Abstract
Rana pipiens oocytes contain two homologues of pancreatic ribonuclease A that are cytostatic and cytotoxic to human cancer cells. Extensively studied Onconase is in advanced Phase IIIb clinical trials against malignant mesothelioma, while Amphinase is a novel enzyme in pre-clinical development. Onconase is the smallest (104 amino acid residues) member of the ribonuclease A superfamily while Amphinase (114 residues) is the largest among amphibian ribonucleases. Both enzymes share the characteristic frog ribonucleases C-terminal disulfide bond but another signature of this group, the N-terminal pyroglutamate, an integral part of Onconase active site is not conserved in Amphinase.
Although Onconase and Amphinase are weak catalysts their enzymatic activities are required for cytostatic and cytotoxic activity. While it was postulated that tRNA is the primary substrate of Onconase in vivo there is also extensive indirect evidence that suggests other RNA species, in particular micro RNAs, may actually be the critical target of these ribonucleases. The cytostatic effects of Onconase and Amphinase are manifested as cell arrest in the G1 cell cycle phase. Apoptosis then follows involving activation of endonucleases(s), caspases, serine proteases and transglutaminase. Onconase was shown to be strongly synergistic when combined with numerous other antitumor modalities. Onconase and Amphinase are highly cationic molecules and their preferential toxicity towards cancer cells (having distinctly higher negative charge compared to normal cells) may depend on increased binding efficiency to the cell surface by electrostatic interactions.
Here we will discuss the structures of Onconase and Amphinase and the molecular basis for their enzymatic and anticancer functions.
Keywords: Onconase, amphinase, cytotoxic ribonucleases, Rana pipiens, structure and function, apoptosis, cell cycle, microRNA
INTRODUCTION
Cationic exchange chromatography of acidic extracts of Rana pipiens (leopard frog) eggs [1,2] reveals three distinct components with antitumor and ribonucleolytic activities. They are, in order of increasing basicity and decreasing content in the source, Onconase (ranpirnase, P-30 Protein) (Onc), its more basic natural variant, and recently characterized Amphinase (Amph). The latter is a mixture of four variants separable by reversed phase HPLC. Thus, two ribonucleases (RNases) present in R. pipiens oocytes in two or four variants, respectively, are apparently responsible for the anti-tumor activity in the eggs. This was originally observed in the frog early embryos1.
Onc and Amph were first isolated and sequenced by Alfacell Corporation; the former nearly two decades ago [1], the latter more recently2 [2]. Onc (ONCONASE®) is presently in advanced Phase III clinical trials for the treatment of unresectable malignant mesothelioma, a lung cancer associated with the exposure to asbestos or similar fibers.
This enzyme has been extensively studied and has been a subject of review articles [3,4]; it was also discussed in reviews on cytotoxic ribonucleases [5-14] and evaluations of clinical trials [15,16].
In this article we discuss structures and functions of both enzymes as well as their mechanisms of toxicity. We focus mainly on the data on Onc published since our previous review [3]; those on Amph are quite recent [2,17].
PRIMARY STRUCTURES
Amino acid sequencing [1,2] revealed that both enzymes belong to the pancreatic ribonuclease A (RNase A) superfamily (reviewed in [18]). Onc with 104 amino acid residues (20 residues less than RNase A) is the smallest known member of the family while Amph variants have 114 residues and are the largest among known amphibian RNases.
Onc isolated from frog eggs turned out to be polymorphic at amino acid position 25. Thr was found at this position during the original sequencing [1] but Ser was recently discovered in about 30% of molecules by peptide mapping (Ardelt, W., unpublished). The polymorphism was not detected by Edman degradation due to the “carryover” effect of the preceding Ser24. The replacement of Thr by Ser does not seem to affect the enzyme's function as natural and recombinant “wild type” Onc (with Thr25) were found to be equivalent in respect of enzymatic and cytotoxic activities. Also, the replacement is conservative and the polymorphic position is sterically distant from the enzyme active site. Most studies on Onc were performed with its recombinant forms. These were obtained by the expression of synthetic cDNAs in bacterial systems [19-21] and had Thr25.
As previously mentioned, a more basic, natural Onc variant was also isolated from the oocytes. In this variant1, Ile11 of Onc is replaced by Val, Asp20 by Asn and Ser103 by Arg. The mutated form is, therefore, I11V, D20N, S103R-Onc.
Cloning from genomic DNA revealed the presence of a gene encoding the wild type Onc with Thr25 [22] as well as another Onc variant: I11L, D20N, K85T-Onc [23]. It seems, therefore, that the R. pipiens genome contains at least four genes encoding various Onc variants with replacements occurring at five polymorphic positions: 11, 20, 25, 85 and 103.
Amph 1-4 variants (numbered according to their elution order from a reversed phase HPLC column) have highly similar amino acid sequences; 95 residues are invariant (83% conservation) [2]. Amph 1 and 2 differ by one residue at position 44 (Val and Ile, respectively) while the other variants differ from one another by 12-15 residues (86.8-89.9% identity) at 19 polymorphic positions. The variants are 38.2-40.0% identical with Onc, 40.7-42.5% with Rana catesbeiana ribonuclease (RC-RNase) and 24.8-28.0% with RNase A.
The N-terminal pyroglutamic acid residue, characteristic for Onc and other frog RNases, is not conserved in Amph variants that have a highly polar N-terminal extension segment of six amino acid residues. Thus, in this respect, Amph is more similar to mammalian than to amphibian homologues.
The catalytic triad of RNase A, His12, Lys41 and His119 [24,25] is strictly conserved in all variants of Onc (His10, Lys31 and His97) and Amph (His15, Lys42 and His107). As suggested by homology and confirmed by crystallographic studies discussed later, three of four disulfide bonds of RNase A (positions 26-84, 40-95 and 58-110) are conserved in Onc at positions 19-68, 30-75 and 48-90 as well as in Amph, at positions 26-79, 41-85 and 59-100. The 65-72 disulfide bond of RNase A is not conserved but, Onc and Amph have another bond linking Cys residues 87 and 104 or 97 and 114, respectively [1,2]. This C-terminal disulfide bond was found in all amphibian RNases.
Although the Onc sequence demonstrates a single putative site for N-glycosylation (Asn 69), no glycosylated forms of the enzyme were isolated from the oocytes. However, a glycosylated recombinant Onc was obtained by the expression of a synthetic gene in the Pichia pastoris system [26]. In contrast to natural Onc, all four variants of Amph are glycosylated at two sites: Asn27 (Asn25 in Amph 3) and Asn91 [2] and they are the only known glycoproteins among amphibian RNases.
Onc and Amph are very basic, single chain, small proteins. Their molecular weights and isoelectric points calculated from the amino acid sequences [1,2] are as follows: Onc (Thr25), 11820 and 9.7; Onc (Ser25), 11806 and 9.7; I11V, D20N, S103R-Onc, 11874 and 9.94; Amph-1, 13063 and 10.16; Amph-2, 13077 and 10.16; Amph-3, 13058 and 9.95; Amph-4, 12968 and 10.10.
Sequence alignments of Onc and other enzymes of the superfamily of RNase A were presented and discussed previously [1,3 (and papers cited therein), 18]. Here, in (Fig. 1), we present the structure based alignment of the amino acid sequences of Amph-2, Onc, two other amphibian RNases and RNase A [2]. More sequence details will be discussed later in relation to the structural basis for catalysis.
Fig. (1).

Structure-based sequence alignment of Amph-2, Onc, RC-RNase, RC-RNase-61 and RNase A. Elements of secondary structure are shaded (α-helices, magenta; (β-strands, cyan) and labeled below. In the RC-RNase and RNase A sequences, residues shown crystallographically to form the B1 and B2 subsites are white and orange, respectively, while those that form the P1 subsite are encircled. Amph-2 residue numbers are given above the sequences and RNase A residue numbers below. The length of each protein sequence is given at its end. <Q, pyroglutamate. Recent crystallography study on Onc in complex with nucleic acid [33]confirmed Lys9, His10, Lys31 and His97 at the enzyme P1 subsite, Lys33, Thr35, Asp67 and Phe98 at the B1 as well as Thr89 and Glu91 in the B2 subsite. Reprinted with permission from [2], modified.
RIBONUCLEOLYTIC ACTIVITY
Catalytic residues of RNase A are strictly conserved in Onc and Amph. This implies that these enzymes catalyze RNA cleavage by similar mechanisms. However, Onc is much less effective enzymatically than RNase A and most other members of the superfamily. Furthermore, Amph is substantially less active than Onc. Catalytic and cytotoxic activities of Onc and Amph can be abolished by alkylation of histidine residues [1-3,19,28] but, the enzymes do not interact with mammalian ribonuclease inhibitor protein (RI) [2,3,19,29,30]. Thus, cytotoxicity of Onc and Amph depends on the catalysis of phosphodiester bonds cleavage inside mammalian cells where both enzymes evade the ribonuclease inhibitor.
Degradation of Highly Polymerized RNA and Synthetic Substrates
Early results with RNA, monotonous polynucleotides and natural dinucleotides as substrates demonstrated that Onc was 102-105-fold less active than RNase A [1,3,19]. Onc was 100-fold more active towards poly(U) than poly(C) and 10-fold more active against uridylyl 3′,5′guanosine (UpG) than cytidylyl 3′,5′guanosine (CpG). Thus, the enzyme demonstrated strong preference for uracil at the pyrimidine (B1) subsite and for guanine at the purine (B2) subsite.
Amph variants turned out to be much less efficient than Onc and degraded highly polymerized RNA over 100-times slower. Enzymatic characterization of such slow RNases was only feasible after the development of hypersensitive fluorogenic substrates [30-32]. The values of Kcat/KM for Amph variants were approximately four orders of magnitude lower than those of RNase A when determined with 6-FAM-dArC(dA)2-6-TAMRA (rCA), 6-FAM-dArU(dA)2-6-TAMRA (rUA) or 6-FAM-dArUdGdA-6-TAMRA (rUG) [2]. Unlike Onc, Amph variants along with RNase A demonstrated very little ability to discriminate between those substrates. The activity of Onc towards rUG (the substrate optimized for this enzyme [30]) was 59- and 850-fold higher than towards rUA or rCA, respectively, but it was still 82-fold lower than that of RNase A [2]. Thus, the known preference of Onc for uracil at B1 and guanine at B2 nucleobase subsites was confirmed with those substrates. The structural basis for this preference was recently explained [33] and will be discussed later.
Degradation of Transfer RNA by Onconase
Transfer RNA was reported as an apparent primary target of Onc in cells [34-37]. However, in vitro determination of the cleavage sites in this substrate brought quite unexpected results. Suhasini and Sirdeshmukh [37] demonstrated that Onc preferentially cleaved natural tRNAs specific for Phe (yeast) or Lys, fMet and Val (E. coli) between two guanine nucleobases in the UGG sequences. These are present in the structurally important regions: the variable loop or the D-arm. The major cleavages were at G42–G43 in tRNAphe, G45–G46 in tRNALys, G19–G20 in tRNAfMet or tRNAVal. Several minor cuts occurred at GU and CU and were in accordance with Onc nucleobase specificity determined with synthetic substrates. The same cuts were observed during the digestion of denatured tRNAs but, amazingly, the GG cleavage sites were no longer preferred. It was, therefore, speculated that Onc recognizes some native structural elements of tRNA for preferential cleavages. The other cleavage sites become available for the enzyme after substrate structure is damaged or lost as a result of preferential cleavages [37]. When tRNALys3, the primer for HIV-1 reverse transcription (not having the UGG triplet anywhere in the sequence) was digested, the major cleavages occurred within the GGG segment [38]. Moreover, analogous cleavages were observed when the mutants of this substrate having AAA or CCCCC motifs instead of GGG were tested. Lee et al. [33] recently synthesized two novel fluorogenic substrates 6-FAM–dUrGdGdA–6-TAMRA and 6-FAM–dArGdGdA–6-TAMRA in order to check whether Onc may cleave the GG bond in a test tube. They did not observe any subsequent enzymatic activity. Thus, this unprecedented phenomenon among the RNase A superfamily enzymes regarding the nucleobase specificity of Onc with tRNA as a substrate is currently difficult to explain. Binding of guanine at Onc B1 subsite seems improbable since this site is highly similar to those of other RNase A homologues [33]. It seems that some yet unknown structural elements of the substrate may be involved [33,37,38].
THREE-DIMENSIONAL STRUCTURES AND CATALYTIC MECHANISMS
First crystallographic structures of Onc [39], Amph 2, and recombinant Amph-2 [2] were determined at 1.7, 1.8 and 1.9Å resolution, respectively. The crystallographic and solution structures of Onc mutants were also reported [40,41]. Quite recently, the first structures of Onc (and its T89N/E91A mutant) in complexes with nucleic acids were refined by crystallography [33].
Molecular Conformations
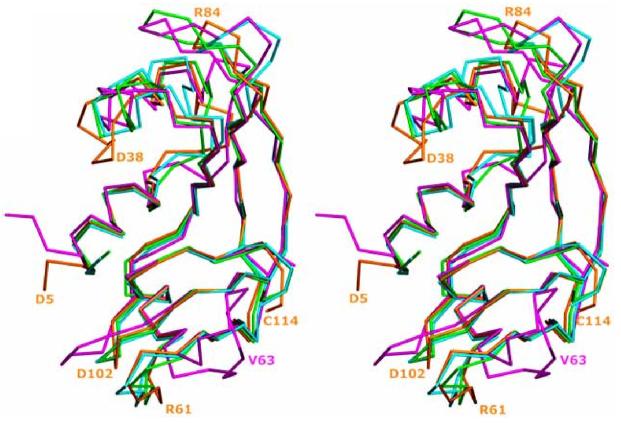
Although amino acid sequences of Onc and Amph variants are only 30 and 24.8 – 28%, respectively, identical to that of RNase A, the general folds of the three enzymes are very similar. Onc [38] and Amph-2 [2] adopt the classic bilobal, V or kidney shaped topology of RNase A [42,43] formed by two 3-stranded antiparallel (β-sheets and three α-helices. Helices α2 and α3 flank the two (β-sheets shielding them from water and helix α1 is located in the middle of the v-motif and makes contacts to both lobes [39]. (Fig. 2) presents the superposition of the Cα traces of Amph-2, Onc, RC-RNase and RNase A [2]. The backbones align well despite differences in the chains lengths which reach 20 amino at id residues for Onc and RNase A. Segments of secondary structure are similar in the four enzymes except that the (β2 strand of RNase A is not conserved in the frog RNases (it is replaced by a short loop) and Amph-2 has an additional, short β-strand designated (β0, adjacent to helix α1 [2] (Fig. 1). As mentioned before, three of four disulfide bonds of RNase A are conserved in the Onc and Amph variants. The fourth (65-72) disulfide bond of RNase A, connecting strands (β-2 and (β3, is missing in known frog RNases. These enzymes have another, C-terminal disulfide (87-104 in Onc and 97-114 in Amph) that significantly contributes (as we discuss later) to Onc conformational stability and cytotoxicity.
Fig. (2).
Superposition of the Cα traces of Amph-2 (gold), ONC (PDB entry 1ONC [39]; cyan), RC-RNase (PDB 1M07 [45]; green) and RNase A (PDB entry 7RSA [27]; magenta), shown in stereo. Selected residues from Amph-2 and RNase A are labeled in their respective colors. Reprinted with permission from [2], modified.
There is another structural distinction between RNase A (along with Amph) and the remaining two frog enzymes compared in (Fig. 1): the presence of the N-terminal Pca1 residue in Onc and RC-RNase. This uncoded residue is a product of the cyclization of Gln. Cyclization occurs spontaneously during the recombinant Onc production [19-21] or may be catalyzed by glutaminyl cyclase that most probably occurs in vivo. Welker et al. [44] demonstrated that this enzyme catalyzed Gln1 cyclization in the unfolded, reduced Onc, but not in the disulfide-intact, folded Onc. The authors hypothesized that in vivo cyclization takes place in the endoplasmic reticulum during the initial stages of oxidative folding and may be cotranslational.
Pca1 was found in frog RNases except Amph and is important for their functions. In Onc, Pca1 together with Asp2 folds back against helix α1 and forms hydrogen bonds with Val96 and Lys9 [39]. It is thus an integral part of the active site. The same was found for RC-RNase [45].
Conformational Stability and Folding of Onconase
Onc is one of the most stable proteins [22,26,40,41,46-51] with midpoints of the thermal [46,48,49,50] or guanidine induced [49,50] transitions around 90°C or 4.4 M, respectively. Respective values for RNase A, 62.4°C [48] and 2.8 M [52] were distinctly lower. As a consequence of high conformational stability Onc is also remarkably resistant to proteolysis [48].
The unusual thermodynamic stability of Onc is largely due to the C-terminal disulfide bond and in lesser degree to the N-terminal network of hydrogen bonds [47,49,50], interactions within the hydrophobic cluster (Val17, Ile22, Met23, Leu27, Phe28, and Phe36) [50,53] and the absence of cis isomers of proline residues [50,54]. Studies of guanidine induced unfolding of Onc and RNase A with intact disulfide bonds demonstrated that the pronounced difference in the conformational stability between the two enzymes can be attributed to the 103-fold slower unfolding rate of the former [50]. Pradeep et al. [54] demonstrated that refolding of Onc denatured with guanidine (with no reduction of disulfide bonds) at pH 8, was substantially faster and less complicated compared to those of RNase A and angiogenin. The authors observed four folding phases of the latter enzymes (fast, medium, slow and very slow) while only two phases (medium and slow) for Onc. Overall folding of Onc was approximately five-fold faster that that of RNase A. Quite recently, Schulenburg et al. [55] observed three kinetic phases of Onc folding at pH 5.5 (fast, medium and slow) with rate constants differing from one another by a factor of about 10. As expected, there were no folding phases involving isomerization of proline residues. Enzymatic activity reappeared at the last folding phase and followed the burial of Trp3 signaling the formation of Onc active site. It seems, therefore, that folding of Onc proceeds with at least two intermediates that, in contrast to those of RNase A, are enzymatically inactive [55].
Studies of Onc reductive unfolding and oxidative refolding by Scheraga and coworkers revealed more differences between this enzyme and RNase A [44,56-61]. While RNase A reductively unfolds through parallel pathways, with either 40-95 or 65-72 disulfide bond cleaved before becoming fully reduced [59], only one intermediate, des-(30-75) was identified for Onc [56,57] as this enzyme lacks a disulfide bond analogous to 65-72. The reduction rate of the 30-75 disulfide bond is 103-fold greater than that of the analogous 40-95 bond in RNase A. The reason for this pronounced difference was elucidated: the 30-75 disulfide bond in Onc was found “less buried” than its counterpart in RNase A and therefore more accessible to the solvent and the reducing agent. In the latter enzyme, Tyr92 shields the 40-95 disulfide by a ring-stacking interaction with Pro93 while in Onc Tyr92 is replaced by Arg72 which projects to the solvent leaving the 30-75 bond unprotected [59]. The des-(30-75) Onc is much less stable than the wild-type enzyme (although almost as stable as native RNase A). This indicates that the 30-75 disulfide bond contributes very significantly to Onc conformational stability as does the C-terminal 87-104 one [46].
Oxidative refolding of Onc is approximately 102-fold faster than that of RNase A [58] and generates three intermediates [59,60]. The most recent study [61] revealed that the same intermediates can also be generated during the reductive unfolding process. In addition to the des-(30-75) intermediate [56,57] identified earlier, two other species, des-(19-68) and des-(30-75, 19-68) were identified. The latter is formed and processed early during oxidative refolding. The remaining structured intermediates follow and des-(19-68) converts into native form slower than des-(30-75). Thus, the detailed mechanism of Onc oxidative folding will soon be fully elucidated.
Molecular dynamic simulations and the NMR data demonstrated high rigidity of Onc [40,41]. This enzyme does not have the characteristic RNase A “breathing” motion of the (β-sheet motif considered to be important for catalytic efficiency [40]. Short loops of Onc were implicated as a possible reason for the rigidity and reduced catalytic activity [40]. However, site specific mutations installing longer loops of RC-RNase to Onc [33] unexpectedly failed to affect the enzyme's catalytic efficiency. Other mutations destabilizing the enzyme did not increase its catalytic or cytotoxic activities [51]. On the other hand, earlier studies demonstrated lowered stability and increased catalytic activity of M23L-Onc [48]. This variant was also more flexible than wild type Onc [40,41].
Active Sites
RNase A homologues interact with RNA substrates through multiple subsites binding phosphoryl groups (P subsites) and nucleobases (B subsites) (reviewed in [24]). The enzymes catalyze cleavage of the P–O5′ bonds in nucleotides bound at the B1–P1–B2 subsites [33].
Catalytic sites (or P1 subsites) of Onc and Amph are located in the clefts of their kidney shapes. In Onc, in addition to the catalytic triad, His1O, Lys31 and His 97, the active site contains Pca1, Lys9, Thr35 and Phe98 [39]. Pca1 forms hydrogen bonds with Lys9 and His97 [39] and its apparent role is to stabilize Lys9 [32]. Both Lys9 and Lys31 contribute 103-fold to kcat/KM values for the degradation of the fluorogenic rUG substrate but do not seem to have a role in substrate binding [32].
Crystallographic structure of the Onc-d(AUGA) complex [33] revealed that catalytic His97 adopts a substantially different conformation than analogous His residues in other RNases and also different than that in free Onc [39]. Imidazole ring of this residue is rotated by almost 180 degrees in a way that its Nδ2 rather than Nδ1 provides a proton for catalysis [33]. The Nδ1 is engaged in a hydrogen bond with Thr89. The classic bell shape of the kcat/KM dependence on pH indicates that both His10 and His 97 participate in catalysis by Onc [33].
According to the sequence homology (Fig. 1) and structural studies, the following residues form the catalytic site in Amph-2: His 15, Lys42 and His107 (the catalytic triad) as well as Lys14 and Phe108 [2]. His15 is located in a hydrophobic pocket and stabilized by a hydrogen bond formed by its Nδ1 form and the main chain oxygen of Thr89. The conformation of His15 side chain is similar to those of some highly active RNases. Unlike in RNase A and in many of its homologues, mobility of the side chain of the second catalytic histidine (His107) in Amph-2 is not restricted by hydrogen bonding. Lys42 and Lys14 of Amph-2 are equivalent to Lys31 and Lys 9 in Onc. Lys9 is stabilized by a hydrogen bond with Pca1. The latter contributes 20-fold to kcat/KM for Onc catalyzed reactions [32] but is not conserved in Amph [2]. Amph-2 Lys 14 is stabilized at different position than Lys9 in Onc by a network of salt bridges and reorients the side chain of Lys42 from the active site to solvent. This together with the mentioned lack of constraints on His107 may contribute to low substrate turnover by this enzyme.
B1 Subsites
Sequence homology (Fig. 1) indicated that Lys33, Thr35, Asp67 and Phe68 constitute the pyrimidine binding subsite in Onc. All those residues but Lys33 are conserved in members of the RNase A superfamily. In the structure of the Onc–d(AUGA) complex those residues are indeed proximal to uracil and Thr35 forms hydrogen bonds with N4 and O2 of this nucleobase [33].
In Amph-2 Ile 44 replaces Lys33 of Onc and Thr78 is equivalent to Onc Asp67 at this subsite. The B1 subsite closely resembles that of RC-RNase [2].
B2 Subsites
Several amino acid residues of RNase A forming the purine nucleobase subsite are deleted from the sequences of frog RNases (Fig. 1). It was therefore expected that in Onc only three residues, Thr89, Glu91, and the catalytic His97 may be involved in binding guanine or adenine nucleobases. The equivalent residues in Amph-2 are Thr99, Arg10l and the catalytic His107. However, as discussed before, both His97 of Onc and His107 of Amph-2 assume different conformations than their counterparts in other members of the superfamily and might not be involved in purine binding. In the Onc-d(AUGA) complex Glu91 forms two hydrogen bonds with guanine; Thr89 is in close proximity to this nucleobase [33]. The authors employed site directed mutagenesis at these two residues to study strong preference of Onc to guanine over adenine at this subsite. Replacement of Thr89 with a positively charged residue further increased the preference for guanine (by a factor of 10 when Arg was placed there). Substitution of Glu91 with residues unable to form hydrogen bonds with guanine, lowered the preference. Lysine introduced at position 91 reversed the preference as this variant was slightly more active towards rUA than rUG substrate. The double variant, T89N/E91A-Onc exerted 2.6-fold preference for adenine nucleobase [33]. Thus, Raines and his group not only successfully explained the purine specificity of Onc but were able to substantially change this specificity by rational site specific mutagenesis.
Highly conserved among RNases, Glu91 of Onc (Glu111 in RNase A) is replaced by Arg1Ol in Amph (Fig. 1). This substitution might favor adenine binding. However, in Amph-2 structure Arg1Ol is positioned away of the B2 subsite [2]. On the other hand, Glu7 that is a new residue, specific for Amph, seems to be capable of interaction with the guanine bound at this subsite. Thus, this residue could possibly take a role of Onc Glu91. In this situation, explanation of purine binding by Amph awaits structure refinement of this enzyme in complex with nucleic acid.
Conclusions: Possible Reasons for Low Catalytic Efficiency
Onc and Amph are distinctly less active than RNase A and most homologues. Molecular basis for this difference has not been fully elucidated. However, the most obvious factors seem to be atypical conformations of catalytic residues, Lys 31 and His 97 in Onc as well as equivalent Lys 42 and His 107 in Amph. These residues, especially in Amph, are less constrained by hydrogen bonding to neighboring residues than their counterparts in highly active members of RNase A superfamily. The substrate binding by both enzymes is impaired. This is mainly due to atypical purine binding (B2) subsites but also to apparent lowered binding abilities of the mentioned catalytic residues. In Onc, the unusual conformational stability and lack of flexibility of the (β-sheet motif may be also considered.
CYTOSTATIC AND CYTOTOXIC PROPERTIES OF ONCONASE AND AMPHINASE
Initial Observations
The initial studies on cytostatic and cytotoxic properties of Onc (“P-30 Protein”) were carried out in vitro on human leukemic HL-60, submaxillary carcinoma A-253 and colon adenocarcinoma Colo 320 CM, the cell lines found to be sensitive to this ribonuclease [62]. The cytostatic effect was manifested by suppression of cell growth rate which was due to a prolongation of (or arrest in) G1 phase of the cell cycle concomitant with a decrease in frequency of DNA replicating (S-phase) cells. The Onc-induced cytotoxicity was seen as an induction of apoptosis, evident by the presence of cells with fractional (“sub-G1”) DNA content detected by flow cytometry, and as reduction of cells reproductive capability manifesting by lowered clonogenicity [62]. The clonogenicity of A-253 cells was reduced by 50 and by 90% at 10 and 100 nM Onc, respectively. The average colony size (cell number per colony) was also dramatically decreased reflecting the slower growth rate of the cells surviving in the presence of Onc. Unlike as most chemotherapeutic agents that act rather rapidly the cytostatic and cytotoxic effects of Onc become apparent after a 24 to 48 hour delay. The intensity of cytostatic and cytotoxic effects was Onc-concentration dependent and increased progressively with time of cells exposure to this ribonuclease [62].
The first in vivo studies revealed a striking effect of Onc on survival of mice with Ml09 Madison carcinoma, with some animal groups having over a 12-fold longer survival time compared to the untreated animal groups [63]. In other early studies a synergism in the cytotoxic effects of Onc was reported when Onc was combined either with tamoxifen or trifluoroperazine (Stelazine) on human lung carcinoma A549 or pancreatic adenocarcinoma ASPC-1 cells [64]. The virtue of these drug combinations is low toxicity of the individual drugs and the apparent enhancement of their cytotoxicity towards tumor cells when combined with Onc. This study prompted a series of subsequent investigations that revealed synergism of Onc in combination with a variety of other relatively non-toxic agents. Thus, the synergistic cytotoxic effects were observed in vitro when Onc was combined with lovastatin [65], vincristine [66], tumor necrosis factor α [67], interferons [68-70], ionizing radiation [71,72], or differentiation-inducing agents [74] and in vivo with tamoxifen [75]. The cytotoxic potency of Onc was also enhanced by combination with mild hyperthermia (39.0°C) [76]. The striking feature is that these synergisms took place despite that Onc was combined with diverse antitumor agents, each characterized by entirely different mechanism of action.
Internalization
Since Onc and Amph act intracellularly, the first step in the cytotoxic pathway is internalization of these ribonucleases. The initial report postulated the presence of high- and low- affinity Onc receptors on the plasma membrane; binding of Onc to these receptors led to its internalization by means of endocytosis [28]. Other studies, however, have shown that Onc binds to the cell surface in a non-saturable manner, consistent with lack of specific receptors [77]. It was recently suggested [78] that electrostatic forces play the key role in Onc binding to plasma membrane. It was also postulated that these electrostatic interactions may provide explanation for the preferential sensitivity of tumor cells [78]. Onc (and Amph) are strongly cationic molecules while the cell surface of majority of tumor types has both, net and per surface unit, distinctly higher negative charge compared to their normal counterparts [79,80]. In fact, cancer cells with high metastatic potential have even stronger electronegative membrane charge than cells with low metastatic capability [81]. Consistent with this mechanism of electrostatic binding are findings that the bacterial RNase binase that is also preferentially cytotoxic to tumor cells and as Onc and Amph, is strongly cationic [82].
Onc binding to the cell surface is followed by internalization, which occurs by energy-dependent endocytosis [77,83,84]. From endocytes Onc gains access to the cytosol most likely bypassing the Golgi or ER organelles; its localization within the intracellular vesicles [77] is consistent with such a pathway. Recent studies provided evidence that endocytosis of Onc is mediated by AP-2/clathrin [84], i.e. the mechanism dependent on clathrin adaptors and the GTPase dynamin [85]. This observation appears to disagree with the earlier report indicating lack of involvement of dynamin in the process of Onc internalization [77]. The apparent discrepancy may be related to the fact that earlier studies [77] were carried out on the cell line stably transfected with the inducible dynanin-K44A dominant-negative mutant, whose expression appears to protect cells against Onc [85]. Interestingly, endosome neutralization was shown to enhance Onc cytotoxicity by nearly 100-fold, likely by facilitating translocation of this protein from endosomes to the cytosol [85].
Intracellular Targets
The evidence is indisputable that the target of Onc (and also of Amph), responsible for cytostatic and cytotoxic effects, is intracellular RNA. Numerous studies have shown that Onc [1,2,19,28,83,86] as well Amph [17], chemically modified to extinguish their enzymatic activity, although were able to enter the cell, were ineffective. The ribonucleolytic activity of Onc within the cell is sustained by two factors. One is its exceptional resistance to RI [87,88], the inhibitor that binds to some enzymes of RNase A family with an extreme (femtomolar range) affinity [89]. It has been shown that cytotoxicity of different RNase A variants inversely correlates with their RI-binding affinity [90]. The second factor is the exceptional conformational stability of this protein, already discussed in this article. The stability allows Onc to escape conformational changes that otherwise could incur after internalization upon binding to other proteins and within the environment of diverse intracellular compartments.
It was initially observed that degradation of cellular 28 S and 18 S rRNA in Onc-treated cells coincided with inhibition of protein synthesis. This observation led the authors to propose that rRNA is the preferential target of Onc and that suppression of translation is the mechanism responsible for its cytostatic and cytotoxic activity [28]. Subsequently, however, the same authors observed degradation of tRNA and postulated that in fact tRNA is the key Onc target and that its degradation is the primary factor responsible for cytotoxic activity of Onc [35]. As mentioned earlier, the Onc-specific cleavage sites on tRNA have been identified as the bonds between UG and GG bases [37,38].
Several observations, however, are incompatible with the view that regardless whether rRNA or tRNA is the preferential target of Onc, the inhibition of protein synthesis is the sole or key mechanism by which this ribonuclease exerts its cytostatic or cytotoxic activity. First, the overall pattern and kinetics of changes observed in the cells treated with Onc are entirely different as compared with the changes in cells treated with protein synthesis inhibitors such as cycloheximide [91]. While the latter kill cells rather rapidly and indiscriminately with regard to the cell cycle phase, Onc initially induces G1 arrest and apoptosis occurs with a 24–48 h delay [86]. Furthermore, the transcription of several genes, some involved in regulation of cell cycle progression, is in fact upregulated in the cells treated with Onc [86]. It is quite evident that the latter observation conflicts with the mechanism in which Onc nonspecifically blocks translation by destruction of tRNA or rRNA. We postulated, therefore, that One of the targets of Onc [92] and Amph [17] is the non-coding RNA (microRNAs) that is involved in regulation of gene expression through RNA interference (RNAi) [93-95].
Targeting RNAi, in fact, may be one of the mechanisms responsible for preferential effectiveness of Onc towards tumors. It was recently reported that development of many tumors is associated with early alterations at the level of microRNA genes and that these genes are located in the genome hot spots associated with cancer [96-98]. The microRNAs are extensively involved in pathogenesis of both leukemias and solid tumors and they promote neoplastic growth by controlling the expression of protein-coding tumor suppressors and oncogenes [96,99]. By targeting RNAi, thus, Onc may be more effective in suppressing growth or killing tumor rather than normal cells.
The intriguing synergism of Onc when combined with different chemotherapeutics, radiation, differentiation agents, interferons, NFκB or proteasome inhibitor [65-76, 100] may be explained by the mechanism in which this ribonuclease targets the family of microRNAs recently found to specifically modify tumor resistance to cytotoxic anticancer therapy via mobilizing the cell defense mechanisms [101]. If this were the case one may expect that this ribonuclease, by enhancing treatment efficiency via lowering tumor cell resistance, will find wide application as a universal adjunct to most kinds of antitumor therapy. Another intracellular potential target of Onc whose destruction is expected to promote sensitivity to antitumor modalities is the family of microRNAs within the p53 tumor suppressor network that modulate inclination of cells to undergo apoptosis [102].
Consistent with the view that Onc and Amph have potential to target RNAi is the presence of these ribonucleases in amphibian eggs and early embryos where their physiological function during embryogenesis is as yet unexplained. It is known, however, that RNAi plays an essential role in gene regulation during development [95,103-105]. In the developing embryo, thus, Onc and Amph could modulate the progress of cell differentiation by targeting RNAi.
In addition to microRNAs, another potentially attractive target of Onc may be RNA associated with telomerase [106,107]. Telomeres are replenished by internal RNA-templated synthesis of telomeric DNA. There is strong evidence for the implication of the status of telomerase RNA in cancer progression and “immortality” of tumor cells [106]. It was shown that extent of telomerase RNA limits the telomere length and even minute changes in the level of telomerase RNA are reflected by profound changes in cell ability to proliferate [107]. Destruction of telomerase RNA by Onc, thus, may lead to cytostasis, which would be compatible with the observed arrest in G1 phase of the cell cycle [86].
Of interest is the observation that tumoricidal effectiveness of Onc on A549 tumors in vivo was distinctly greater following multiple small doses administration rather than larger single-dose treatment [109]. The multiple, frequent Onc administration, thus, may be the preferred method of treatment regardless of whether the drug is used alone or as an adjunct, in combination with other antitumor modalities.
Cell Cycle Effects and Induction of Apoptosis
Most in vitro studies on different cell lines provided evidence that the suppression of the cellular growth rate by Onc was a result of either prolongation of G1 phase of the cell cycle or cell arrest at that phase [62,67,74,86]. Similarly, accumulation of HL-60 cells in G1 was also seen upon treatment with Amph [17]. The most detailed studies on the mechanism of cell arrest in G1 were carried out on human histiocytic lymphoma U937 cells [91]. These cells, when treated with 157 nM Onc for 24 h, showed a downregulation of cyclin D expression concomitant with distinct upregulation of cyclin-dependent kinase inhibitors p16INK4A, p21WAF1/CIP1, and p27KIP1. The retinoblastoma susceptibility gene protein pRb was hypo-phosphorylated in the Onc-arrested G0/1 cells but was phosphorylated in the cells that were still progressing through S and G2 in the presence of Onc [86]. The cytostatic effect of Onc in these cells, thus, was mediated by suppression of Cdks activity leaving the cells at the restriction point controlled by Cdk4/6 and D type cyclins [110].
An intriguing possibility has been discussed in this study [86], namely that one of the targets of Onc is dsRNA which is a cofactor of the dsRNA-dependent protein kinase R (PKR), the enzyme that phosphorylates IκB [108] and activates the ubiquitous transcription factor NFκB. Among numerous genes activated by NFκB are the genes that regulate cell growth [111,112]. If dsRNA, the PKR cofactor, is one of the Onc targets, its degradation would lead to suppression of cell proliferation manifested as arrest in G0/1. Consistent with this mechanism is the observation of downregulation and suppression of NFκB activity of Jurkat cells treated with Onc, concomitant with their arrest in the cell cycle [113].
Regardless of the tumor cell type, prolonged cells exposure to Onc [62,67,114-117] or to Amph [17] leads to apoptosis. The Onc-induced apoptosis is manifested by characteristic morphologic changes such as cell shrinkage, chromatin condensation, nuclear fragmentation and formation of apoptotic bodies, changes considered to be the “gold standard” for identification of the apoptotic mode of cell death [118]. Activation of caspases [114] is accompanied by activation of serine proteases [115] and activation of transglutaminase [116]. Activation of endonucleases(s) is reflected by typical internucleosomal DNA fragmentation, with appearance of cells characterized by fractional (“sub-G1”) DNA content and multiplicity of DNA strand breaks that can be labeled with fluorochrome-tagged deoxynucleotides using the TUNEL assay [119]. Demise of cells treated with Amph resembles in many respects apoptosis induced by Onc, since it also involves activation of endonucleases(s) that leads to extensive DNA fragmentation, and activation of caspases, serine proteases and transglutaminase [17]. It should be noted, however, that in some cell types such as neuroblastoma, Onc was reported to induce caspase-independent cell death, with features resembling autophagy [120]. Of particular interest in the context of induction of apoptosis by Onc is already discussed possibility of targeting of dsRNA that activates PKR and subsequently NFκB. Namely, prevention of NFκB activation by this mechanism may in turn preclude the induction of the cell survival factors that otherwise provide drug-resistance [121,122]. This mechanism is also consistent with the findings of synergism of Onc with variety of anti-cancer modalities [65-76].
Conclusions: Antitumor Activity, Synergisms and Clinical Application
As discussed above there is undisputable evidence that Onc and Amph are able to enter the cells and that their target therein is RNA; its destruction manifests by the observed cytostatic and cytotoxic effects. The evidence is also compelling that Onc shows preference to tumor cells.
In advanced clinical trials in malignant mesothelioma Onc, while having a favorable safety profile, was shown to prolong periods of stable disease and to offer potential survival benefit when compared with the alternative (doxorubicin) treatment (reviewed in [15,16]). A confirmatory Phase IIIb clinical trial of Onc combined with doxorubicin versus doxorubicin alone, in treatment of malignant mesothelioma, is currently nearing completion [16]. A Phase I/II trial in non-small cell lung cancer is in progress.
The mechanism responsible for Onc and Amph preference for tumor cells, which is the most important question to be addressed, is not yet fully elucidated. One of the mechanisms may be related to cationic properties of Onc and Amph that favor their electrostatic binding to the plasma membrane of cancer cells [78]. We presented the evidence from literature providing rationale that rRNA and/or tRNA may not be the exclusive Onc targets and that other RNA species may be targeted as well. Another mechanism, thus, may be related to targeting microRNAs, specifically the families of microRNAs known to engage cell defenses against stress and protect from induction of apoptosis. This mechanism would explain striking synergism observed when Onc was combined with any of a variety of antitumor modalities of different mechanisms of action. If further studies confirm the property of Onc (and/or Amph) to specifically weaken tumor cell resistance, a tantalizing possibility to use these ribonucleases as universal adjunct to increase effectiveness in treatment with diverse antitumor modalities has to be considered. The antitumor activity Onc is also compatible with the notion that in addition to microRNAs, telomerase RNA and dsRNA, the activator of PKR and NFκB, may also be targeted by this ribonuclease. Clearly, both Onc as well as Amph represent a new class of antitumor agents with entirely different mechanisms of action than drugs currently used. Their full potential still remains to be elucidated.
ABBREVIATIONS
- Onc
Onconase
- Amph
amphinase
- RNase
Ribonuclease
- RC-RNase
Rana catesbeiana ribonuclease
- Pca
Pyroglutamic acid
- poly(U)
Polyuridylc acid
- poly(C)
Polycytidylic acid
- UpG
Uridylyl 3′,5′guanosine
- CpG
Cytidylyl 3′,5′guanosine
- 6-FAM
6-carboxyfluorescein
- 6-TAMRA
6-carboxytetramethylrhodamine
- rCA
6-FAM-dArC(dA)2-6-TAMRA
- rUA
6-FAM-dArU(dA)2-6-TAMRA
- rUG
6-FAM-dArUdGdA-6-TAMRA
- RI
Ribonuclease inhibitor protein
- RNAi
RNA interference
- Ds
Double-stranded
- PKR
dsRNA-dependent protein kinase R
Footnotes
Shogen, K. and Yoon, W.K.. The 27th Annual Eastern Colleges Science Conference, Pennsylvania State University, April 28, 1973.
Ardelt, W.; Vidunas, E.; Saxena, S.K.; Lee, H.-S.; Saxena, A.; Viera, A. and Shogen, K. The 6th International Meeting on Ribonucleases, Bath, U.K. June 19-23, 2002.
Ardelt, W., Lee, H.-S., Randolph, G., Viera, A., Mikulski, S.M. & Shogen, K. The 4th International Meeting on Ribonucleases: Chemistry, Biology and Biotechnology, Groningen, The Netherlands June 19-23, 1996.
Tsai, C.J.; Liu, J.H.; Liao, Y.D.; Chen, L.Y.; Cheng, P.T. and Sun, Y.J. (2003). Protein Data Bank (unpublished).
REFERENCES
- 1.Ardelt W, Mikulski SM, Shogen K. J. Biol. Chem. 1991;266(1):245–251. [PubMed] [Google Scholar]
- 2.Singh UP, Ardelt W, Saxena SK, Holloway DE, Vidunas E, Lee HS, Saxena A, Shogen K, Acharia KR. J. Mol. Biol. 2007;371(1):93–111. doi: 10.1016/j.jmb.2007.04.071. [DOI] [PubMed] [Google Scholar]
- 3.Saxena SK, Shogen K, Ardelt W. Lab. Med. 2003;34(5):380–387. [Google Scholar]
- 4.Lee JE, Raines RT. BioDrugs. 2008;22(1):53–58. doi: 10.2165/00063030-200822010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youle RJ, D'Alessio G. In: Ribonucleases: Structures and Functions. D'Alessio G, Riordan JF, editors. Academic Press; New York: 1997. pp. 491–514. [Google Scholar]
- 6.Shein CH. Nat. Biotechnol. 1997;15(6):529–536. doi: 10.1038/nbt0697-529. [DOI] [PubMed] [Google Scholar]
- 7.Raines RT. In: Enzymatic Mechanisms. Frey PA, Northrop DB, editors. IOS press; Washington, DC: 1999. pp. 235–249. [Google Scholar]
- 8.Rybak SM, Newton DL. Exp. Cell. Res. 1999;253(2):325–335. doi: 10.1006/excr.1999.4718. [DOI] [PubMed] [Google Scholar]
- 9.Leland PA, Raines RT. Chem. Biol. 2001;8(5):405–413. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matoušek J. J. Comp. Biochem. Physiol. 2001;129(3):175–91. doi: 10.1016/s1532-0456(01)90202-9. [DOI] [PubMed] [Google Scholar]
- 11.Makarov AA, Ilinskaya ON. FEBS Lett. 2003;540(13):15–20. doi: 10.1016/s0014-5793(03)00225-4. [DOI] [PubMed] [Google Scholar]
- 12.Benito A, Ribó M, Vilanova M. Mol. Biosyst. 2005;1(4):294–302. doi: 10.1039/b502847g. [DOI] [PubMed] [Google Scholar]
- 13.Arnold U, Ulbrich-Hofmann R. Biotechnol. Lett. 2006;28(20):1615–1622. doi: 10.1007/s10529-006-9145-0. [DOI] [PubMed] [Google Scholar]
- 14.Tafech A, Basset T, Sparnese D, Lee CH. Curr. Med. Chem. 2006;13(8):683–881. doi: 10.2174/092986706776361021. [DOI] [PubMed] [Google Scholar]
- 15.Costanzi J, Sidransky D, Navon A, Goldsweig H. Cancer Invest. 2005;23(7):643–650. doi: 10.1080/07357900500283143. [DOI] [PubMed] [Google Scholar]
- 16.Pavlakis N, Vogelzang NJ. Expert Opin. Biol. Ther. 2006;6(4):391–399. doi: 10.1517/14712598.6.4.391. [DOI] [PubMed] [Google Scholar]
- 17.Ardelt B, Ardelt W, Pozarowski P, Kunicki J, Shogen K, Darzynkiewicz Z. Cell Cycle. 2007;6(24):3097–3102. doi: 10.4161/cc.6.24.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyer KD, Rosenberg HF. Mol. Divers. 2006;10(4):585–597. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- 19.Boix E, Wu Y-N, Vasandani VM, Saxena SK, Ardelt W, Ladner J, Youle RJ. J. Mol. Biol. 1996;257(5):992–1007. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- 20.Leland PA, Schultz LW, Kim BM, Raines RT. Proc. Natl. Acad. Sci. USA. 1998;95(18):10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Natomista E, Cafaro V, Fusiello R, Bracale A, D'Alessio G, Di Donato A. FEBS Lett. 1999;463(3):211–215. doi: 10.1016/s0014-5793(99)01623-3. [DOI] [PubMed] [Google Scholar]
- 22.Liao YD, Wang SC, Leu YJ, Wang CF, Chang ST, Hong YT, Pan YR, Chen C. Nucleic Acids Res. 2003;31(18):5247–5255. doi: 10.1093/nar/gkg746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S-I, Le S-Y, Newton DL, Maizel JV, Rybak SM. Nucleic Acids Res. 2000;28(12):2375–2382. doi: 10.1093/nar/28.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raines RT. Chem. Rev. 1998;98(3):1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 25.Raines RT. In: Artificial Nucleases. Zenkowa MA, editor. Springer–Verlag; Heidelberg: 2004. pp. 19–32. [Google Scholar]
- 26.Kim B-M, Kim H, Raines RT, Lee Y. Biochem. Biophys. Res. Commun. 2004;315(4):976–983. doi: 10.1016/j.bbrc.2004.01.153. [DOI] [PubMed] [Google Scholar]
- 27.Wlodawer A, Svensson LA, Sjölin L, Gilliland GL. Biochemistry. 1988;27(8):2705–2717. doi: 10.1021/bi00408a010. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y, Mikulski SM, Ardelt W, Rybak SM, Youle RJ. J. Biol. Chem. 1993;268(14):10686–10693. [PubMed] [Google Scholar]
- 29.Leland PA, Schultz LW, Kim BM, Raines RT. Proc. Natl. Acad. Sci. 1998;95(18):10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelemen BR, Klink TA, Behlke MA, Eubanks SR, Leland PA, Raines RT. Nucleic Acids Res. 1999;27(18):3696–3701. doi: 10.1093/nar/27.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park C, Kelemen BR, Klink TA, Sweeney RY, Behlke MA, Eubanks SR, Raines RT. Methods Enzymol. 2001;341:81–94. doi: 10.1016/s0076-6879(01)41146-3. [DOI] [PubMed] [Google Scholar]
- 32.Lee JE, Raines RT. Biochemistry. 2003;42(39):11443–11450. doi: 10.1021/bi035147s. [DOI] [PubMed] [Google Scholar]
- 33.Lee JE, Bae E, Bingman CA, Philips GN, Jr, Raines RT. J. Mol. Biol. 2008;375(1):165–177. doi: 10.1016/j.jmb.2007.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin JJ, Newton DL, Mikulski SM, Kung HF, Youle RJ, Rybak SM. Biochem. Biophys.Res.Commun. 1994;204(1):156–162. doi: 10.1006/bbrc.1994.2439. [DOI] [PubMed] [Google Scholar]
- 35.Saxena SK, Sirdeshmukh R, Ardelt W, Mikulski SM, Shogen K, Youle RJ. J. Biol. Chem. 2002;277(17):15142–15146. doi: 10.1074/jbc.M108115200. [DOI] [PubMed] [Google Scholar]
- 36.Iordanov MS, Ryabinina OP, Wong J, Dinh TH, Newton DL, Rybak SM, Magun BE. Cancer Res. 2000;60(7):1983–1984. [PubMed] [Google Scholar]
- 37.Suhasini AN, Sirdeshmukh R. J. Biol. Chem. 2006;281(18):12201–12209. doi: 10.1074/jbc.M504488200. [DOI] [PubMed] [Google Scholar]
- 38.Suhasini AN, Sirdeshmukh R. Biophys. Res. Commun. 2007;363(2):304–309. doi: 10.1016/j.bbrc.2007.08.157. [DOI] [PubMed] [Google Scholar]
- 39.Mosimann SC, Ardelt W, James MNG. J. Mol. Biol. 1994;236(4):1141–1153. doi: 10.1016/0022-2836(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 40.Merlino A, Mazzarella L, Carannante A, Di Fiore A, Di Donato A, Notomista E, Sica F. J. Biol. Chem. 2005;280(18):17953–17960. doi: 10.1074/jbc.M501339200. [DOI] [PubMed] [Google Scholar]
- 41.Gorbatyuk VY, Tsai CK, Chang CF, Huang TH. J. Biol. Chem. 2004;279(7):5772–5780. doi: 10.1074/jbc.M311233200. [DOI] [PubMed] [Google Scholar]
- 42.Richardson JS. Adv. Prot. Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 43.Raines RT. Chem. Rev. 1998;98(3):1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 44.Welker E, Hathaway L, Xu G, Narayan M, Pradeep L, Shin H-C, Scheraga HA. Biochemistry. 2007;46(18):5485–5493. doi: 10.1021/bi602495a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leu YJ, Chern SS, Wang SC, Hsiao YY, Amiraslanov I, Liaw YC, Liao YD. J. Biol. Chem. 2003;278(9):7300–7309. doi: 10.1074/jbc.M206701200. [DOI] [PubMed] [Google Scholar]
- 46.Leland PA, Schultz LW, Kim BM, Raines RT. Proc. Natl. Acad. Sci., USA. 1998;95(18):10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leland PA, Staniszewski KE, Kim B, Raines RT. FEBS Lett. 2000;477(3):203–207. doi: 10.1016/s0014-5793(00)01804-4. [DOI] [PubMed] [Google Scholar]
- 48.Notomista E, Catanzano F, Graziano G, Dal Piaz F, Barone G, D'Alessio G, Di Donato A. Biochemistry. 2000;39(30):8711–8718. doi: 10.1021/bi000415x. [DOI] [PubMed] [Google Scholar]
- 49.Notomista E, Catanzano F, Graziano G, Di Gaetano S, Barone G, Di Donato A. Biochemistry. 2001;40(3):9097–9103. doi: 10.1021/bi010741s. [DOI] [PubMed] [Google Scholar]
- 50.Arnold U, Schulenburg C, Schmidt D, Ulbrich-Hofmann R. Biochemistry. 2006;45(11):3580–3587. doi: 10.1021/bi0525223. [DOI] [PubMed] [Google Scholar]
- 51.Schulenburg C, Ardelt B, Ardelt W, Arnold U, Shogen K, Ulbrich-Hofmann R, Darzynkiewicz Z. Cancer Biol. Ther. 2007;6(8):1233–1239. doi: 10.4161/cbt.6.8.4423. [DOI] [PubMed] [Google Scholar]
- 52.Leich F, Köditz J, Ulbrich-Hofman R, Arnold U. J. Mol. Biol. 2006;358(5):1305–1313. doi: 10.1016/j.jmb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 53.Kolbanovskaya EY, Trewisscha van Scheltinga AC, Mukhortov VG, Ardelt W, Beintema JJ, Karpeysky MY. Cell. Mol.Life Sci. 2000;57(89):1306–1316. doi: 10.1007/PL00000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pradeep L, Shin HC, Scheraga HA. FEBS Lett. 2006;580(21):5029–5032. doi: 10.1016/j.febslet.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 55.Schulenburg C, Martinez-Senac MM, Löw C, Golbik R, Ulbrich-Hofmann R, Arnold U. FEBS J. 2007;375(1):165–177. doi: 10.1111/j.1742-4658.2007.06106.x. [DOI] [PubMed] [Google Scholar]
- 56.Xu G, Narayan M, Welker E, Scheraga HA. Biochemistry. 2004;43(11):3246–3254. doi: 10.1021/bi036215d. [DOI] [PubMed] [Google Scholar]
- 57.Narayan M, Xu G, Ripoli DR, Zhai H, Breuker K, Wanjalla C, Leung HJ, Navon A, Welker E, McLafferty FW, Scheraga HA. J. Mol. Biol. 2004;338(4):795–809. doi: 10.1016/j.jmb.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 58.Gahl RF, Narayan M, Xu G, Scheraga HA. Biochem. Biophys. Res. Commun. 2004;325(3):707–710. doi: 10.1016/j.bbrc.2004.10.088. [DOI] [PubMed] [Google Scholar]
- 59.Xu G, Narayan M, Kurinov I, Ripoli DR, Welker E, Khalili M, Ealick SE, Scheraga HA. J. Am. Chem. Soc. 2006;128(4):1204–1213. doi: 10.1021/ja055313e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu G, Narayan M, Welker E, Scheraga HA. Biochem. Biophys. Res. Commun. 2003;311(2):514–517. doi: 10.1016/j.bbrc.2003.10.039. [DOI] [PubMed] [Google Scholar]
- 61.Gahl RF, Narayan M, Xu G, Scheraga HA. Protein. Eng. Des. Sel. 2008 Jan. 31 doi: 10.1093/protein/gzm093. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darzynkiewicz Z, Carter SP, Mikulski SM, Ardelt W, Shogen K. Cell Tissue Kinet. 1988;21(3):169–182. doi: 10.1111/j.1365-2184.1988.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 63.Mikulski SM, Ardelt W, Shogen K, Bernstein EH, Menduke H. J. Natl. Cancer. Inst. 1990;82(2):151–153. doi: 10.1093/jnci/82.2.151-a. [DOI] [PubMed] [Google Scholar]
- 64.Mikulski SM, Viera A, Ardelt W, Menduke H, Shogen K. Cell Tissue Kinet. 1990;23(3):237–346. doi: 10.1111/j.1365-2184.1990.tb01119.x. [DOI] [PubMed] [Google Scholar]
- 65.Mikulski SM, Viera A, Darzynkiewicz Z, Shogen K. Brit. J. Cancer. 1992;66(2):304–310. doi: 10.1038/bjc.1992.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rybak SM, Pearson JW, Fogler WE, Volker K, Spence SE, Newton DL, Mikulski SM, Ardelt W, Riggs CW, Kung HF, Longo DL. J. Natl. Cancer Inst. 1996;88(11):747–753. doi: 10.1093/jnci/88.11.747. [DOI] [PubMed] [Google Scholar]
- 67.Deptala A, Halicka HD, Ardelt B, Ardelt W, Mikulski SM, Shogen K, Darzynkiewicz Z. Int. J. Oncol. 1998;13(1):11–16. doi: 10.3892/ijo.13.1.11. [DOI] [PubMed] [Google Scholar]
- 68.Vasandani VM, Castelli JC, Hott JS, Saxena S, Mikulski SM, Youle RJ. J. Interferon Cytokine Res. 1999;19(5):445–454. doi: 10.1089/107999099313884. [DOI] [PubMed] [Google Scholar]
- 69.Tsai SY, Hsieh TC, Ardelt B, Darzynkiewicz Z, Wu JM. Int. J. Oncol. 2002;20(5):891–896. [PubMed] [Google Scholar]
- 70.Tang CH, Hu CC, Wei CW, Wang JJ. FEBS Lett. 2005;579(1):265–270. doi: 10.1016/j.febslet.2004.11.086. [DOI] [PubMed] [Google Scholar]
- 71.Lee I, Lee YH, Mikulski SM, Lee J, Shogen K. Anticancer Res. 2000;20(2A):1037–1040. [PubMed] [Google Scholar]
- 72.Kim DH, Kim EJ, Kalota A, Gewirtz AA, Glickson J, Shogen H, Lee I. Adv. Exp. Biol. Med. 2007;599:53–59. doi: 10.1007/978-0-387-71764-7_8. [DOI] [PubMed] [Google Scholar]
- 73.Lee I, Kim DH, Sunar U, Magnitsky S, Shogen K. In vivo. 2007;21(5):721–728. [PubMed] [Google Scholar]
- 74.Halicka HD, Murakami T, Papageorgio CN, Mittelman A, Mikulski SM, Shogen K, Darzynkiewicz Z. Cell Proliferat. 2000;33(6):407–417. doi: 10.1046/j.1365-2184.2000.00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee I, Lee YH, Mikulski SM, Shogen K. Adv. Exp. Med. Biol. 2003;530:187–196. doi: 10.1007/978-1-4615-0075-9_18. [DOI] [PubMed] [Google Scholar]
- 76.Halicka HD, Ardelt B, Shogen K, Darzynkiewicz Z. Int. J. Oncol. 2007;30:841–847. [PubMed] [Google Scholar]
- 77.Haigis MC, Raines RT. J. Cell Sci. 2003;116(Pt 2):313–324. doi: 10.1242/jcs.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson RJ, Chao T-Y, Lavis LD, Raines RT. Biochemistry. 2007;46(36):10308–10316. doi: 10.1021/bi700857u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abercrombie E, Ambrose EJ. Cancer Res. 1962;22(5P1):525–548. [PubMed] [Google Scholar]
- 80.Márquez M, Nilsson S, Lennartsson L, Liu Z, Tammela T, Raitanen M, Holmberg AR. Anticancer Res. 2004;24(3A):1347–51. [PubMed] [Google Scholar]
- 81.Carter HB, Partin AW, Coffey DS. J. Urol. 1989;142(5):13338–13348. doi: 10.1016/s0022-5347(17)39093-6. [DOI] [PubMed] [Google Scholar]
- 82.Ilinskaya ON, Zelenikhin PV, Petrushanko IY, Mitkevich VA, Prassolov VS, Makarov AA. Biochem. Biophys. Res. Commun. 2007;361(4):1000–1005. doi: 10.1016/j.bbrc.2007.07.143. [DOI] [PubMed] [Google Scholar]
- 83.Wu Y, Saxena SK, Ardelt W, Gadina M, Mikulski SM, De Lorenzo XC, D'Alesio G, Youle RJ. J. Biol. Chem. 1995;270(29):17476–17481. doi: 10.1074/jbc.270.29.17476. [DOI] [PubMed] [Google Scholar]
- 84.Rodriguez M, Torrent G, Bosch M, Dubremetz J-F, Ribo M, Benito A, Vilanova M, Beaumelle B. J. Cell Sci. 2007;120(Pt 8):1405–1411. doi: 10.1242/jcs.03427. [DOI] [PubMed] [Google Scholar]
- 85.Cooner BP, Schmid SL. Nature. 2003;422(6927):37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 86.Juan G, Ardelt B, Mikulski SM, Shogen K, Ardelt W, Mittelman A, Darzynkiewicz Z. Leukemia. 1998;12(8):1241–1248. doi: 10.1038/sj.leu.2401100. [DOI] [PubMed] [Google Scholar]
- 87.Haigis MC, Kurten EL, Raines RT. Nucleic Acids Res. 2003;31(3):1024–1032. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dickson KA, Haigis MC, Raines RT. Prog. Nucleic. Acid Res. Mol. Biol. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johnson RJ, McCoy JG, Bingman CA, Phillips GN, Raines RT. J. Mol. Biol. 2007;277(2):434–449. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leland PA, Staniszewski KE, Kim BM, Raines RT. J. Biol. Chem. 2001;276(46):43095–43102. doi: 10.1074/jbc.M106636200. [DOI] [PubMed] [Google Scholar]
- 91.Gong J, Li X, Darzynkiewicz Z. J. Cell Physiol. 1993;157(2):263–270. doi: 10.1002/jcp.1041570208. [DOI] [PubMed] [Google Scholar]
- 92.Ardelt B, Ardelt W, Darzynkiewicz Z. Cell Cycle. 2003;2(1):22–24. doi: 10.4161/cc.2.1.232. [DOI] [PubMed] [Google Scholar]
- 93.Mattick JS. J. Exp. Biol. 2007;210(Pt 9):1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 94.Mattick JS, Makunin IV. Hum. Mol. Genet. 2006;15(1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 95.Mattick JS. Nat. Rev. Genet. 2004;5(4):316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 96.Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yunaihara N, Lanza G, Scarpa A, Veccione A, Negrini M, Harris CC, Croce CM. Proc. Natl. Acad. Sci. USA. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hernando E. Clin. Transl. Oncol. 2007;9(3):155–60. doi: 10.1007/s12094-007-0029-0. [DOI] [PubMed] [Google Scholar]
- 98.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen G, Hermeking H. Cell Cycle. 2007;6(13):1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 99.He X, He L, Hannon GJ. Cancer Res. 2007;67(23):11099–11105. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 100.Mikulski SM, Viera A, Deptala A, Darzynkiewicz Z. Int. J. Oncol. 1998;13(4):633–644. doi: 10.3892/ijo.13.4.633. [DOI] [PubMed] [Google Scholar]
- 101.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, Slacl FJ. Cancer Res. 2007;67(23):11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.He X, He L, Hannon GJ. Cancer Res. 2007;67(23):11099–11105. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 103.Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Curr. Biol. 2006;16(21):2135–42. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. J. Biol. Chem. 2005;280(10):9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 105.Looijenga LHJ, Gillis AJM, Stoop H, Hersmus R, Oosterhuis JW. Int. J. Androl. 2007;30(4):304–15. doi: 10.1111/j.1365-2605.2007.00765.x. [DOI] [PubMed] [Google Scholar]
- 106.Li S, Blackburn EH. Cold Spring Harb. Symp. Quant. Biol. 2006;71:211–215. doi: 10.1101/sqb.2006.71.009. [DOI] [PubMed] [Google Scholar]
- 107.Greider CW. Cold Spring Harb. Symp. Quant. Biol. 2006;71:225–229. doi: 10.1101/sqb.2006.71.063. [DOI] [PubMed] [Google Scholar]
- 108.McKenna SA, Lindhout DA, Kim I, Liu CW, Gelev VM, Wagner G, Puglisi JD. J. Biol. Chem. 2007;282(15):11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- 109.Lee I, Shogen K. Cancer Chemother. Pharmacol. 2007 Dec 5; doi: 10.1007/s00280-007-0637-y. Epub. [DOI] [PubMed] [Google Scholar]
- 110.Darzynkiewicz Z, Gong J, Juan G, Ardelt B, Traganos F. Cytometry. 1996;25(1):1–13. doi: 10.1002/(SICI)1097-0320(19960901)25:1<1::AID-CYTO1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 111.Baldwin AS., Jr. Ann. Rev. Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 112.Liu J, Lin A. Oncogene. 2007;26(22):3267–3278. doi: 10.1038/sj.onc.1210417. [DOI] [PubMed] [Google Scholar]
- 113.Tsai SY, Ardelt B, Hsieh TC, Darzynkiewicz Z, Shogen K, Wu JM. Int. J. Oncol. 2004;25(6):1745–1752. [PubMed] [Google Scholar]
- 114.Hu CC, Tang CH, Wang JJ. FEBS Lett. 2001;503(1):65–68. doi: 10.1016/s0014-5793(01)02691-6. [DOI] [PubMed] [Google Scholar]
- 115.Grabarek J, Ardelt B, Du L, Darzynkiewicz Z. Exp. Cell Res. 2002;278(1):61–71. doi: 10.1006/excr.2002.5568. [DOI] [PubMed] [Google Scholar]
- 116.Grabarek J, Ardelt B, Kunicki J, Darzynkiewicz Z. Cytometry. 2002;49(2):83–89. doi: 10.1002/cyto.10150. 2002. [DOI] [PubMed] [Google Scholar]
- 117.Halicka DH, Pozarowski P, Ita M, Ardelt W, Mikulski SM, Shogen K, Darzynkiewicz Z. Int. J. Oncol. 2002;21(6):1245–1250. doi: 10.3892/ijo.21.6.1245. [DOI] [PubMed] [Google Scholar]
- 118.Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz MA, Lassota P, Traganos F. Cytometry. 1992;13(8):795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 119.Gorczyca W, Bruno S, Darzynkiewicz RJ, Gong J, Darzynkiewicz Z. Int. J.Oncol. 1992;1:639–648. doi: 10.3892/ijo.1.6.639. [DOI] [PubMed] [Google Scholar]
- 120.Michaelis M, Cinatl J, Anad P, Rothweiler F, Kotchetkov R, von Deimling A, Doerr HW, Shogen K, Cinatl J., Jr. Cancer Lett. 2007;250:107–116. doi: 10.1016/j.canlet.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 121.Mayo MW, Wang C-Y, Cogswell PC, Rogers-Graham KS, Lowe SW, Der CL, Baldwin AS., Jr. Science. 1997;287(5344):1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 122.Van Antwerp DJ, Martin SJ, Verma IM, Green DR. Trends Cell Biol. 1998;8(3):107–111. doi: 10.1016/s0962-8924(97)01215-4. [DOI] [PubMed] [Google Scholar]