Abstract
Histone modifications have important roles in transcriptional control, mitosis and heterochromatin formation. G9a and G9a-like protein (GLP) are euchromatin-associated methyltransferases that repress transcription by mono- and dimethylating histone H3 at Lys9 (H3K9). Here we demonstrate that the ankyrin repeat domains of G9a and GLP bind with strong preference to N-terminal H3 peptides containing mono- or dimethyl K9. X-ray crystallography revealed the basis for recognition of the methylated lysine by a partial hydrophobic cage with three tryptophans and one acidic residue. Substitution of key residues in the cage eliminated the H3 tail interaction. Hence, G9a and GLP contain a new type of methyllysine binding module (the ankyrin repeat domains) and are the first examples of protein (histone) methyltransferases harboring in a single polypeptide the activities that generate and read the same epigenetic mark.
Methylation of histone H3 at Lys9 (H3K9) in euchromatin is associated with transcriptional repression of potentially active genes. G9a and GLP function as heterodimers that are responsible for most mono- and dimethylation of H3K9 (H3K9me1 and H3K9me2)1-3. G9a and GLP are found in co-repressor complexes associated with DNA binding proteins, and repression requires the methyltransferase activity located in their C-terminal SET domain1,4-7. G9a also functions as a coactivator for nuclear receptors7. G9a and GLP have large N-terminal regions including six centrally located ankyrin repeats (Supplementary Fig. 1a online). Here we show that the ankyrin repeat domains of G9a and GLP bind with strong preference and specificity to N-terminal histone H3 peptides containing mono- or dimethylated K9, suggesting that G9a and GLP are involved in both making and reading the histone code.
RESULTS
G9a and GLP ankyrin repeats are methyllysine binding modules
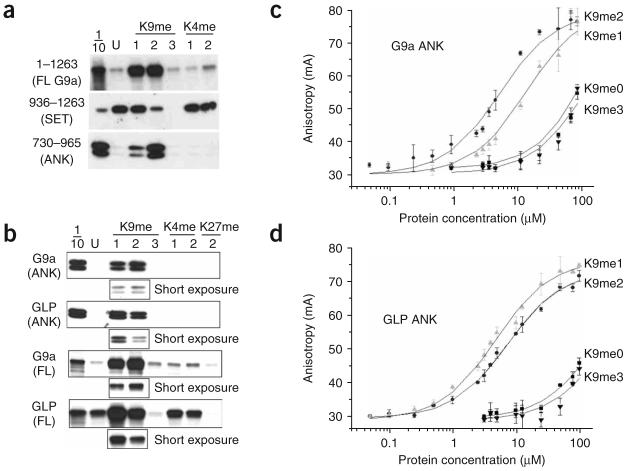
To further investigate how G9a and GLP function together to regulate H3K9 methylation, we studied the binding of G9a to either unmodified or methylated H3 tail peptides. As expected, the G9a SET domain alone bound effectively to unmodified and H3K9me1 peptides (residues 1–21), which are substrates for methylation by the G9a SET domain8, and bound poorly to H3K9me2 and H3K9me3 peptides (Fig. 1a). Notably, full-length G9a had a far greater affinity for H3K9me1 and H3K9me2 peptides than for unmodified H3 peptides. Deletion analysis identified the G9a ankyrin repeats (residues 730–965) as the H3K9me2 binding module (Supplementary Fig. 1). The G9a ankyrin repeats bound specifically to K9-methylated peptides and had no interaction with either K4- or K27-methylated peptides (Fig. 1a,b). The patterns of binding to various modified H3 peptides by full-length GLP and its ankyrin repeats were similar to the binding patterns of the corresponding full-length G9a and its ankyrin repeats, with the exception that GLP bound more tightly to H3K9me1 than to H3K9me2 (Fig. 1b). From fluorescence polarization studies, the dissociation constants (Kd) were 14 ± 3 μM for H3K9me1 and 6 ± 2 μM for H3K9me2 with G9a ankyrin repeats, and 5 ± 0.4 μM for H3K9me1 and 7 ± 1 μM for H3K9me2 with GLP ankyrin repeats (Fig. 1c,d). A single methyl group decrease (H3K9me0) or increase (H3K9me3) resulted in Kd values greater than 150 μM. To rule out the effect of the N-fluorescein modification, we carried out competition experiments using unmodified peptides and found only modest changes in the values of Kd but no change in relative affinities (data not shown).
Figure 1.
G9a and G9a-like protein (GLP) bind to mono- and dimethylated histone H3. (a,b) Peptide pull-down assays with peptides that were unmodified (U) or mono-, di- or trimethylated (1, 2 or 3) at K4 or K9 (H3 1–21) or at K27 (H3 21–44) using G9a full-length (FL, residues 1–1263) or fragments containing the SET domain (residues 936–1263 of G9a) or the ankyrin repeats (ANK, residues 730–965 of G9a and 734–968 of GLP). Proteins were detected by SDS-PAGE and autoradiography. 1/10, 10% input. Shorter exposures of H3K9me1 and H3K9me2 binding are shown below each panel. (c,d) Binding of G9a and GLP ankyrin repeats with N-terminal fluoresceinated H3 peptides (residues 1–15), as determined by fluorescence polarization.
Structural basis for H3K9me2 recognition by ankyrin repeats
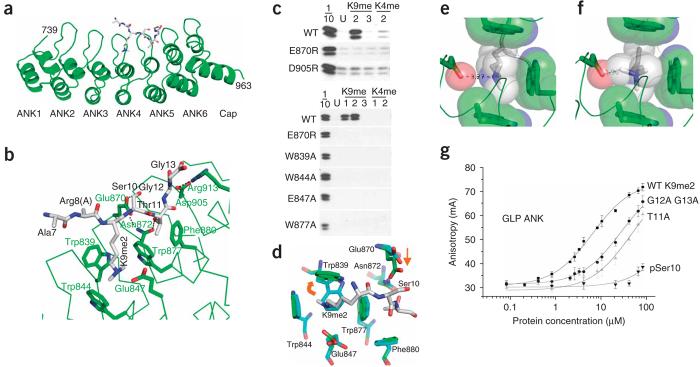
Sequence comparison between the ankyrin repeats of G9a and GLP revealed a high degree of similarity (Supplementary Fig. 2 online). We solved the structures of the GLP ankyrin repeats in the absence and presence of a bound H3K9me2 peptide (residues 1–15) to a resolution of 3 Å (Table 1). Ankyrin repeats are helix-turn-helix-β-turn structures (Fig. 2a). The helices stack, and the β-turns project out at right angles. The H3 peptide is sandwiched between β-turns and helices of the fourth and fifth repeats. H3 peptide residues 7–13 are visible with the absence of the side chain of Arg8. K9me2 is bound in a partial hydrophobic cage formed by three tryptophans (Trp839, Trp844 and Trp877) and one acidic residue (Glu847) (Fig. 2b). The K9me2 methyl moieties point toward the hydrophobic residues, and the lone proton of the Ne atom bridges to the acidic Glu847. Mutations of the G9a residues involved in the methyllysine cage formation (W839A, W844A, W877A and E847A) abrogate peptide binding (Fig. 2c). Although there is no overall conformational change in the ankyrin repeats upon peptide binding (with the r.m.s. deviation of 0.17 Å comparing 224 Cα atoms), the indole ring of Trp839 undergoes large rotations of two torsion angles to allow the binding of methyllysine in the cage (Fig. 2d).
Table 1.
Data collection and refinement statistics
| Crystal 1 (H3 peptide) |
Crystal 2 (native) |
Crystal 3 (selenium) |
||
|---|---|---|---|---|
| Data collection | ||||
| Space group | C2221 | |||
| Cell dimensions | ||||
| a (Å) | 59.5 | 59.8 | 60.1 | 59.9 |
| b (Å) | 152.4 | 151.3 | 151.6 | 150.9 |
| c (Å) | 167.3 | 167.9 | 168.4 | 167.5 |
| Beamline (SERCAT) | APS 22-BM | APS 22-ID | ||
| Wavelength (Å) | 0.97625 | 0.97625 | 0.97923 (peak) | 0.97174 (remote) |
| Resolution (Å)a | 24.91–2.99 | 28.75–2.99 | 35–3.49 | 34.4–2.99 |
| (3.1–2.99) | (3.1–2.99) | (3.61–3.49) | (3.1–2.99) | |
| Ramerge | 0.050 (0.697) | 0.123 (0.535) | 0.087 (0.574) | 0.103 (0.317) |
| I/σIa | 16.4 (4.2) | 13.7 (5.7) | 9.0 (4.6) | 14.1 (6.2) |
| Completeness (%)a | 97.1 (93.1) | 97.9 (94.6) | 99.9 (100) | 99.7 (99.7) |
| Redundancya | 11.2 (11.5) | 11.9 (11.0) | 6.5 (5.8) | 5.0 (4.9) |
| Observed reflections | 178,378 | 190,388 | 67,499 | 78,942 |
| Unique reflectionsa | 15,898 (1559) | 15,965 (1564) | 10,414 (1015) | 15,845 (1550) |
| Refinement | ||||
| Resolution (Å) | 2.99 | 2.99 | ||
| No. reflections | 15,898 | 15,965 | ||
| Rwork/Rfree | 0.195 / 0.238 | 0.205 / 0.261 | ||
| Number of atoms | ||||
| Protein | 3,546 | 3,467 | ||
| Heterogen (sulfate) | 50 (10 ions) | 40 (8 ions) | ||
| Water | 29 | 28 | ||
| B-factors (Å2) | ||||
| Protein | 46.7 | 47.2 | ||
| Sulfate | 120.5 | 111.4 | ||
| Water | 41.1 | 36.6 | ||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.010 | 0.009 | ||
| Bond angles (°) | 1.5 | 1.5 | ||
Highest-resolution shell is shown in parenthesis.
Figure 2.
Structural basis for ankyrin repeat recognition of dimethylated histone H3 lysine 9 (H3K9me2). (a) The helix-turn-helix-β-turn structure of G9a-like protein (GLP) ankyrin repeats. The H3 peptide (gray) binds between the fourth and fifth repeats. For convenience, G9a residue numbers are shown (Supplementary Fig. 2). (b) H3K9me2 binds in a partial hydrophobic cage. Peptide binding is further specified by the interactions with H3 Ser10, Thr11, Gly12 and Gly13. (c) Peptide pull-down assays with H3 peptides (residues 1–21) using G9a wild-type (WT) or mutant ankyrin repeats. H3 peptides were either unmodified (U), or mono-, di- or trimethylated (1, 2 and 3). 1/10, 10% input. (d) Structural comparison of GLP ankyrin repeats with (green) and without (blue) bound peptide. The Trp839 indole ring rotated along the side chain torsion angles χ1 (∼20°) and χ2 (∼80°). In addition, the carboxyl group of Glu870 moved approximately 2.5 Å toward Ser10. (e) Dimethyllysine from the crystal structure of GLP ankyrin repeats is satisfactorily accommodated in the cage. Spheres represent approximate van der Waals radii of atoms. (f) Trimethyllysine is excluded from the GLP ankyrin repeat cage. Modeling a trimethyllysine in the cage causes steric clashes. The conformation with the fewest clashes is shown. (g) Binding of GLP ankyrin repeats with wild-type (WT) or mutated H3 peptides (residues 1–15) containing K9me2, as determined by fluorescence polarization.
The crystal structure of GLP ankyrin repeats shows that dimethyllysine is satisfactorily accommodated in the cage (Fig. 2e). The slight preference of GLP for H3K9me1 versus H3K9me2, which is reversed in G9a, could be explained by small differences in the width of the cage induced by regional packing, where a wider cage accommodates H3K9me2 slightly better than a narrow cage, which accommodates H3K9me1 best. Modeling a trimethyllysine in the cage causes steric clashes (Fig. 2f). Even in this rotamer, which slightly withdraws the methyl groups from the pocket, a severe clash with Glu847 is apparent. This residue could rotate out of the way, but the charge-charge interaction would be lost. The E847A mutant loses binding (Fig. 2c), underscoring the importance of this interaction.
H3Ser10 is contacted by a glutamic acid (Glu870) (Fig. 2b), and charge reversal of this residue (E870R) or phosphorylation of H3Ser10 in the context of K9me2 completely eliminates peptide binding (Fig. 2c,g). H3Thr11 resides in a distinctive pocket—half of which is formed by the back side of Trp877 and Phe880 (Fig. 2b)—that accommodates the methyl moiety of the H3Thr11 side chain. The Thr11 hydroxyl moiety forms a polar contact with Asn872, which in turn forms a hydrogen bond with Glu870. Mutation of Thr11 to Ala (T11A) in the context of K9me2 decreases binding approximately seven-fold (Kd from 7 ± 1 μM to 54 ± 16 μM). H3Gly12 and H3Gly13 form a sharp turn (∼90°) and run up a wall formed by the salt bridge between Asp905 and Arg913. Mutation of both glycines to the next smallest amino acid (G12A G13A) was sufficient to reduce binding (Kd) from 7 ± 1 μM to 32 ± 12 μM (Fig. 2g) and the mutation D905R blocked peptide binding (Fig. 2c). The charge-charge repulsion generated between D905R and Arg913 could cause the wall that the peptide climbs to buckle. The recognition of H3K9me2, Ser10, Thr11 and two glycines is sufficient to explain the specificity of the GLP as well as the G9a ankyrin repeats, as all residues involved in peptide binding are identical between the two (Supplementary Fig. 2).
Cage mutations do not affect co-regulator–related functions
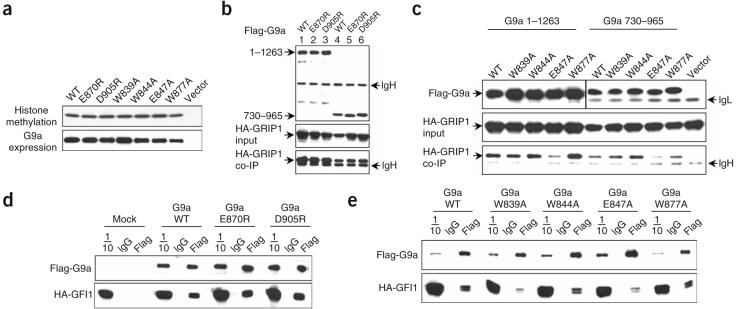
To determine whether recognition of mono- and dimethyllysine by the G9a ankyrin repeats is necessary for methyltransferase activity, we expressed wild-type and methyllysine binding– deficient G9a proteins in Cos cells and assayed immunopreciptated proteins for their ability to transfer methyl groups to core histones. The wild-type and methyllysine binding–deficient G9a proteins were expressed at comparable levels and had comparable methyltransferase activity (Fig. 3a). Thus, lack of methyllysine recognition through the ankyrin repeats has no effect on methyltransferase activity in vitro. In addition to harboring mono- and dimethyllysine targeting activity, the G9a ankyrin repeats interact with the nuclear receptor coactivator GRIP1, and G9a (in cooperation with GRIP1) can function as a coactivator for several nuclear receptors7. In coimmunoprecipitation assays, full-length G9a protein (Fig. 3b, lanes 1–3), as well as the ankyrin repeats (lanes 4–6), containing either the E870R or D905R mutation, interacted as effectively as the wild-type protein with the N terminus of GRIP1. Although mutation of Glu847 caused some reduction in the interaction of both full-length G9a and the ankyrin repeats with GRIP1, all four cage mutants still interacted with GRIP1 (Fig. 3c), demonstrating that these mutations did not disrupt overall structural integrity but caused selective loss of binding to the H3K9me1 and H3K9me2 peptides. Notably, all of the ankyrin mutants are expressed at levels comparable to the wild-type protein (Fig. 3b,c). We also carried out interaction studies between G9a and the DNA binding repressor growth factor independent 1 (GFI1), which has been previously shown to interact with G9a (ref. 9). All of the methyllysine binding–deficient ankyrin mutants were still able to associate with GFI1 in coimmunoprecipitation studies, again demonstrating the selective loss of methyllysine binding and maintenance of the overall structural integrity of the mutant proteins (Fig. 3d,e).
Figure 3.
Analysis of G9a ankyrin mutants that lack H3K9me1 and H3K9me2 peptide binding. (a) Expression and methyltransferase activity of full-length G9a proteins. Immunoprecipitated (IP) proteins, derived from transfected Cos cells, were analyzed for either methyltransferase activity using core histones as a substrate (above) or immunoblotted with anti-Flag antibody (below). Wild-type G9a (WT), indicated point mutants or Flag vector control (vector) are shown. (b,c) Coimmunoprecipitation (Co-IP) of wild-type G9a (WT) or the indicated mutant Flag-G9a full-length (1–1263) or Flag-G9a ankyrin region (730–965) with HA-GRIP1 (5–479). Above, IP and immunoblot of G9a with Flag antibodies. Middle, immunoblot of input GRIP1 with HA antibodies. Below, Co-IP of GRIP1, immunoblotted with HA antibodies. IgG heavy (IgH) and light (IgL) chains are indicated. (d,e) Coimmunoprecipitation of WT or mutant full-length Flag-G9a with HA-GFI1 delta zinc finger mutant (ΔZF). Cos7 cells were cotransfected with plasmids encoding Flag-G9a and HA-GFI1 ΔZF. Above, anti-Flag blots of IP proteins. Below, anti-HA blots of Co-IP proteins. Mock, no G9a; 1/10, 10% input; IgG, IP with normal mouse IgG; Flag, IP with anti-Flag.
DISCUSSION
The binding and X-ray crystallographic data presented here show that the ankyrin repeats of G9a and GLP are capable of binding methylated histone tails. This is the first example of such a function by an ankyrin repeat domain. The specificity for mono- and dimethyllysine of the G9a and GLP ankyrin repeats is comparable to that of other methyl binding protein modules, such as the chromodomain, tudor domain and PHD finger10-14, which can preferentially bind lysines methylated to specific degrees at specific histone tail residues. The organization of the methyllysine binding hydrophobic cage structure with one acidic residue in the ankyrin repeat domains of G9a and GLP is also similar to those found in the H4K20me2 binding double tudor domain of 53bp1 and, to a lesser extent, in the H3K4me3,2 binding PHD finger of BPTF (Supplementary Fig. 3 online).
The histone code hypothesis suggests that specific epigenetic marks on histones can be translated into distinct biological outcomes through effectors that are recruited to these marks and subsequently act on the local chromatin structure or transcriptional machinery15,16. Several histone-methylating complexes contain components to both synthesize and bind a specific histone mark, such as Suv39h–HP1 (for H3K9me3)17 and LSD1–BHC80 (for H3K4me0)18, where the components that make (or remove) and recognize a specific histone mark are separate proteins. In these examples, both components are necessary for the overall function of the complex. G9a and GLP are the first example of histone-methylating proteins that contain modules, within the same polypeptide, for both making (via the SET domain) and recognizing (via the ankyrin repeats) a given methyl mark. The presence of a G9a–GLP heterodimer2, however, may allow one enzyme to create the modification whereas the other enzyme binds this modification. In addition, the opposite approximately two-fold preference for either H3K9me1 or H3K9me2 by GLP and G9a, respectively, may better enable the G9a–GLP heterodimer to bind both of the H3K9me1 and H3K9me2 marks.
Recruitment of complexes containing G9a–GLP to regulated genes may be accomplished through direct interaction with sequence-specific DNA binding proteins or through the interaction with coactivator or co-repressor proteins. The studies presented here provide an alternative method of recruitment of G9a–GLP complexes through direct ankyrin repeat–mediated interaction with K9 methylation marks on histone tails. Although it is tempting to speculate that, similarly to the paired protein examples given above, G9a and GLP contribute these dual histone-methylating and methyl histone binding roles to the coactivator and co-repressor functions of complexes, further studies will be needed to elucidate the functional significance of this newly identified mode of histone tail recognition.
METHODS
Peptide interaction assays
We produced mouse G9a and human GLP proteins using the TnT T7 transcription and translation system (Promega) and 35S-methionine (ICN). Template plasmids pSG5-Flag G9a and pSG5-Flag G9a ankyrin repeats (residues 730–965) were either made as described7 or generated by PCR. Plasmid pSG5-HA GLP was generated by inserting the complete human GLP coding sequence into the vector pSG5-HA. The GLP expression vector pSG5-Flag GLP (residues 734–968) was generated by PCR. Biotinylated histone H3 peptides (residues 1–21 or 21–44, Upstate), 0.5 μg per sample, were bound to streptavidin-Sepharose beads (Amersham) for 1 h. Beads were then washed three times with binding and washing buffer (25 mM Tris, pH 8.0, 140 mM NaCl, 3 mM KCl and 0.1% (v/v) Nonidet P-40) and incubated with 35S-labeled G9a or GLP proteins overnight at 4 °C in a total volume of 300 μl of binding and washing buffer. Beads were then washed three separate times each in 1 ml binding and washing buffer at 4 °C. Bound proteins were eluted directly into SDS-PAGE loading buffer and analyzed by SDS-PAGE and autofluorography.
Generation of ankyrin repeat point mutations
We generated point mutations in G9a in the vector pSG5-Flag G9a (residues 730–965), using the QuikChange II XL system (Stratagene). Reinserting the mutant fragment into the plasmid encoding full-length G9a created mutant full-length Flag-tagged G9a expression constructs. The G9a H1166K mutant was generated as described7. All mutations were confirmed by sequencing.
Coimmunoprecipitations
We assayed the interaction of Flag-tagged G9a with either the hemagglutinin (HA)-tagged GRIP1 N terminus or the HA-tagged GFI1 N terminus with cleared lysates from transfected Cos-7 cells, using the plasmids pSG5-HA GRIP1 (residues 5–479; ref. 19) and pSG5.HA GFI1 delta zinc fingers (ΔZF). The plasmid pSG5-HA GFI1 ΔZF was generated by inserting GFI1 residues 1–271 into the vector pSG5-HA. Briefly, 3 × 106 Cos-7 cells were electroporated with a total of 5 μg plasmid DNA in 300 μl growth medium contained in a 0.4-cm cuvette using a 240-V and 960-μf pulse. Cells were subsequently plated on 10-cm dishes, and whole-cell extracts were made 48 h later using 500 μl 25 mM Tris, pH 8.0, 140 mM NaCl, 3 mM KCl, 1% (v/v) Nonidet P-40, 0.2% (v/v) SDS and protease inhibitor cocktail (Roche). Cleared lysates were immunoprecipitated with anti-Flag antibody (M2, Sigma) and protein A/G–Sepharose beads (Santa Cruz Biotechnology) for 4 h at 4 °C, washed two times with binding and washing buffer and subsequently analyzed by SDS-PAGE and anti-HA (3F10, Roche) immunoblotting.
Methyltransferase assays
We immunoprecipitated Flag-tagged G9a from transfected Cos cells as above. Bead-bound G9a was incubated with core histones (Sigma) and 3H-labeled AdoMet (NEN), and radioactive products were subsequently analyzed by standard denaturing SDS gel electrophoresis and autofluorography.
Fluorescence anisotropy
We carried out fluorescence anisotropy measurements at 25 °C on a Beacon 2000 Fluorescence Polarization System (PanVera). A constant amount of synthetic N-fluoresceinated peptide (5 nM) was incubated for 30 min with increasing amounts of proteins. No change in fluorescence intensity was observed with binding. Each curve was reproduced in triplicate. Curves were individually fit using Origin 7.0 software (OriginLab Corporation). Binding constants, calculated as described previously20, were averaged and s.d. of the three curves are reported. For competition experiments, 5 nM H3K9me1 N-fluoresceinated peptide was incubated with a concentration of protein sufficient to give about 60% of the total shift in anisotropy, and unlabeled peptides were titrated in.
Expression and purification for crystallization
We generated two hexahistidine-SUMO tagged constructs: pXC571 (mouse G9a residues 730–965) and pXC587 (human GLP residues 734–968). There is a 4-amino-acid difference in numbering between GLP and G9a, and so pXC587 and pXC571 encode the ankyrin repeat domain of the two proteins. Relative to G9a, GLP (pXC587) was isolated with a better yield and purity. Furthermore, GLP crystallized, whereas G9a failed to do so. All proteins were expressed in Escherichia coli BL21(DE3)-Gold cells (Stratagene) that were transformed in house with the RIL-Codon plus plasmid to compensate for the expression of rare tRNAs. For selection and maintenance throughout expression, cells were cultured with 100 μg ml−1 kanamycin. Plates and starter cultures used MDAG repressive (glucose-containing) media21. Expression cultures were grown at 37 °C in LB medium until D = 0.8 at 600 nm; the temperature was then shifted to 16 °C, and the cultures were induced overnight with 0.4 mM isopropyl β-d-thiogalactoside.
Cells were lysed as a 20% (v/v) suspension in 50 mM potassium phosphate, pH 8.0, 250 mM NaCl, 5% (v/v) glycerol, 0.25 mM DTT and 40 μg ml−1 PMSF by two passes through an ice-cold French pressure cell press. The lysate was clarified by centrifugation twice at 50,000g for 30 min. Hexahistidine-SUMO fusion proteins were isolated on nickel-charged HiTrap Chelating HP (GE Healthcare). Following imidazole elution, the fusion protein was cleaved with Ulp1 protease at 25 U ml−1 in overnight dialysis at 4 °C. Only two extraneous N-terminal amino acids (HisMet) were left as a result of the NdeI restriction site. For the GLP ankyrin repeat protein, the digestion products could be passed back through the nickel column to capture the tag and protease; however, the G9a ankyrin repeat protein remained associated with the cleaved SUMO tag. Hydrophobic interaction chromatography was used, and this displaced the SUMO tag efficiently. The protein was further purified by ion exchange (HiTrap-Q) and gel filtration chromatography (Superdex-75, GE Health-Care). A maximum yield of 7 mg l−1 for the G9a ankyrin repeat protein and 25 mg l−1 for the GLP ankyrin repeat protein was obtained. Both are monomeric and show little aggregation in gel filtration in the presence of 5% (v/v) glycerol and 2 mM DTT. The purified ankyrin repeat proteins tolerate snap freezing in liquid nitrogen (particularly if small volumes are dropped directly into the liquid nitrogen and stored as pellets) and are stable for about 2 weeks at 4 °C. In addition, the complexes of GLP ankyrin repeat proteins with H3K9me2 peptides were copurified over an analytical gel filtration column. H3K9me0 peptide does not copurify with the ankyrin repeat proteins (data not shown).
Crystallography
We obtained crystals of GLP ankyrin repeat protein derived from pXC587 (human GLP 734–968), and copurified complexes of ankyrin repeat–H3 peptide (residues 1–15) under the conditions of 2.0 M LiSO4, 4% (v/v) polyethylene glycol 550 and 0.1 M Tris, pH 8.5. The crystals appeared in various morphologies, but only diamond-like plates were single crystals. X-ray diffraction data were collected from three crystals, cryoprotected by xylitol mixed with mother liquor at 35% (w/v), at the SER-CAT beamline, and processed using HKL2000 (ref. 22; Table 1).
We used molecular replacement to obtain crystallographic phases. The closest homolog of GLP ankyrin repeats with a known structure is a synthetic ankyrin repeat construct (PDB 1N0R), which contains 4 ankyrin repeats, as opposed to GLP's six repeats plus a cap, and has only 32% sequence identity with GLP. It failed as a search model for molecular replacement. Therefore, we chose to produce search models using homology modeling for both G9a and GLP sequences, using the PHYRE server23 (http://www.sbg.bio.ic.ac.uk/phyre). Among the models with high confidence scores, a homology model of G9a readily gave a solution using the molecular replacement program PHASER24. Even though G9a and GLP ankyrin repeats have only ∼50% sequence identity, the model against G9a succeeded as a search model for GLP because of an excellent structural alignment of backbone residues across a greater distance than could be achieved using any other model (Supplementary Fig. 4a online). The resulting electron density map was easily interpretable (Supplementary Fig. 4b), using the model-building program O25. The accuracy of the molecular replacement phases was evident by correct locations for nine (out of 14) selenium positions—six per molecule and one from the tag with two molecules per asymmetric unit—in an anomalous difference Fourier map (Supplementary Fig. 4c) using data collected from a selenium-incorporated crystal (Table 1). Refinement proceeded using CNS26 scripts, and the statistics shown in Table 1 were calculated for the entire resolution range. The Rfree and Rwork values were calculated for 5% (randomly selected) and 95%, respectively, of observed reflections. Following completion of the peptide-free structure, a difference Fourier map was calculated against data from a peptide-containing crystal. The peptide electron density was readily apparent (Supplementary Fig. 4d), and the cocrystal structure was built and refined (Table 1). The crystals contain two molecules, of which one contains peptide, per crystallographic asymmetric unit (Supplementary Fig. 4e).
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Ruiz and D. Gerke for technical assistance. The Emory University School of Medicine supported the use of SER-CAT beamlines. This work was supported by grants DK55274 to M.R.S. and GM068680 to X.C. from the US National Institutes of Health.
Footnotes
Supplementary information is available on the Nature Structural & Molecular Biology website.
Accession codes
The X-ray structures of GLP, with and without bound H3 peptide, have been submitted to PDB as 3B7B and 3B95, respectively.
References
- 1.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tachibana M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3–K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueda J, Tachibana M, Ikura T, Shinkai Y. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J. Biol. Chem. 2006;281:20120–20128. doi: 10.1074/jbc.M603087200. [DOI] [PubMed] [Google Scholar]
- 4.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 5.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl. Acad. Sci. USA. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 7.Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyl-transferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem. 2006;281:8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins RE, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. GFI1 coordinates epigenetic repression of p21(Cip/WAF1) by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 2005;25:10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 11.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 12.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4–K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 14.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 17.Grewal SIS, Jia ST. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 18.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YH, Campbell HD, Stallcup MR. Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol. Cell. Biol. 2004;24:2103–2117. doi: 10.1128/MCB.24.5.2103-2117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson KD. Quantitative analysis of protein-protein interactions. Methods Mol. Biol. 2004;261:15–32. doi: 10.1385/1-59259-762-9:015. [DOI] [PubMed] [Google Scholar]
- 21.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Kelley LA, MacCallum RM, Sternberg MJE. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:501–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 24.Storoni LC, Mccoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron-density maps and the olcation of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT, et al. Crystallography & NMR system: a new software suite for macro-molecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.