Abstract
We present a high-resolution mass spectrometric footprinting approach enabling the identification of amino acids in the protein of interest interacting with cognate RNA. This approach is particularly attractive for studying large nucleoprotein complexes that are less amenable to crystallographic or NMR analysis. Importantly, our methodology allows examination of protein-RNA interactions under biologically relevant conditions using limited amounts of protein and nucleic acid samples.
Keywords: Mass Spectrometry, Protein, RNA, Nucleoprotein Complex, Footprinting, Structure
1. Introduction
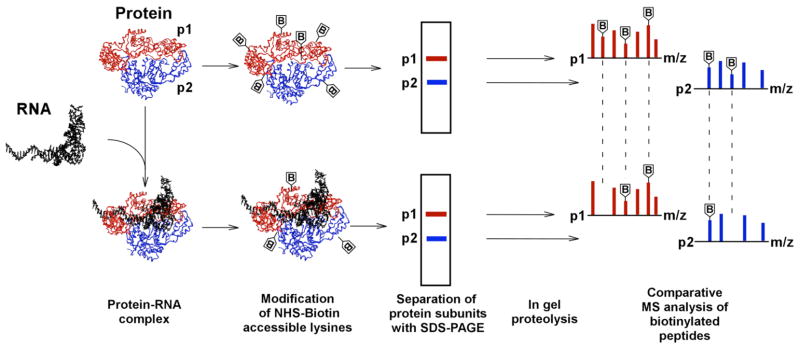
The structures for many RNA processing proteins or separate protein subunits are available at atomic resolution (1–9). However, biologically relevant large protein-RNA structures are often less amenable to crystallographic and/or NMR analysis. Therefore, there is a need for devising new and complementary approaches that enable rapid and accurate mapping of protein-RNA contacts. Here, we describe a mass spectrometric footprinting methodology that allows us to identify amino acids in the protein of interest that interact with cognate RNA (10). The experimental strategy is depicted in Figure 1. The method exploits differential accessibility of the primary amine modifying reagent N-hydroxysuccinimide (NHS)-biotin to lysine residues in the free protein versus the protein-RNA complex. Subsequent mass spectrometric analysis enables accurate identification of these residues. Monitoring lysine accessibility is a logical choice, as lysine-phosphate backbone contacts play a key role in formation of many nucleoprotein complexes. Introducing SDS-polyacrylamide gel electrophoresis (PAGE) and in-gel proteolysis prior to mass spectrometry is important for the following reasons. SDS-PAGE allows separation of individual protein subunits based on their molecular weight differences. Thereafter, contact lysines can be accurately assigned to individual components of a multi-subunit complex. For our analysis of HIV-1 reverse transcriptase (RT), this was of particular importance, since both subunits are derived from the same gene but differentially processed by the viral protease (10). Subsequent in-gel proteolysis produces short peptide fragments amenable to MS and MS/MS analysis. The biotinylated peptide peaks can be readily identified from MS data and the modified sites accurately assigned to appropriate lysine residues by MS/MS analysis. Comparative examination reveals lysines readily modified in the free protein but protected in the context of the nucleoprotein complex (Figure 2). The methodology can be expanded to probe other RNA interacting amino acids such as Arg, Trp, Tyr, His and Cys using corresponding commercially available reagents (11).
Figure 1.
Protein footprinting strategy. Biotinylation reactions of free protein comprised of separate protein subunits and the preformed protein-RNA complexes are carried out in parallel. Surface exposed lysines are modified by NHS-biotin in free protein, while those coordinating RNA become shielded from modification in the nucleoprotein complex. Individual protein subunits are separated by SDS-PAGE and then subjected to in-gel proteolysis. Comparative MS analysis of the peptide fragments enables us to identify lysines shielded by RNA contacts from those remaining susceptible to modification in the complex. The experimental scheme is adapted from Kvaratskhelia et al. (10)
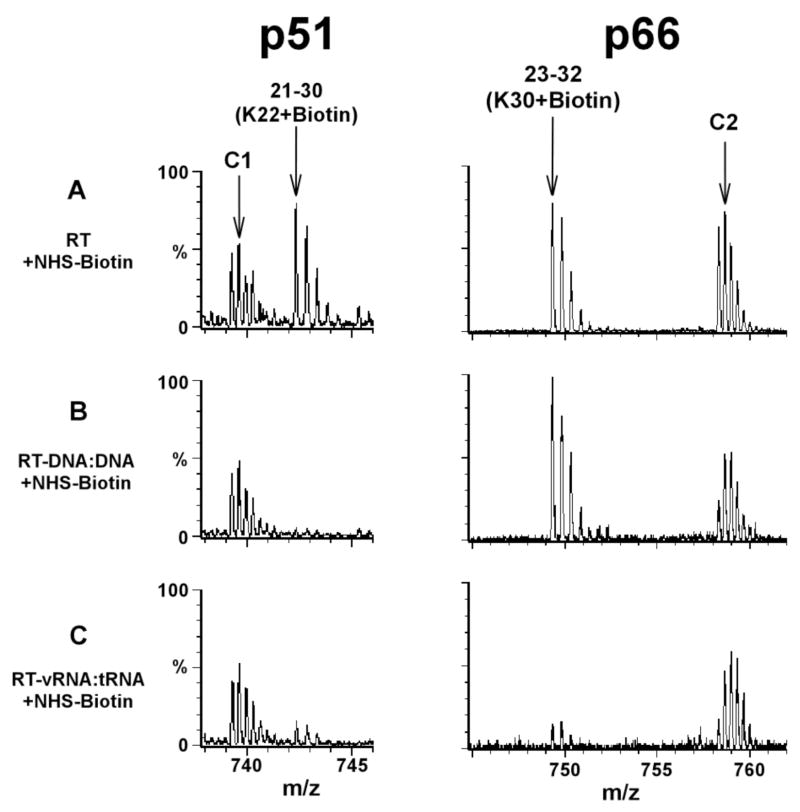
Figure 2.
Mass spectrometric data showing similarity (left panel) and differences (right panel) between HIV-1 RT when complexed with DNA:DNA and a viral RNA:tRNA duplex. RT is comprised of two protein subunits p66 and p51. Treatment of free RT with NHS-biotin resulted in modification of K22 of p51 yielding the peak corresponding to the 21–30 peptide stretch, plus one biotin molecule (A, left panel). In the context of the RT-DNA:DNA (B, left panel) or RT-viral RNA:tRNA complex (C, left panel) this peak was significantly diminished, most likely due to protection by the cognate nucleic acid contacts. The modification of K30 in p66 is shown in the right panel. The 23–32(K30+Biotin) peak persisted in RT-DNA:DNA (B, right panel) but diminished significantly in the RT-viral RNA:tRNA complex (C, right panel). These data indicate that K22 of p51 is coordinating both DNA:DNA and viral RNA:tRNA, while K30 of p66 contacts specifically the viral RNA:tRNA duplex and not DNA:DNA. Unmodified RT peptide peaks C1 and C2 serve as internal controls. Each multiply-charged peptide ion resulted in a clearly resolved peak cluster, indicating monoisotopic resolution in our mass spectrometric analysis. Data adapted from Kvaratskhelia et al. (10)
2. Materials
2.1. Modification of lysine residues
50 mM HEPES buffer, pH 7.5, 50 mM NaCl (Note 1).
NHS-Biotin (Pierce).
Quenching solution: 100 mM Tris-HCl buffer, pH 8.0 containing 100 mM lysine.
2.2. SDS-PAGE and in-gel proteolysis
Staining solution: 0.5% w/v Comassie Brilliant Blue (BioRad), 50% methanol, 10 % acetic acid, 40 % HPLC pure water.
Destain solution: 50% methanol, 10 % acetic acid, 40 % HPLC pure water.
50 mM NH4HCO3 buffer, pH 8.0.
Acetonitrile (Fisher).
SpeedVac (Thermo Savant, Holbrook, NY).
Platform shaker (New Brunswick Scientific, Edison, NJ).
Scalpel (Fisher).
2.3. Mass Spectrometric Analysis
Matrix: 5 mg/ml solution of α-cyano-4-hydroxy-cinnamic acid (Sigma) in 75 % acetonitrile/25 % water.
MALDI-ToF instrument equipped with a curved field reflectron feature (Kratos Analytical Instruments, Manchester, U.K.).
Waters Q-ToF-II instrument (Manchester, U.K.) equipped with an electrospray source and a Waters cap-LC (Waters Symmetrie300 precolumn and a Micro-Tech Scientific (Vista, CA) ZC-10-C18SBWX-150 column).
3. Methods
3.1. Modification of lysine residues
Prepare and analyze the following two reaction mixtures in parallel: free protein at 10–100 ng/μl final concentration, and protein-RNA complex containing the same amount of protein and 2- to 10-fold molar excess of RNA.
Add freshly prepared stock solution of NHS-biotin to the both reactions to obtain the final concentration of 200 to 1000 μM (Note 2). Incubate the reactions at room temperature for 30 minutes.
Terminate both reactions by adding the quenching solution to the reaction mixture in 1/10 ratio.
If required, concentrate the reaction mixes by vacuum dessication (SpeedVac) at medium heat (45°C) until the sample volume is reduced to 20 μl (Note 3).
3.2. SDS-PAGE and in-gel proteolysis
Add SDS-PAGE loading buffer to the sample and fractionate protein subunits via SDS-PAGE.
Following electrophoresis, stain the gel with Coomassie blue for 5 min at room temperature.
Destain the gel for at least two hours (or until the protein bands are distinctly visible), exchanging the destaining solution several times.
Excise the bands with a clean scalpel (Notes 4) and slice the protein bands in 3 to 4 pieces. Place the sliced gel pieces into 1.5 ml micro-centrifuge tubes.
Add 1 ml destain solution to each tube and shake overnight at 200 rpm.
Carefully remove the destain solution using a pipette without touching the gel slice. Add 1 ml fresh destain solution and shake samples for an additional 1 hr.
Centrifuge samples in a micro-centrifuge and carefully remove the destain solution with a pipette.
Add 1 ml 50 mM NH4HCO3 solution and shake the tubes for 15 min. Centrifuge the samples and remove ammonium bicarbonate solution with a pipette. Repeat this step.
Add 50% H20/50% acetonitrile solution to the gel pieces and shake samples for 1 hr at room temperature.
Remove the solution and add 200 μl of 100% acetonitrile to the gel pieces. Shake samples for 15 min.
Remove acetonitrile and desiccate the gel pieces with SpeedVac for 15 min at medium heat (45°C).
Prepare a stock solution (0.2 μg/μl) of trypsin in 10 mM HCl (Note 5). Immediately prior to use dilute the stock solution 10-fold with 50 mM ammonium bicarbonate buffer.
Add 50 μl trypsin solution to each micro-centrifuge tube (Note 6) and shake at 200 rpm overnight at room temperature.
Add 150 μl pure acetonitrile to each digestion and immediately vortex samples.
Centrifuge the tubes and carefully pipette out 180 μl supernatant without touching the gel slice (Note 7). Transfer the solution into 500 μl micro-centrifuge tubes.
Dry peptide mixtures completely using vacuum dessication at medium heat (45°C).
Add 15 μl HPLC grade water to each sample. At this stage, the samples are ready for Mass Spectrometric analysis.
3.3. Mass Spectrometric Analysis
Divide each sample in two portions for MALDI-ToF (0.5 to 2 μl) and Q-ToF (6 to 12 μl) analysis.
Apply 0.5 μl sample onto the MALDI plate and immediately mix with 0.5 μl matrix solution by pipetting the mix up and down for several times. Air dry samples at room temperature (~15 min). These sample can now be used for MALDI-ToF analysis.
Analyze samples in the reflectron mode. Once the peptide peaks are indentified, activate the post source decay feature of the MALDI to determine amino acid sequencing of the peptides.
Place 6–12 μl sample into the vials for Q-ToF analysis. Perform two sequential linear gradients of 5–40% acetonitrile 35 min and 40–90% acetonitrile for 10 min. Operate the instrument in the MS/MS mode.
Analyze the data with MASCOT engine (http://www.matrixscience.com).
For quantitative comparison of lysine modifications in free protein and protein-RNA complexes use at least two unmodified proteolytic peptide peaks as controls.
4. Notes
Use non-amine buffers, pH 7–9. Should protein or RNA stock solutions contain amines (for example, Tris), dialyze the preparations against non-amine buffers such as phosphate, HEPES, borate and carbonate.
Prior to proceeding to the footprinting experiments optimize the NHS-biotin concentration and check that the integrity of the protein-RNA complex is fully preserved under the experimental conditions. For this, incubate the protein with increasing concentration of NHS-biotin and monitor the ability of the protein to bind cognate RNA using conventional assays such as gel retardation or filter binding analysis. For footprinting experiments, choose the lowest concentration of NHS-biotin at which the ability of the protein to bind cognate RNA is fully impaired. Use this concentration of NHS-biotin to test the integrity of the protein-RNA complex. Pre-form the protein-RNA complex first and then expose the complex to NHS-biotin. The complex should be stable enough that it does not dissociate upon modification with NHS-biotin. For quantitative comparison examine unmodified protein-RNA complex in parallel experiment.
If the sample is completely dried, re-dissolve the protein by adding 20 μl H2O.
Wear gloves during all experiments. Human keratin is a main contamination observed during mass spectrometric analysis.
Store the remaining stock solution of trypsin at −20° C. Re-use only once immediately after thawing and discard the remaining stock. Alternatively, store trypsin in aliquots corresponding to those required for proteolysis.
Allow the gel slices to fully re-hydrate (10–15 min). Prior to overnight hydrolysis, check that the hydrated gel slices are fully submerged in the buffer. If needed, add an additional 10 to 20 μl ammonium bicarbonate buffer to the reaction mix.
The peptide extraction step can be repeated to increase the yield by 20 to 30%. In particular, add 50 μl ammonium bicarbonate buffer to the gel slice and incubate the mix at room temperature for 15 min. Then add 150 μl acetonitrile and vortex the mix immediately. Remove 180 μl supernatant without touching the gel slices and combine with previous extraction of the same sample.
Contributor Information
Mamuka Kvaratskhelia, Center for Retrovirus Research and Comprehensive Cancer Center, College of Pharmacy, The Ohio State University, 500 West 12th Avenue, Room 238 L.M. Parks Hall, Columbus, OH 43210. Phone: (614)-292-6091; Fax: (614)-292-7766; e-mail: kvaratskhelia.1@osu.edu.
Stuart F.J. Le Grice, RT Biochemistry Section, HIV Drug Resistance Program, National Cancer Institute at Frederick, Building 535, Room 312, P.O. Box B, Frederick, MD 21702-1201, Phone: 301-846-5256; Fax 301-846-6013. e-mail: slegrice@mail.ncifcrf.gov
Literature
- 1.Ma JB, Ye K, Patel DJ. Nature. 2004;429:318. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Opalka N, et al. Cell. 2003;114:335. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 3.Monzingo AF, Gao J, Qiu J, Georgiou G, Robertus JD. J Mol Biol. 2003;332:1015. doi: 10.1016/s0022-2836(03)00970-7. [DOI] [PubMed] [Google Scholar]
- 4.Lau CK, Diem MD, Dreyfuss G, Van Duyne GD. Curr Biol. 2003;13:933. doi: 10.1016/s0960-9822(03)00328-2. [DOI] [PubMed] [Google Scholar]
- 5.Krasilnikov AS, Yang X, Pan T, Mondragon A. Nature. 2003;421:760. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 6.Augustin MA, et al. J Mol Biol. 2003;328:985. doi: 10.1016/s0022-2836(03)00381-4. [DOI] [PubMed] [Google Scholar]
- 7.Calero G, et al. Nat Struct Biol. 2002;9:912. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 8.Nagai K, et al. Biochem Soc Trans. 2001;29:15. doi: 10.1042/0300-5127:0290015. [DOI] [PubMed] [Google Scholar]
- 9.Cramer P, Bushnell DA, Kornberg RD. Science. 2001;292:1863. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 10.Kvaratskhelia M, Miller JT, Budihas SR, Pannell LK, Le Grice SF. Proc Natl Acad Sci U S A. 2002;99:15988. doi: 10.1073/pnas.252550199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundbland RL, editor. Techniques in ProteinModification. CRC Press LLC; Boca Raton, FL: 1995. [Google Scholar]