 149.Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci U S A. 2005;102:15965–15970. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell. 2000;12:1551–1568. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Engels WR, Preston CR. Hybrid dysgenesis in Drosophila melanogaster: the biology of female and male sterility. Genetics. 1979;92:161–174. doi: 10.1093/genetics/92.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kidwell MG. Hybrid dysgenesis in Drosophila melanogaster: the genetics of cytotype determination in a neutral strain. Genetics. 1981;98:275–290. doi: 10.1093/genetics/98.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bingham PM, Kidwell MG, Rubin GM. The molecular basis of P-M hybrid dysgenesis: the role of the P element, a P-strain-specific transposon family. Cell. 1982;29:995–1004. doi: 10.1016/0092-8674(82)90463-9. [DOI] [PubMed] [Google Scholar]
- 154.Vrana PB, Fossella JA, Matteson P, del Rio T, O'Neill MJ, Tilghman SM. Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nat Genet. 2000;25:120–124. doi: 10.1038/75518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.