Abstract
Domain mobility plays essential role in the biological function of multidomain systems. The characteristic times of domain motions fall into the interval from nano- to milliseconds, amenable to NMR studies. Proper analysis of NMR relaxation data for these systems in solution has to account for interdomain motions, in addition to the overall tumbling and local intradomain dynamics. Here we propose a model of interdomain mobility in a multi-domain protein, which considers domain reorientations as exchange/interconversion between two distinct conformational states of the molecule, combined with fully anisotropic overall tumbling. Analysis of 15N-relaxation data for Lys48-linked di-ubiquitin at pH4.5 and 6.8 showed that this model adequately fits the experimental data and allows characterization of both structural and motional properties of di-ubiquitin, thus providing information about the relative orientation of ubiquitin domains in both interconverting states. The analysis revealed that the two domains reorient on a time scale of 9–30 ns, with the amplitudes sufficient for allowing a protein ligand access to the binding sites sequestered at the interface in the closed conformation. The analysis of possible mechanism controlling the equilibrium between the interconverting states in di-ubiquitin points toward protonation of His68, which results in three different charged states of the molecule, with zero, +e, and +2e net charge. Only two of the three states are noticeably populated at pH4.5 or 6.8, which assures applicability of the two-state model to di-ubiquitin at these conditions. We also compare our model with the “extended model-free” approach and discuss possible future developments of the model.
Introduction
Many proteins in the cell have modular architecture, i.e. they are composed of several well-folded regions (domains). Relative orientation of the domains and interdomain dynamics often play key regulatory role in functional regulation and molecular recognition events in these systems 1–3. NMR is a powerful tool for studying molecular motions, and recent developments based on spin-relaxation and residual dipolar coupling measurements offer a unique possibility to characterize the conformation and dynamics of multi-domain proteins. Despite significant progress in the determination of interdomain orientation by these methods 4–9, the process of interdomain motion is largely unexplored.
Reorientational motion detected in NMR experiment in solution results from a combination of the overall tumbling and inter- and intradomain dynamics. Deconvolution and separate characterization of these motions from experimental data is non-trivial. A full analysis, accounting for coupling between various modes of motion in a protein, is extremely complex and could involve a many-body stochastic treatment of rotational motions, e.g. like that of Freed and co-workers 10,11. Recent approaches 12–18 based on mode-coupling ideas have been focused on the effect of the overall tumbling on intradomain dynamics. Adequate treatment of multidomain systems, however, requires account for the effect of domain motion on the shape/overall tumbling of the molecule and vice versa, and is currently not available.
The only quantitative treatment of interdomain motions in a protein thus far was the analysis of domain dynamics in Ca2+-ligated calmodulin by Tjandra and co-authors 19,20, based on a variant of the so-called “extended model-free” approach 21. In this model, the overall tumbling and interdomain dynamics are considered independent from each other, and the latter is characterized by an order parameter that measures the angular amplitude of domain motions. The structure of calmodulin in the Ca2+-ligated form is characterized by the absence of a definitive interdomain conformation, thus the “extended model-free” or “wobbling-in-a-cone” models 19,20,22 considering a continuum of available interdomain orientatons seem adequate for that protein. While acceptable in describing domain dynamics in calmodulin, these models have limited applicability when the distribution of domain orientations is asymmetric or discontinuous, like in the case of Lys48-linked di-ubiquitin (Ub2) considered here. Indeed, NMR data23 clearly indicate that in solution Ub2 is in fast dynamic equilibrium between a “closed” conformation with a well-defined Ub/Ub interface and one of more “open” conformations with no direct contact between Ub domains. Models that could adequately represent this situation are currently missing.
Here we present a more structurally detailed than the “model-free” yet still simple model that describes interdomain dynamics in a dual-domain system as interconversion (exchange24) between two distinct conformational states (ITS) of the molecule. The two-state-exchange paradigm is often used as the simplest model of a system undergoing transitions between distinct states. With regard to NMR applications to protein dynamics, exchange or random jump models were applied to fast internal rotations in a molecular label 25 and in amino acid side chains 26. Inspired by these papers, we applied similar ideas to describe reorientational dynamics of protein domains. We show that analysis of 15N relaxation data for Lys48-linked di-ubiquitin based on the ITS model allows determination of both the motional characteristics and the structure of the molecule in the two interconverting states. A preliminary analysis of Ub2’s dynamics has been published elsewhere 27, here we focus on theoretical aspects of the model, its justification, and further development.
Theory
NMR relaxation measurements directly probe the spectral density function (e.g. ref.28), J(ω), which is a cosine Fourier transform of the time-dependent angular autocorrelation function C(t) describing reorientational motion of an internuclear vector under the observation (throughout this paper we focus on 15N relaxation, hence the 15N-1H-bond motion):
| (1) |
where
| (2) |
Here is an element of the Wigner rotation matrix 29 (Supporting Information), the asterisk means complex conjugation, and ΩL→I = {αL→I, βL→I, γL→I} denotes a set of three Euler angles (0 ≤ αL→I ≤ 2π, 0 ≤ βL→I ≤ π , and 0 ≤ γ L→I ≤ 2π) which specify the orientation of a unit vector µ in the direction of a given NH vector with respect to the laboratory coordinate frame (L), aligned with the static magnetic field (see Fig.1 ). Note that, strictly speaking, only two Euler angles are required to define the orientation of a vector, i.e. the value of the angle γL→I is arbitrary. However, for consistency with the other definitions of coordinate frames (see below), we define an “instantaneous” coordinate frame (I) with its z-axis along the instantaneous direction of the NH bond, and the x,y-axes defined such that, for simplicity, γL→I = 0.
Figure 1.
Schematic representation of the various reference frames introduced here and the rotational transformations between them.
Note that the general expression for the correlation function in the right-hand side of Eq. 2 explicitly contains the index q. However, in the case of protein tumbling in isotropic solutions considered in this work, the correlation functions (Eq.2) for different values of q are all equal – this is a consequence of the orientational (ensemble) averaging in the isotropic medium28. Thus the correlation function C(t) and the corresponding spectral density J(ω) both are independent of the actual value of q.
The angular brackets 〈…〉L→I in Eq. 2 denote an equilibrium ensemble-averaging with respect to all possible initial, , and final, , orientations of µ, separated by the time interval t:
| (3) |
where is the equilibrium probability to have the initial orientation of µ in the angular interval is the conditional probability that at the time moment t the orientation of µ is within the interval provided that the initial orientation (at t=0) was .
Thus the problem of describing protein dynamics is reduced to the determination of the probability density functions, , which enter Eq. 3. In principle, these functions could be obtained from the corresponding stochastic dynamics equation (for example, a Fokker-Planck or Smoluchowski equation) that should account for all interactions, both within a protein and with the solvent. However, the complexity of proteins as molecular systems possessing a variety of motions with different amplitudes and time scales makes such ab initio theoretical description impossible. This leads to the necessity of developing a model that would account for the motions of interest.
The model
In this paper we consider motions relevant to spin relaxation in a NH group in protein backbone, and therefore consider only orientational dynamics leading to a change in orientation of a given NH-bond vector. To simplify the problem, we consider three dynamic modes:
the overall tumbling of the protein as a whole;
the interdomain-mobility mode, that is the reorientation of one domain with respect to the other; and
the local dynamics mode, which includes a wobbling-type motion of a given NH bond about its average orientation with respect to a coordinate frame associated with each individual domain.
To separate and analyze the contributions from these dynamics modes to the NMR-observed picture of the NH-bond motion, we consider the orientation of the NH vector with respect to the static magnetic field as a set of subsequent rotations: from the laboratory frame L to the protein reference frame (P) which describes rotation of a protein as a whole (e.g. the principal axes frame of the rotational diffusion tensor), then to the coordinate frame (D) for each individual domain (e.g. PDB coordinate frame), then to the residue-specific frame (R) associated with the average (over the time interval longer than the correlation time of the local motion) orientation of a given NH-vector within each individual domain, and, finally, to the instant frame (I) attached to the NH-vector (Fig. 1). These rotations are characterized by the four sets of Euler angles: angles: ΩL→P, ΩP→D, ΩD→R, and ΩR→I, respectively.
Using properties of Wigner rotation matrices (see Supporting Information) Eq. 2 can then be rewritten in the following form
| (4) |
The model considered here rests on two assumptions:
Postulate I
All three dynamics modes: the overall, interdomain, and local, are statistically independent from each other. This allows factorization of each term in Eq. 2 into a product of separate correlation functions corresponding to these modes (see below).
Postulate II
On a time scale slower than the characteristic time of local NH-bond (wobbling) motion the tertiary structure of each domain holds constant, therefore the Euler angles ΩD→R that describe the averaged orientation of a NH bond with respect to the domain’s reference frame are time-independent.
The usual justification for an assumption like Postulate I is the large separation of the characteristic time scales of the motions. Indeed, the experimentally observed correlation time for local (internal) dynamics in proteins (τlocal) is typically in the 1–100 ps range (e.g. 30), whereas the overall tumbling time (τoverall) is of the order of several nanoseconds and longer (~8 ns for Ub2 23). Thus the statistical independence of these two modes of motion seems a reasonable approximation and is typically assumed, as e.g. in the model-free approach 31,32, although the effect of their coupling on local motion has recently been included in data analysis 14–18.
The situation with interdomain motion is more complex, and one can envision several scenarios. The characteristic time scale of the interdomain dynamics (τinterdomain) could either fall in-between (i.e. the hierarchy of the time scales is formulated as τoverall >> τinterdomain >> τlocal) or could significantly exceed the overall tumbling time, τinterdomain>> τoverall >> τlocal. Moreover, the difference between the time scales of the overall and interdomain motions could be less pronounced, as e.g. in the case of calmodulin 19, where τinterdomain ≈ 3 ns compared to τoverall ≈ 9 ns. If domain reorientation involves tumbling within some energy well, rather than crossing of substantial energy barriers, one might expect that the ratio of τinterdomain and τoverall should reflect the ratio of the rotational friction coefficients (in solution) for the domain and for the whole protein molecule.
Rigorous mathematical treatment of such a system is further complicated by the fact that domain motion can alter the shape of the molecule, hence the assumption of time-independent rotational diffusion tensor might not be valid in the general case. Unlike the restricted local (intradomain) backbone dynamics that are not expected to affect the rotational diffusion, the interdomain dynamics and the overall tumbling could have mutual effect. Adequate treatment of this phenomenon is currently not available. This problem, however, does not exist in a hypothetical case of spherical-shape domains, when their only motion involves rotations around the (fixed) centers of mass of each individual domain, because these rotations will not change the overall shape or the diffusion tensor of the molecule. Our analysis of data (below) suggests that, to a good approximation, Ub2 falls into this category.
The hierarchy of time scales could also imply causality, in that the properties of slower time-scale dynamics are determined by motion on faster time-scales. In contrast, Postulate I assumes no causality between the dynamic modes under consideration, which are therefore completely decoupled from each other (see also below).
Under these assumptions the correlation function Eq. 4 simplifies:
| (5) |
This equation implies that each dynamic mode can be described by its own correlation function:
| (6a) |
| (6b) |
| (6c) |
Each of these correlation functions has its own pair of probability density functions (cf. Eq. 3), for overall tumbling, for interdomain mobility, and and for local motion; and every pair of probability densities is assumed to be independent from the others. Thus, in the framework of this model the description of NMR-relevant dynamics in a multidomain protein reduces to the problem of deriving the corresponding equilibrium and conditional probability densities. These functions are considered here for each of the dynamics modes specified above.
The overall tumbling mode
For the overall tumbling we used the model of rotational diffusion of a fuly anisotropic rigid body 33,34. Following the derivation by Favro 33, we write
| (7) |
where ar,m are decomposition coefficients and Er are the corresponding rate constants which depend solely on the principal values {Dx, Dy, Dz} of the rotational diffusion tensor D (see Supporting Information).
Note that the representation in Eq. 7 assumes a diagonal form of the diffusion tensor, which implies that protein reference frame P is the principal axes frame (PAF) of the diffusion tensor. Another point worth mentioning here is that Eq. 7 implies that the principal values of the diffusion tensor remain constant; this follows from Postulate I, in that the characteristics of the overall tumbling are considered independent of the other dynamics modes in a protein. This approximation might not hold for those proteins where interdomain mobility could alter the overall diffusion tensor.
The domain mobility mode
The simplest model of domain mobility considered here involves interconversion between two states (ITS) of the protein, states A and B, characterized by distinct orientations of each domain with respect to the protein’s global coordinate frame P. These orientations are given for each domain by two sets of Euler angles, and , and the transitions from A to B and backwards are characterized by the rate constants kAB and kBA, respectively. We also assume here that the domain structure and intradomain (backbone) dynamics are the same in both states (see Postulate II).
In this case the continuous probability density functions, that determine the statistical averaging of the correlation function, Eq. 6b, have to be replaced with discrete occupation and transition probabilities:
| (8) |
where the conditional probabilities
| (9) |
are given by the solutions to the corresponding kinetic equations (Supporting Information), and the occupation probabilities
| (10) |
are normalized, pA + pB = 1, and satisfy the condition of detailed balance, pA/kBA = pB/kAB, where kAB and kBA are the forward and backward transition rates, respectively. The characteristic correlation time for this interconversion mode, τITS = 1/(kAB+kBA), is the reciprocal of the interconversion rate, K = kAB+kBA.
Combining Eq. 8–Eq. 10, the correlation function describing the interdomain mobility mode can be recast in a “model-free”-like form (cf. Eq(12)):
| (11a) |
where matrix
| (11b) |
replaces the unity matrix and
| (11c) |
is a matrix analog of the (scalar) squared order parameter.
The local mobility mode
Detailed description of the correlation function of local mobility, Eq. 6c, requires proper selection of the model of motion (not known a priori) for a given group of atoms (for a review see e.g. ref. 35). This problem can be circumvented by using the so-called “model-free” approach 31,32 that assumes a simple form for the correlation function:
| (12) |
where δa,b is the Kronecker delta, τloc is the correlation time for local motion, and S is the generalized order parameter: where the equilibrium ensemble averaging 〈…〉 = ∫ …Peq (Ω) sin β dβ is performed with respect to the Euler angle β. The order parameter is a model-independent measure of the degree of spatial restriction for the local motion: S = 0 when this motion uniformly samples all possible directions in the three-dimensional space, while S = 1 in the case of completely restricted local motion.
The “model-free” form, Eq.12, is just one of the possible models for the correlation function of local motion; examples of explicit mechanical models can be found in refs. 25,26,35,36. It is worth mentioning here that the analysis below is focused on structurally well-defined regions in the protein, where the amplitudes of local backbone dynamics are restricted. In this case the results presented in this paper do not directly depend on the particular form of the correlation function for the local dynamics mode.
Methods
Representation of relaxation data
The analysis of 15N relaxation data here is based on the approach developed in 5 which, instead of dealing with actual NMR relaxation rates, is focused on their ratio
| (13) |
where R1 and R2 are the 15N longitudinal and transverse relaxation rates, respectively, and the primes indicate that the measured rates were modified to subtract the contributions from high-frequency components of the spectral density (see e.g. 37):
| (14) |
where Here γH and γN are the gyromagnetic ratios for 1H and 15N, respectively, ωN is the Larmor frequency of 15N, and d and c represent the strengths of the 1H-15N dipolar coupling and of the 15N chemical shift anisotropy (CSA) (c = − ωN ·CSA/3). As shown in refs.5,38 the ratio ρ, Eq. 13, is independent, to the first approximation, of the site-specific values of the 15N CSA and the NH-distance. Moreover, for well-ordered regions in a protein, where the local backbone mobility is restricted (S2 ≥ 0.8) and fast (τloc << τoverall), J(0) and J( ωN) both scale approximately as S2, so the order parameters in the numerator and denominator of Eq. 13 cancel out, thus rendering the ratio ρ independent of the characteristics of local motion38, to a good approximation39.
The spectral density J( ω) for the ITS model (which takes into consideration all the dynamic modes discussed above) can be obtained as analytical Fourier transform of Eq. 5, combining Eq. 7, Eq. 11, and 12. However, because for the well-defined structural regions, considered here, the ratio ρ is insensitive to the local backbone dynamics, in the subsequent analysis we set the order parameter of local backbone motion in Eq.12 to S = 1. This essentially eliminates the contributions from local motion to the resulting spectral density function, and hence to ρ. Thus in the following analysis we replace the spectral density J(ω) in Eq.13 with its reduced form (designated here as Jρ(ω)):
| (15) |
which was obtained directly from Eq.5 as described above; an explicit expression for this spectral density can be found in our earlier paper 40. The applicability of this treatment to Ub2 is supported by the observation 6 that the backbone dynamics in the core of each Ub domain in Ub2 are restricted and fast and the corresponding order parameters are essentially identical to those in monomeric Ub.
After this paper was submitted, we have learned that a similar, “model-free” like functional form of the spectral density in Eq.15 was recently derived by Bernatowicz et al.41 for an anisotropically tumbling molecule with the intramolecular dynamics modeled using a two-site exchange. Although our paper and that of Bernatowicz et al. both consider essentially the same physical picture of random exchange superimposed by a fully anisotropic overall tumbling, it is instructive to discuss differences between the specific models utilized here and in ref. 41. In that paper, the orientations of each individual spin pair (as, e.g., N-H group in our case) in the exchanging states are not correlated with the other pairs, and the quantities analogous to our matrices andQmk,nl (Eqs.11) are simply scalars (see Eqs. 20–22 in ref. 41). This simplifies the equations, but inevitably limits the structural information available from these parameters. The treatment applied here (see also refs. 27, ref 40) is conceptually different in that we consider random exchange between orientations of the whole domain, thus the P→D transformation is applied to all spin pairs within each domain, while the local intradomain dynamics are neglected. Moreover, the use of stepwise transformations between the corresponding reference frames (Fig.1), allowed us to isolate the dependence of J(ω) on domain orientation, which is one of the principal results of our paper. The dependence of and Qmk,nl on the indices k, l links J(ω) to NH-bond orientations within a given domain, which thus allowed us to determine the orientation of each domain in both interconverting states (see below); this would not be possible if and Qmk,nl were scalars.
Fitting procedure and parameters
All parameters in the ITS model can be separated into two types: global parameters that describe motion of the whole protein and domain parameters that are specific for every domain. Thus, the principal values Dx, Dy, and Dz of the diffusion tensor and the transition rates kAB and kBA are the same for all NH vectors in the protein, as they describe reorientation of the whole protein and correlated changes in the mutual orientation of the domains. The sets of Euler angles and which specify orientation of the domain’s coordinate frame (D) with respect to the global protein coordinate frame are domain-specific, i.e. apply only to a subset of NH vectors belonging to a particular domain. The Euler angles ΩD→R that are also present in Eq. 15 and specify the averaged orientation of each NH bond within the domain, are defined by the three-dimensional structure (e.g. PDB coordinates) of each domain and therefore are fixed throughout the analysis. Thus, when considering a protein system of n domains that simultaneously change their mutual orientation between two distinct conformations of the whole protein, the ITS model will require 5 global parameters (Dx, Dy, Dz, kAB and kBA) and 6n domain-specific parameters (6 Euler angles, and , for each domain). Thus, the total number of ITS parameters is Np=17 in the case of Ub2.
The parameters of the ITS model were obtained by minimizing the following target function:
| (16) |
where for each of the N residues included in the sum, is the value of ρ derived directly from the experimental data according to the first equality in Eq. 13 and is obtained by substituting into the second equality in Eq. 13 the value of Jρ(ω) calculated from Eq. 15 for the current values of the Np fitting parameters F = {X1,X2, …X Np}. Here σj is the experimental uncertainty in extimated as , where δR1j' and δR2j' are the experimental uncertainties in the relaxation rates R1j' and R2j', respectively, for residue j.
The minimization of the target function, Eq. 16, was performed using in-house Matlab program based on the simplex algorithm. Multiple runs with different starting conditions were performed to assure that the resulting set of fitting parameters is the global minimizer. The confidence intervals for the fitting parameters were obtained using the method of constant- χ2 boundaries and the bootstrap method42. The latter method, becoming increasingly popular for confidence interval estimations in complex systems, is based on the idea of using the original experimental data set to generate a number of synthetic data sets in which a certain fraction of the original data points (1/e≈37%) is replaced by randomly chosen duplicates of the remaining data. Thus, these synthetic data sets should have the same statistical properties as the original one. At least 200 sets of synthetic data were generated in each case. These synthetic data were analyzed in the same way as the original data, and the confidence intervals reported here were obtained from the resulting distributions of the values of fitting parameters.
Degeneracy of interdomain orientation
As indicated above, the Euler angles and define the orientation of a given domain with respect to the global protein frame. However, determining the interdomain orientation from these angles requires caution, because of the degeneracy of the problem, which stems from the fact that the diffusion tensor has no directionality, i.e. it is not sensitive to the inversion of one or more of its principal axes (two axes if the right- or left-handedness has to be preserved). This results in a four-fold degeneracy (e.g. 4) of the eigenvectors of the diffusion tensor (assuming they form a right-handed system), such that if one set of the vectors is known, the other three can be obtained by rotations specified by the Euler angles { π,0,0}, {0,π,0}, and {0,π,π}.
Applied to domain orientation procedure, this means that a given set of Euler angles or , obtained from the fit) determines four different domain orientations which, however, have equivalent spin-relaxation properties. Thus, the fitted values of two sets of Euler angles and for one domain and sets of similar angles for another domain could belong to different branches of solution. However, a pair of and for a given protein domain should belong to the same solution branch, because the correlation function of the interdomain dynamics, Eq. 11, is independent of the overall diffusion and, therefore, has no such degeneracy. Which of all possible (degenerate) domain orientations is selected should be based on additional information, for example, chemical linkage, chemical shift perturbation mapping, spin-labeling data or other considerations 6. When the branches of solution for all domains are chosen, the appropriate angles should be used to orient the domain’s coordinate frame with respect to the PAF of the overall diffusion tensor and to obtain the conformation for the state with the occupation probability pA. The sets of should be used in the same way to get the conformation with occupation probability pB.
Results and Discussion
Analysis of 15N relaxation data for di-ubiquitin
To illustrate the ITS model, we apply it here to interdomain dynamics in Lys48-linked Ub2. The 15N relaxation data, R1, R2, and steady-state {1H}-15N NOE were collected at 14.1 Tesla and 24°C in segmentally 15N-labeled Ub2 at pH 4.5 and 6.8. Details of the measurements are reported elsewhere 23. The ratio of relaxation rates (ρexp, Eq.13) for the two Ub units in Ub2 is shown in Fig.2. Throughout this paper, the Ub unit that carries the free C-terminus is defined as the proximal Ub domain. The other Ub unit in Ub2 is defined as the distal domain. Thus, the two Ub molecules in Ub2 are linked via an isopeptide bond between the C-terminal Gly76 of the distal Ub and Lys48 of the proximal Ub.
Figure 2.
Comparison between the experimental data and the various fitting models. Symbols represent ρexp, the ratio of relaxation rates for the experimental data included in the fit. Lines represent the values of ρcalc for all amides in Ub back-calculated for all three approaches to data analysis considered here: the ITS model (red), the IDT model, where anisotropic diffusion was applied separately to each domain (IDT) (blue), and are for the SID model that corresponds to domain dynamics being turned off (green). The horizontal bars on the top indicate the location of the secondary structure elements (black for the β-strands; magenta for the helices).
The ratio of relaxation rates (ρexp, Eq.13) for the two Ub units in Ub2 is shown in Fig.2. The observed average value of ρexp~7·10−2 corresponds to the overall correlation time of ~8.5 ns, i.e. twice that for monomeric Ub, which indicates that to a good approximation the two domains tumble as a single molecular entity rather than completely independent “beads on a flexible string”. However, differences in the levels of ρexp between the two Ub units evident from this Figure indicate that Ub2 still possesses some degree of interdomain mobility, which has differential effect on the relaxation data measured for the two domains. Note that the backbone dynamics in the two Ub units are essentially identical (and similar to those in monomeric Ub)6; this excludes the possibility that the observed differences in ρexp between the two Ubs could be caused by differences in their local intradomain motions. Ignoring interdomain dynamics in Ub2 results in a striking discrepancy between the characteristics of the diffusion tensor of Ub2 reported by the two domains27 (see also below).
The chemical shifts and residual dipolar couplings indicate that the tertiary structure of both Ub domains in Ub2 is essentially the same as in monomeric Ub 6,23. Therefore the solution structure of monomeric Ub (PDB code 1D3Z 43) is used here as the structure model for each Ub unit in Ub2. Because the structural deviations within the ensemble of 10 NMR structures are small, the first conformer from 1D3Z was used here as a representative model of Ub structure.
Altogether, 71 and 91 residues from both domains were included in the analysis of relaxation data at pH 6.8 and 4.5, respectively. At pH 6.8 these comprise 36 backbone amides from the proximal Ub (residues 2–6, 11–17, 21, 28, 32–34, 36, 39–45, 49, 50, 57–59, 61, 64–67, 69) and 35 from the distal Ub (2–4, 13–18, 26–30, 32–36, 40–43, 45, 48–50, 54, 57, 59, 65–69). The corresponding numbers for pH 4.5 were 43 residues from the proximal Ub (2, 3, 5–7, 13–18, 20–22, 26–30, 32, 34–36, 39–44, 50, 52, 54–58, 61–67) and 48 from the distal Ub (2–5, 13, 14, 16–18, 20–22, 26–36, 39, 40, 42–50, 54, 55, 57–68). These amides belong to well-defined structural regions in Ub. Residues in the flexible regions and those amides showing conformational exchange 6 were excluded from the analysis, along with those amides that could not be reliably quantified due to signal overlap in the spectra.
Models for data treatment
Based on the chemical shift perturbations observed in Ub2 upon pH titration, it has been suggested that Ub2 is in dynamic equilibrium between two states, corresponding to the closed and open conformations 23. Lowering the pH from 7.5 to 4.5 results in a transition from a predominantly closed to the predominantly open conformation of Ub2. The observation of a single set of signals in the 1H-15N HSQC spectra in a broad pH range (from 4.5 up to 8.0), indicates that the interconversion is fast on the NMR chemical shift time scale. The estimated relative population of the closed conformation at pH6.8 is ≥0.85. This makes Ub2 an appropriate molecular system for testing the ITS model of domain motion. The fitting parameters included (see Methods) the principal values Dx, Dy, Dz of the diffusion tensor, the rates of interconversion, kAB and kBA, all of which were considered global parameters, and two sets of Euler angles, and , per domain, altogether 17 adjustable parameters. The results of the fit are presented in Table 1. In order to validate these results, we used two additional data treatment procedures described below, as controls.
Table 1.
Parameters of the ITS model for Lys48-linked Ub2 derived from 15N relaxation data measured at pH 6.8
| pH 6.8 | pH 4.5 | |||
|---|---|---|---|---|
| Dxa | 1.53 (0.32) | 1.61 (0.13) | ||
| Dya | 1.73 (0.06) | 1.71 (0.06) | ||
| Dza | 2.20 (0.08) | 2.20 (0.06) | ||
| τITS b | 9.3 (4.8) | 31.9 (9.8) | ||
| pA c | 0.90 (0.06) | 0.82 (0.06) | ||
| Proximal | Distal | Proximal | Distal | |
| αA d | 218 (35) | 91 (28) | 147 (30) | 213 (24) |
| β A d | 109 (11) | 58 (7) | 112 (16) | 80 (8) |
| γ A d | 140 (4) | 321 (19) | 322 (8) | 350 (12) |
| αB d | 203 (38) | 156 (33) | 191 (25) | 151 (29) |
| βB d | 110 (9) | 96 (38) | 122 (14) | 51 (20) |
| γB d | 72 (8) | 356 (33) | 45 (13) | 328 (23) |
Numbers in the parentheses represent estimated uncertainties in the parameters.
Principal components, Dx, Dy, and Dz, of the overall diffusion tensor, in 107 s−1 (ordered as Dx ≤ Dy ≤ Dz).
Characteristic time constant for the interconversion between the two states, τITS=1/K, in 10−9 s.
Occupation probability for the more populated state (this state is called state A); the occupation probability for state B is pB =1- pA.
The Euler angles, in degrees, specify for the states A and B (indicated by the superscripts) the orientation of the PDB frame for each Ub domain with respect to the PAF of the overall diffusion tensor. Protein atom coordinates in this study were from the solution structure of monomeric Ub (PDB entry 1D3Z 43, model 1). The use of these coordinates for the individual domains in Ub2 has been validated by RDC measurements 6. The original Ub coordinates were rotated by {90°, 90°, 180°} to avoid having β ~ 0 or 180° when the angles α and γ cannot be accurately separated.
The same relaxation data were analyzed earlier 23 applying fully anisotropic rotational diffusion model to each Ub domain separately. In that treatment, the interdomain mobility was not explicitly taken into account, i.e. the effects of the overall and interdomain dynamics were not separated from each other. Instead, both these motions contributed to the resulting effective diffusion tensor. Thus it is instructional to compare the results of that approach (henceforth called the “individual-domain treatment”, IDT) with those from the ITS model. For this purpose we repeated the analysis of ref.23, using the same list of residues as for the ITS model. The IDT requires six fitting parameters per domain: the principal values of the effective diffusion tensor and three Euler angles, altogether 12 adjustable parameters for Ub2. The results of the analysis are presented in Table 2.
Table 2.
Values of the fitting parameters for the IDT model applied to Lys48-linked Ub2. The residuals of fit per degree of freedom, were 1.9 and 16.3, for the 15N relaxation data measured at pH 6.8 and pH 4.5, respectively.
| pH 6.8 | pH 4.5 | ||||
|---|---|---|---|---|---|
| Proximal | Distal | Proximal | Distal | ||
| Dxa | 1.68 (0.09) | 1.73 (0.10) | 1.78 (0.04) | 1.69 (0.08) | |
| Dya | 2.06 (0.06) | 1.86 (0.06) | 2.00 (0.05) | 1.77 (0.06) | |
| Dza | 2.30 (0.09) | 2.69 (0.13) | 2.15 (0.05) | 2.50 (0.12) | |
| τov b | 8.28 | 7.96 | 8.44 | 8.39 | |
| αd→D c | 89 (8) | 127 (28) | 114 (13) | 139 (27) | |
| βd→D c | 145 (13) | 126 (4) | 66 (13) | 112 (4) | |
| γ d→D c | 139 (9) | 134 (6) | 148 (9) | 145 (4) | |
Numbers in the parentheses represent estimated uncertainties in the parameters.
Principal components, Dx, Dy, and Dz, of the overall diffusion tensor, in 107 s−1 (ordered as Dx ≤ Dy ≤ Dz).
Apparent overall correlation time, τ ov, in 10−9 s.
Euler angles, in degrees, determine orientation of the PDB coordinate frame for each Ub domain with respect to the PAF of the overall diffusion tensor. Note that here we rotate the reference frame, whereas the coordinate vectors instead are being rotated in the ROTDIF program 50 used in ref 6; both methods use the so-called “y-convention” for the reference frame rotations (Supporting Information). Therefore the rotation matrices for the same transformation here and in ROTDIF are transposed to each other.
As one can see from Fig. 2, the overall levels of ρ are different for the two domains. Not surprisingly, when analyzed separately, the two Ub domains yield slightly different effective diffusion tensors, likely reflecting the contributions from domain dynamics, as discussed above. As also shown above, the ITS model also nicely fits the experimental data – in this case both domains are analyzed together. Therefore, in another control fit, the interdomain mobility was turned off by setting 1/kBA=0, which resulted in a single fixed orientation per domain, hence a single Ub2 conformation. In this case, termed suppressed interdomain dynamics (SID) here, the interconversion is completely suppressed by pushing the equilibrium entirely towards one of the two interconverting states (in this case A). This fit served as a test of whether an alternative treatment that involves simultaneous analysis of both domains but no interdomain dynamics could fit the data. It is worth noting that, in contrast to the IDT, the SID model implies a single overall diffusion tensor for both protein domains. Thus, the analysis includes 9 adjustable parameters: Dx, Dy, Dz as global parameters and a set of three Euler angles ΩP→D for each domain. The results of this analysis are presented in Table 3.
Table 3.
Values of the fitting parameters for suppressed interdomain dynamics (SID) model for relaxation data measured at pH 6.8 and pH 4.5 .
| pH 6.8 | pH 4.5 | |||
|---|---|---|---|---|
| Dxa | 1.81 (0.03) | 1.78 (0.02) | ||
| Dya | 2.01 (0.03) | 1.92 (0.02) | ||
| Dza | 2.43 (0.03) | 2.27 (0.02) | ||
| τovb | 8.00 | 8.38 | ||
| Proximal | Distal | Proximal | Distal | |
| αd→ D c | 231 (17) | 9 (8) | 111 (10) | 172 (10) |
| βd→D c | 16 (5) | 51 (3) | 71 (3) | 113 (4) |
| γd→D c | 152 (14) | 347 (8) | 151 (3) | 177 (6) |
Numbers in the parentheses represent estimated uncertainties in the parameters.
Principal components, Dx, Dy, and Dz, of the overall diffusion tensor, in 107 s−1 (ordered as Dx ≤ Dy ≤ Dz).
Apparent overall correlation time, τov, in 10−9 s.
Euler angles, in degrees, specify the orientation of the PDB coordinate frame for each Ub domain with respect to the PAF of the overall diffusion tensor.
The quality of fit
Because all these models use different number of fitting parameters, we first compare the residuals of fit per degree of freedom, χ2/(N-Np) as an objective criterion of the quality of data fit to each model. The data (Table 1– Table 3) suggest that at both pH values, the worst agreement between the model and the experiment was for the SID treatment. The two other models, ITS and IDT, have approximately the same values of χ2/(N-Np) and both exhibit a remarkably better agreement with the experimental data (see Fig. 2). Also the correlation between the experimental and back-calculated values of ρ (Fig. 3) indicates that the suppressed interdomain dynamics model is the least adequate for Ub2. The Pearson’s correlation coefficient 42 for both IDT and ITS was 0.95 (pH6.8) and 0.94 (pH4.5), while turning off the interdomain dynamics reduced it to 0.84 and 0.86 at these pH. These results indicate that the ITS model is a significant improvement, and thus support the necessity of including interdomain dynamics into Ub2 data analysis.
Figure 3.
The agreement between experimentally measured (ρexp) and back-calculated (ρcalc) values of the ratio of relaxation rates, Eq.13, for the ITS, IDT, and SID models. The top and bottom rows correspond to pH6.8 and 4.5, respectively. The lines are guides-for-eye representing the case of absolute agreement. The values of the Pearson’s correlation coefficient r are indicated.
The interdomain orientations in Ub2
The reconstruction of interdomain orientation in the IDT is based on the idea of aligning the axes of the diffusion tensor of Ub2 experienced by the two Ub domains, as detailed in 6. The domain alignment in the ITS model is relatively straightforward, because the Euler angles ΩP→D determine the orientation of each domain with respect to the common, global coordinate frame (P). It should be emphasized that only the relative orientation of the domains not their position with respect to each other is available from the Euler angles. Therefore, the domains are positioned here somewhat arbitrarily in the three-dimensional space. In addition, one should bear in mind the issue of orientational degeneracy discussed above. Thus, additional information is necessary in order to pinpoint the proper conformation. One such piece of information is the chemical link between the two Ub domains, which requires that the C-terminus of the distal Ub is located close to Lys48 of the proximal Ub. In addition, chemical shift perturbations observed in Ub2 at pH6.8 23 indicate a close contact between the β-sheets of the two Ub units. Fig. 4 depicts the conformations of Ub2 derived using these considerations for all three fitting models.
Figure 4.
Conformations of Lys48-linked Ub2 obtained from the analyses using various models considered here. The left panels correspond to pH6.8, the right panels to pH4.5. In all structures, the proximal domain is colored green, the distal is blue. The principal axes of the overall diffusion tensor are shown as cyan rods. Panels a through d show two distinct conformations of Ub2 obtained using the ITS model; the red rods represent the rotation axes for each domain (Supporting Information). Panels e and f depict the Ub2 conformations obtained for anisotropic diffusion model applied to each domain separately (IDT). Panels g and h represent Ub2 conformations obtained by the simultaneous analysis of both domains assuming that the interdomain dynamics are turned off (SID model). The location of the hydrophobic patch residues Leu8, Ile44, Val70 on each Ub domain is represented by spheres (Cβ atoms, colored gold), the side chain of Lys48 of the proximal domain is shown in stick representation, colored orange. All molecular drawings here were made using MolMol 49.
As expected, the ITS model yields two different (interconverting) conformations of Ub2 for both pH6.8 and 4.5 data sets (see Figs. 4A and B). For these structures, the transition between the two orientations of each domain (states A and B), derived from the Euler angles and , can be represented as a rotation about some fixed axis (Supporting Information). It is remarkable that, while the actual Ub2 conformations are significantly different between the two pH values, the spatial orientation of the rotation axes, as well as the rotation angles are quite similar for both pH. Note that the rotation axes for both domains go through the Ub-Ub linker region, consistent with the expectation that the linker acts as a hinge in the interdomain motion in Ub2.
The Ub2 conformations obtained for the IDT model, applied separately to proximal and distal domains, are shown in Figs. 4e and 4f. These conformations are virtually identical to the ones reported in 23, where it has been proposed that they correspond to the “closed” (Fig. 4e, pH 6.8) and “open” (Fig. 4f, pH 4.5) conformations of Ub2. There is a remarkable similarity between these conformations from the IDT analysis and the most populated conformations for the ITS model: the closed conformation is similar to that in state A at pH6.8 (pA = 0.90) and the open conformation resembles that in state A at pH4.5 (pA = 0.82). In contrast, the conformations obtained from the SID treatment are ambiguous: the conformation at pH 4.5 (Fig. 4h) looks similar to the open conformation whereas that at pH 6.8 (Fig. 4g) is different from all others.
pH dependence of the Ub2 conformations
As mentioned earlier, NMR data 23 indicate the disappearance of a defined Ub/Ub interface as pH is titrated from 7.5 to 4.5. The Ub/Ub interface is stabilized by a balance between the hydrophobic effect pushing the two domains together and their electrostatic repulsion caused by positively charged side chains surrounding the hydrophobic patches on both Ub domains. Lowering the pH from 7.5 to 4.5 is expected to cause the protonation of His68 (pKa = 5.5 44) adjacent to the hydrophobic patch on both Ub units.
The accompanying increase in the Coulomb repulsion between the two domains is perhaps the reason that the closed state (Fig. 4a) becomes energetically unfavorable at low pH. From the ratio of the occupation probabilities of states A and B, the difference in their Gibbs free energies is ΔG = GA–GB ≈ −5.5 kJ/mol (pH6.8) and −3.8 kJ/mol (pH4.5), where the lower-energy state (state A) corresponds to the closed conformation at pH 6.8 (Fig. 4a) and the open one at pH 4.5, shown in Fig.4d.
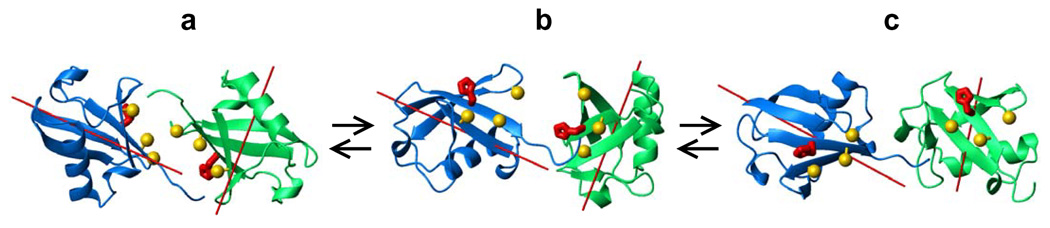
The picture of interconverting Ub2 conformations obtained here (Fig.4a–d) appears more complex than a simple, seemingly intuitive model in which the limiting states, open and closed, would be the same at both pH values and only their relative population would change with pH. In this regard it is worth noting a striking similarity between the Ub2 conformations in the weakly populated state B for different pH (Figs.4b and c). While further studies are necessary to understand these results in detail, there is a simple scheme that can explain the relations between Ub2 conformations obtained here for different pH. The observed three conformational states of Ub2: the closed (Fig.4a), the open (Fig.4d), and the intermediate one (Figs. 4b,c), can be explained when taking into account that the stable conformations of Ub2 are determined by the balance between the hydrophobic effect (favoring the interdomain interface) and electrostatic repulsion between the two Ub units. For simplicity, consider that in the pH range from 4.5 to 6.8, the only residue in Ub that is expected to undergo a significant change in the charge state of its side chain is His68. Located in the middle of the hydrophobic patch on Ub (Fig.5), His68 side chains from both Ub domains face each other in the closed conformation. With the distance of approximately 8.3Å between protonatable nitrogens of the two imidazole rings in the crystal structure of Ub2, one could expect a strong electrostatic repulsion between Ub domains in the case when both rings are protonated (at low pH). Depending on its protonation state, His68 on each Ub unit could have a charge of either zero or +e (for simplicity, we are not considering states with partial charges here). Therefore, Ub2 can be in one of the three possible states:
The state of zero net charge, Q=0, (the histidines on both domains of Ub2 are neutral) – this state corresponds to the closed state in Fig 4a, because the hydrophobic effect is predominant.
The state with the intermediate net charge, Q=+e, when only one of the two histidines is protonated. In this state the electrostatic repulsion (between His68 on one Ub and the positively charged side chains surrounding the hydrophobic patch on the other Ub unit) is stronger than in the neutral state. This could drive Ub2 into the intermediate conformation (Fig.4b,c), in which a new balance is reached between the hydrophobic effect and the electrostatic repulsion.
The state of maximal net charge, Q=+2e, when both histidines are protonated – here the electrostatic repulsion is the strongest and could drive Ub2 into an open conformation (Fig.4d), thus keeping the His-containing hydrophobic patches on the two Ub units apart from each other.
Figure 5.
Available conformational states of Lys48-linked Ub2 in solution: (a) closed, (b) intermediate, and (c) open. The spheres (colored gold) indicate the location of the hydrophobic patch residues Leu8, Ile44, Val70 on each Ub unit; the side chains of the histidines (His68) are shown in stick representation, colored red. The distal Ub is colored blue; the proximal Ub is green. The experimental and predicted occupation probabilities for these conformations are shown in Table 4.
Given the pKa value of His68 in Ub2 is 5.5 44, the fraction of protonated histidines at pH6.8 is 0.048, such that the fractions of the Ub2 chains in the closed (I), intermediate (II), and open (III) states are pIpH6.8=0.907, pIIpH6.8=0.091, and pIIIpH6.8=0.002, respectively. A reverse situation is expected at pH 4.5, where each histidine side chain carries a positive charge of +e with the probability of 0.909, and the fraction numbers for the three states are pIpH4.5=0.008, pIIpH4.5=0.165, and pIIIpH4.5=0.826, respectively. These numbers suggest that only two out of the three possible charge states/conformations of Ub2 are significantly populated and, therefore, can be experimentally detected at the pH values considered here: states I and II at pH6.8 and states II and III at pH4.5. The occupation probabilities for these states are in excellent agreement with the pA and pB values obtained from our relaxation data analysis at both pH values (Table 4). This then supports the applicability of the two-state model at the pH values considered here. Obviously, a more sophisticated, three-state model might be necessary at intermediate pH values, where the occupation probabilities of all three states could be comparable. Further refinement of this model of the charged states of Ub2 could involve consideration of the partial charges on all titratable side chains (especially Glu and Asp at pH4.5), which is beyond the scope of this study. Relaxation studies at intermediate pH values also might be required to provide further details on the Ub2 conformations.
Table 4.
Occupation probabilities of various conformational states of Lys48-linked Ub2 at the two pH values used in this study. The experimental values are from the ITS model (numbers in the parentheses indicate experimental uncertainties), the predicted values are based on the populations of the possible charged states of His68 (see text).
A posteriori validation of the ITS model for Ub2
The critical assumption (Postulate I) in the ITS model is that the interdomain and overall motions are statistically independent from each other. As discussed above, because the interdomain dynamics happen on the time scale comparable with the overall tumbling, this assumption needs validation. Statistical independence of these motions implies that domain reorientations do not affect the shape/diffusion tensor of the molecule. Indeed, the overall diffusion tensors of Ub2 at pH 6.8 (predominantly closed conformation) and pH 4.5 (predominantly open conformation) are essentially identical (Table 1), thus posteriori supporting the validity of this assumption. To further test this assumption, we compared the derived Ub2 structures (Figs. 4a–d) for the interconverting states at a given pH. As reported earlier27, hydrodynamic calculations based on the Ub2 structures yielded very similar principal values of the overall rotational diffusion tensor for the states A and B. In addition, the Ub2’s radius of gyration, another parameter sensitive to the shape of the molecule45, is identical for both interconverting states at a given pH (Supporting Information). Thus the likely reason the analysis based on Postulate I works well in the case of Ub2 is that the overall diffusion tensors of the various conformational states of this molecule are quite similar.
The derived structures are also in good agreement with other available experimental data. In particular, the “closed” state (Fig. 4a) is consistent with the chemical shift perturbations observed in the hydrophobic patch residues at pH6.8 23, when this conformation is predominant. This conformation also agrees both with the crystal structure of Ub2 46 and with solution Ub2 structure47 obtained based on the chemical shift perturbations and residual dipolar couplings. It is worth emphasizing that no information about chemical shift perturbations was included in the calculation of the structures shown in Fig.4. The “open” conformation (Fig.4d), predominant at pH4.5, is also in agreement with the NMR data, as no direct interdomain contacts were observed at this pH. Note also that the Ub2 structures at pH6.8 agree well with the paramagnetic spin-labeling data 27.
Interestingly, the Ub2 conformations in the less populated states (Figs.4b,c) are very similar to each other. The interdomain orientation in these conformations is also very similar to Ub2 structure in the complex with UBA2 domain 48: they superimpose with the backbone RMSD of 1.7Å. This observation has functional significance because it suggests that the partly open conformation (Figs.4b,c) represents some intermediate, binding-competent conformation of Ub2 and thus indicates that the amplitudes of interdomain motions at pH6.8 are sufficient for allowing ligands as big as UBA domain access to the hydrophobic patches on Ub units.
Comparison with the extended mode-free approach
It is instructional to discuss the relationship between the ITS model and the “extended model-free” approach to domain mobility, the only other model for domain mobility currently available. Proposed as a model for domain dynamics in calmodulin 19,22, this approach treats the effect of domain motions using the “extended model-free” form 21 for the correlation function of local motion, originally developed to account for “fast” and “slow” backbone dynamics in a protein:
| (17) |
The original 21 interpretation of dynamic parameters entering Eq. 17 is such that the order parameters, Ss and Sf, as well as the correlation times, τs and τf, represent the “slow” and “fast” ( τs >> τf) modes of local motion of a particular NH bond. The extension of this model to domain dynamics in calmodulin 19,22 treats only S f and τ f as local parameters representing the mobility of a NH bond, while the other pair of parameters, S s and τ s , is attributed to domain’s mobility. Note that in this case, as well as in the original “extended model-free” model, the fast and slow motions, i.e. the local and domain mobility modes, are assumed decoupled from each other and from the overall tumbling.
Following the notations of Eq. 4,Eq. 5, the total correlation function corresponding to the “extended model-free” treatment of 19,22 in the case of fully anisotropic overall tumbling can then we written as follows:
| (18) |
In the spirit of 19,22, here we introduced the Euler angles ΩP→R that define the (time-averaged) orientation of the residue-specific frame R with respect to the PAF of the diffusion tensor and thus replace two consecutive transformations, P→D and D → R. Note that ΩP→R (as well as ΩP→D) are now time-independent, because domain motion is already included in , Eq. 17.
Our ITS model for domain reorientations, Eq. 11, provides a more detailed structural picture while preserving a mono-exponential, “model-free”-like form of C(t). Indeed, the “extended model-free” approach provides information about time scales of domain reorientation and the related order parameters (i.e. amplitudes of motions) only. In contrast, the ITS parameters, and Qmk,nl (Eq.11), bear dependency on domain orientations, and . Thus, the ITS treatment yields the times scales of domain reorientations and particular domain arrangements in different conformations -- not only amplitude of motion -- together with the occupation probabilities for each state. This provides structural “snapshots” of the system in different conformations, unavailable from the “extended model-free” approach. It should be pointed out that in the case of the Ub2 molecule characterized by preferred domain orientations, the “extended model-free” model is inadequate. Indeed, in this approach (Eq.18) domain dynamics are characterized by a scalar order parameter (corresponding to a continuum of available interdomain orientations), while tensorial quantities, and Qmk,nl, are required even for a simple two-site exchange model, Eq.11.
To have a quantitative comparison with the “extended model-free” treatment, we also analyzed our relaxation data using Eq.18. The results of this analysis (Supporting Table 1) show that for Ub2 this approach leads to a markedly worse description of experimental data than the one provided by the ITS model. Thus, this analysis resulted in a 2-fold increase (compared to the ITS model) in the residuals of fit per degree of freedom at both pH values and in a worse correlation between the experimental and back-calculated data: Pearson’s r was 0.89 at pH 6.8 and 0.86 at pH 4.5, compared to 0.95 and 0.94, respectively, for the ITS model. Moreover, in contrast to the ITS analysis, the “extended model-free” treatment produced physically unreasonable values of the fitting parameters. Here the overall tumbling time of Ub2 was 17 ns (pH 6.8) and 11 ns (pH 4.5) and the squared order parameters (Ss2) associated with the interdomain mobility were 0.02 for the distal and 0.3 for the proximal domain at pH 6.8 and 0.2 and 0.56, respectively, at pH 4.5.
Conclusions
In this paper, we present a framework for building a motional model for a multi-domain protein that takes into account interdomain dynamics, together with other motional modes, including the overall tumbling and local intradomain dynamics. Our approach is based on the assumption of statistical independence of the various dynamic modes. Specifically, we developed a model of motion in a dual-domain protein that treats interdomain dynamics as an exchange, or interconversion, between two conformational states of the protein. We demonstrate that this model provides a reasonable first-order approximation to experimentally observed dynamics in Lys48-linked di-ubiquitin at both neutral and acidic pH. The analysis of NMR data using this model revealed many unknown details of domain mobility in di-ubiquitin, including structural information about the interconverting conformations of the molecule and the rates and energies associated with this process. The assumption of statistical independence between the overall motion and domain dynamics is a strong one and can be inadequate in the case when domain motion alters the overall shape of the molecule (hence its overall diffusion tensor). In the case of di-ubiquitin this assumption is justified by the close-to-spherical shape of ubiquitin monomers. Thus, domain reorientations in di-ubiquitin do not significantly alter the overall shape of the molecule and its overall rotational diffusion tensor. The similarity of the diffusion constants for various conformational states of di-ubiquitin is the likely reason the analysis works well for this system. Note that the assumption of statistical independence of the overall and interdomain motions is a separate one from the ability to accurately and independently extract their characteristics from experimental data. In fact, the characteristic time of the interdomain mobility in di-ubiquitin happens to be comparable to that for the overall tumbling, which necessarily limits the precision of the derived rates of interconversion.
The analysis of Ub2 data presented in this paper shows that in this particular system the characteristic time of the interdomain motion (τITS) is comparable to the overall tumbling time: τITS ~τoverall. It is worth pointing out that the model of domain motion described here requires no a priori assumption regarding the relationship between τoverall and τITS. Thus, for example, the above equations also describe the situation when domain motion is significantly slower than the overall tumbling (τITS >> τoverall), yet faster than the spin relaxation. In this case it can be shown that the relevant spectral density (e.g. Eq. 15) becomes independent of τITS and is a weighted average of the spectral densities describing overall tumbling in the individual conformational states of the molecule, in full agreement with the expectation that the measured relaxation rate be an average of the corresponding relaxation rates for these states.
The particular treatment of domain dynamics utilized here can be improved in many ways. While the two-state model is sufficient for Ub2 at the pH conditions used here (see Table 4 and the discussion above), a three-state model might be required at the intermediate pH values. Moreover, the application of our model to other protein systems might also require considering exchange between more than two states. The extension of the ITS model to n>2 is relatively straightforward and will require modification of the corresponding equations, Eqs.8–9, for and for conditional probabilities (see e.g. 25). Other modifications, beyond the simple n-site exchange model, might be required to describe domain dynamics in other multi-domain systems. These could include random domain rotations (diffusion) about a fixed axis, analogous to the model applied by D. Wallach 25 to describe the effect of internal rotations in a molecular label attached to a macromolecule. Furthermore, when domain’s diffusion is limited to some angular interval, it might be necessary to consider restricted rotational diffusion about a fixed axis. Such a model was originally introduced for the description of restricted internal rotations in the side chains by R. J. Wittebort and A. Szabo 26. The correlation functions for the interdomain mobility mode corresponding to these two cases of domain’s axial diffusion can be found in Supporting Information. However, further conceptual development of the approach presented here is necessary in order to take into account possible coupling between domain motion and the overall tumbling.
Supplementary Material
A table with the results of the “extended model-free” analysis of the Ub2 data; a table with the predicted values of the radius of gyration for Ub2 conformations; the procedure of defining the axis of rotation between two orientations of a molecule, and a summary of the relevant information on Wigner rotation matrices and Euler rotations, probabilities and correlation functions for anisotropic rotational diffusion, the conditional probabilities for the two-site exchange model, and the correlation functions of domain dynamics mode for axial diffusion models.
Acknowledgments
Supported by NIH grant GM065334 to D.F. We thank Dr. Attila Szabo for very insightful discussion of our ITS model and extremely helpful remarks. Ya.R. also acknowledges very useful comments of Dr. Ranjani Varadan concerning biochemical properties of Ub2, discussions with Dr. Olivier Walker, and the valuable help with computational software from Dr. Jennifer B. Hall.
References and Footnotes
- 1.Sicheri F, Kuriyan J. Curr Opin Struct Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 2.Pickford AR, Campbell ID. Chem Rev. 2004;104:3557–3566. doi: 10.1021/cr0304018. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Zuiderweg ER. Proc Natl Acad Sci U S A. 2004;101:10272–10277. doi: 10.1073/pnas.0401313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruschweiler R, Liao X, Wright PE. Science. 1995;268:886–889. doi: 10.1126/science.7754375. [DOI] [PubMed] [Google Scholar]
- 5.Fushman D, Xu R, Cowburn D. Biochemistry. 1999;38:10225–10230. doi: 10.1021/bi990897g. [DOI] [PubMed] [Google Scholar]
- 6.Fushman D, Varadan R, Assfalg M, Walker O. Progress NMR Spectroscopy. 2004;44:189–214. [Google Scholar]
- 7.Fischer MWF, Losonczi JA, Weaver LJ, Prestegard JH. Biochemistry. 1999;38:9013–9022. doi: 10.1021/bi9905213. [DOI] [PubMed] [Google Scholar]
- 8.Skrynnikov N, Goto N, Yang D, Choy W, Tolman J, Mueller G, Kay L. J.Mol.Biol. 2000;295:1265–1273. doi: 10.1006/jmbi.1999.3430. [DOI] [PubMed] [Google Scholar]
- 9.Hwang PM, Skrynnikov NR, Kay LE. J. Biomol. NMR. 2001;20:83–88. doi: 10.1023/a:1011226512421. [DOI] [PubMed] [Google Scholar]
- 10.Freed JH. J. Chem. Phys. 1977;66:4183–4199. [Google Scholar]
- 11.Polimeno A, Freed JH. Adv. Chem. Phys. 1993;83:89–206. [Google Scholar]
- 12.La Penna G, Mormino M, Pioli F, Perico A, Fioravanti R, Gruschus JM, Ferretti JA. Biopolymers. 1999;49:235–254. doi: 10.1002/(SICI)1097-0282(199903)49:3<235::AID-BIP5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 13.La Penna G, Fausti S, Perico A, Ferretti JA. Biopolymers. 2000;54:89–103. doi: 10.1002/1097-0282(200008)54:2<89::AID-BIP2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 14.Tugarinov V, Liang Z, Shapiro Y, Freed JH, Meirovitch E. J.Am.Chem.Soc. 2001;123:3055–3063. doi: 10.1021/ja003803v. [DOI] [PubMed] [Google Scholar]
- 15.Tugarinov V, Shapiro YE, Liang Z, Freed JH, Meirovitch E. J Mol Biol. 2002;315:155–170. doi: 10.1006/jmbi.2001.5231. [DOI] [PubMed] [Google Scholar]
- 16.Shapiro YE, Kahana E, Tugarinov V, Liang Z, Freed JH, Meirovitch E. Biochemistry. 2002;41:6271–6281. doi: 10.1021/bi012132q. [DOI] [PubMed] [Google Scholar]
- 17.Meirovitch E, Polimeno A, Freed JH. J Phys Chem B Condens Matter Mater Surf Interfaces Biophys. 2006;110:20615–20628. doi: 10.1021/jp061403+. [DOI] [PubMed] [Google Scholar]
- 18.Meirovitch E, Shapiro YE, Polimeno A, Freed JH. J Phys Chem A Mol Spectrosc Kinet Environ Gen Theory. 2006;110:8366–8396. doi: 10.1021/jp056975t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baber JL, Szabo A, Tjandra N. J.Amer.Chem.Soc. 2001;123:3953–3959. doi: 10.1021/ja0041876. [DOI] [PubMed] [Google Scholar]
- 20.Chang SL, Szabo A, Tjandra N. J. Am. Chem. Soc. 2003;125:11379–11384. doi: 10.1021/ja034064w. [DOI] [PubMed] [Google Scholar]
- 21.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. J.Amer.Chem.Soc. 1990;112:4989–4991. [Google Scholar]
- 22.Chang SL, Tjandra N. J Am Chem Soc. 2001;123:11484–11485. doi: 10.1021/ja016862x. [DOI] [PubMed] [Google Scholar]
- 23.Varadan R, Walker O, Pickart C, Fushman D. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 24.Strictly speaking, this is a two-site exchange model. We termed it interconversion in order to avoid confusion with the conformational or chemical exchange on a µs-ms time scale that is often observed in protein NMR and could reflect intradomain dynamics.
- 25.Wallach D. J. Chem. Phys. 1967;47:5258–5268. [Google Scholar]
- 26.Wittebort RJ, Szabo A. J. Chem. Phys. 1978;69:1722–1736. [Google Scholar]
- 27.Ryabov Y, Fushman D. Proteins. 2006;63:787–796. doi: 10.1002/prot.20917. [DOI] [PubMed] [Google Scholar]
- 28.Abragam A. The Principles of Nuclear Magnetism. Oxford: Clarendon Press; 1961. [Google Scholar]
- 29.Wigner EP. Group theory and its application to the quantum mechanics of atomic spectra; Expanded and improved ed. New York: Academic Press; 1959. [Google Scholar]
- 30.Fushman D, Cowburn D. In: Structure, Motion, Interaction and Expression of Biological Macromolecules. Sarma R, Sarma M, editors. Albany, NY: Adenine Press; 1998. pp. 63–77. [Google Scholar]
- 31.Lipari G, Szabo A. J.Amer.Chem.Soc. 1982;104:4546–4559. [Google Scholar]
- 32.Lipari G, Szabo A. J.Amer.Chem.Soc. 1982;104:4559–4570. [Google Scholar]
- 33.Favro DL. Phys. Rev. 1960;119:53–62. [Google Scholar]
- 34.Woessner D. J.Chem.Phys. 1962;37:647–654. [Google Scholar]
- 35.Luginbuhl P, Wuthrich K. Prog Nucl Mag Res Sp. 2002;40:199–247. [Google Scholar]
- 36.Woessner D. J.Chem.Phys. 1961;36:1–4. [Google Scholar]
- 37.Fushman D, Tjandra N, Cowburn D. J. Am. Chem. Soc. 1999;121:8577–8582. [Google Scholar]
- 38.Fushman D, Cowburn D. In: Protein NMR for the Millenium (Biological Magnetic Resonance Vol 20) Krishna NR, B L, editors. Kluwer: 2002. pp. 53–78. [Google Scholar]
- 39.This follows from the fact that when S2 ~ 1 and τloc << τoverall, the spectral density J(ω) describing “model-free” local dynamics in the presence of overall tumbling (considered isotropic in this example, for simplicity) for ω = 0 and ωN can be approximated as such that the leading term in J(ω) is proportional to S2 and does not contain any information about τloc. In the case of J(ωN) this conclusion might not hold when the term (ωNτoverall)2 in the denominator becomes sufficiently large, i.e. for very large proteins and high magnetic fields. Note also that when S2 ~ 1, i.e. S2 >> 1-S2, the requirement that τloc << τoverall is not very stringent.
- 40.Ryabov Y, Fushman D. Magn Reson Chem. 2006;44:143–151. doi: 10.1002/mrc.1822. [DOI] [PubMed] [Google Scholar]
- 41.Bernatowicz P, Kowalewski J, Szymanski S. J Chem Phys. 2006;124:024108. doi: 10.1063/1.2149858. [DOI] [PubMed] [Google Scholar]
- 42.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in C. NY: Cambridge University Press; 1992. [Google Scholar]
- 43.Cornilescu G, Marquardt JL, Ottiger M, Bax A. J. Amer. Chem. Soc. 1998;120:6836–6837. [Google Scholar]
- 44.Fujiwara K, Tenno T, Sugasawa K, Jee JG, Ohki I, Kojima C, Tochio H, Hiroaki H, Hanaoka F, Shirakawa M. J Biol Chem. 2004;279:4760–4767. doi: 10.1074/jbc.M309448200. [DOI] [PubMed] [Google Scholar]
- 45.Cantor CR, Schimmel PR. Biophysical Chemistry. New York: W. H. Freeman & Co; 1980. [Google Scholar]
- 46.Cook WJ, Jeffrey LC, Carson M, Zhijian C, Pickart CM. J.Biol.Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 47.van Dijk ADJ, Fushman D, Bonvin AM. Proteins. 2005;60:367–381. doi: 10.1002/prot.20476. [DOI] [PubMed] [Google Scholar]
- 48.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 49.Koradi R, Billeter M, Wuthrich K. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 50.Walker O, Varadan R, Fushman D. J. Magn. Reson. 2004;168:336–345. doi: 10.1016/j.jmr.2004.03.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A table with the results of the “extended model-free” analysis of the Ub2 data; a table with the predicted values of the radius of gyration for Ub2 conformations; the procedure of defining the axis of rotation between two orientations of a molecule, and a summary of the relevant information on Wigner rotation matrices and Euler rotations, probabilities and correlation functions for anisotropic rotational diffusion, the conditional probabilities for the two-site exchange model, and the correlation functions of domain dynamics mode for axial diffusion models.