Abstract
Onconase (Onc) is a novel amphibian cytotoxic ribonuclease with antitumor activity and is currently in a confirmatory phase III clinical trial for the treatment of malignant mesothelioma. It was recently reported that Rana pipiens oocytes contain still another ribonuclease, named Amphinase (Amph). Amph shows 38-40% amino acid sequence identity with onconase, presents as four variants varying between themselves from 87-99% in amino acid sequence identity and has a molecular mass ∼13,000. In the present study we describe the effects of Amph on growth of several tumor cell lines. All four variants demonstrated cytostatic and cytotoxic activity against human promyelocytic HL-60-, Jurkat T-cell- and U-937 monocytic leukemia cells. The pattern of Amph activity to certain extent resembled that of Onc. Thus, cell proliferation was suppressed at 0.5-10.0 µg/ml (38-770 nM) Amph concentration with distinct accumulation of cells in G1 phase of the cell cycle. In addition, the cells were undergoing apoptosis, which manifested by DNA fragmentation (presence of “sub-G1” cells, TUNEL-positivity), caspases and serine proteases activation as well as activation of transglutaminase. The cytotostatic and cytotoxic effects of Amph required its ribonuclease activity: the enzymatically inactive Amph-2 having histidine at the active site alkylated was ineffective. The effectiveness and cell cycle specificity was generally similar for all four Amph variants and at the equimolar concentrations was somewhat more pronounced than that of Onc. The observed cytostatic and cytotoxic activity of Amph against tumor cell lines suggests that similar to Onc this cytotoxic ribonuclease may have antitumor activity and find an application in clinical oncology.
Keywords: cytotoxic ribonuclease, onconase, ranpirnase, apoptosis, anti-cancer properties, cell cycle, G1 arrest, caspase activation, serine protease activation, transglutaminase activation
INTRODUCTION
Several members of the superfamily of pancreatic ribonuclease A (RNase A; EC 3.1.27.5) are characterized by cytotoxic properties, with preference to tumor cells (reviewed in refs. 1-6). The most researched of them, Onconase is an amphibian RNase of about 12,000 molecular mass, isolated from oocytes of Northern Leopard frog (Rana pipiens).7,8 Onc demonstrates toxicity towards a variety of tumor cells in vitro9-11 as well as in vivo12-14 and is synergistic with many antitumor modalities, markedly enhancing their effectiveness.14-18 Effectiveness of Onc is also enhanced under conditions of mild hyperthermia.19 The preferred toxicity towards tumor cells in vitro and in vivo activity in mice tumor models prompted the use of Onc in clinical trials, which is currently in a phase III confirmatory clinical trial for the treatment of malignant mesothelioma.20,21
It was recently reported that oocytes of Rana pipiens contain another homologue of RNase A, with molecular mass ∼13,000, which was named Amphinase (Amph).22,23 Four variants (Amph 1-4) were isolated and sequenced. Sequence identities among the Amph variants themselves are 86.8-99.1 % while identities of the Amph variants with respect to Onc are 38.2-40.0 %. Amph variants, in analogy to Onc, show relatively weak ribonucleolytic activity that is not inhibited by recombinant human ribonuclease inhibitor protein (RI). The catalytic efficiency, substrate specificity and glycosylation state of Amphinases are found to be novel, distinct from the other well-characterized amphibian ribonucleases. The three-dimensional structure of one of the variants (Amph-2) was determined by X-ray crystallography at 1.8 Ä resolution.23 In the present study we explored the cytostatic and cytotoxic properties of Amph against several tumor cell lines.
The Amph-1, -2, -3 and -4 protein sequence data will appear in the UniProt Knowledgebase under accession numbers P85072, P85073, P85074 and P85075, respectively. The coordinates/structure factors of Amph-2 and rAmph-2 will appear in the RCSB Protein Data Bank under accession codes 2P7S and 2P6Z respectively.
MATERIALS AND METHODS
Cells and culture conditions
Human Jurkat T-cell leukemia cells were kindly provided by Dr. Douglas R. Green of La Jolla Institute for Allergy and Immunology, Human promyelocytic HL-60 and monocytic U-937 leukemia cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA). The cells were grown in 25 ml FALCON flasks (Becton Dickinson Co., Franklin Lakes, NJ) in RPMI 1640 supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 µg/ml streptomycin and 2 mM L-glutamine (all from GIBCO/BRL Life Technologies, Inc., Grand Island, N.Y.) at 37°C in an atmosphere of 5% CO2 in air. At the onset of the experiments, there were fewer than 5 × 105 cells per ml in culture such that the cells were at an exponential and asynchronous phase of growth. The cells were treated with preparations of Onc or Amph dissolved in culture medium, at different concentration as presented in figure legends.
The alkylated Amph-2 has been used as a control representing the enzyme in which the ribonucleolytic activity has been reduced. This form of Amph-2, with alkylated histidine at the active site, was prepared as described originally by Crestfield et al. in reference 24, with our modification.7 In brief, Amph-2 was incubated with a 140-fold molar excess of sodium iodoacetate (pH 5.5, 23°C, 8 h), desalted by size exclusion chromatography in dilute formic acid, and lyophilized. The ribonucleolytic activity of the alkylated Amph-2 was reduced by over 98%.23 Duration and other conditions of cell treatment with Amph are presented in figure legends and in the Results section.
Analysis of cytostatic and cytotoxic effects of Amph
Cell growth rate and viability in control and Amph-treated cultures was estimated by cell count combined with the trypan blue exclusion assay. The cell cycle distribution was assessed by staining cells with the DNA fluorochrome diamidino-2-phenylindole (DAPI; Sigma Chemical Co., St. Louis, MO) as described in reference 25. Apoptosis was detected by several methods, including the detection of DNA fragmentation assessed by the presence of cells with fractional DNA content25 and by DNA strand break labeling with exogenous terminal transferase, as described before in reference 26. Activation of caspases27 and serine proteases28,29 was detected using fluorochrome-tagged high affinity inhibitors reactive with active center of these enzymes. Activation of tissue transglutaminase (tTG) that induces extensive crosslinking of cytoplasmic proteins in the course of apoptosis was assessed by measuring the resistance of cells to nonionic detergent Triton X-100 as described by us previously in reference 28. Fluorescence intensity of individual cells tagged with the fluorochromes used in the assays described above was measured using EPICS/ELITE flow cytometer (Beckman Coulter, Miami, FL) and employing UV and 488 nm lasers to excite these fluorochromes. At least 106 cells were measured per sample. The DNA content frequency histograms were deconvoluted using MultiCycle software (Phoenix Flow Systems, San Diego, CA). Each experiment was run in duplicate or triplicate. Other details are given in figure legends.
RESULTS
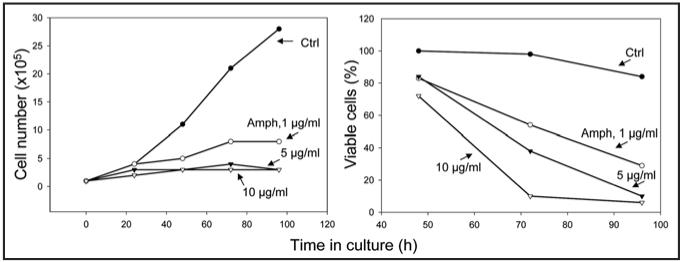
Figure 1 illustrates the effect of Amph-2 on growth and viability of HL-60 cells. It is quite evident that even at Amph-2 concentration as low as 1 µg/ml (80 nM) cell growth was dramatically suppressed, while at 5 and 10 µg/ml concentration essentially there was no evidence of proliferation at all. Cell viability was also markedly affected by Amph-2. Thus, after 72 h incubation with 1 µg/ml Amph-2 over 50% of cells were unable to exclude trypan blue. Even more dramatic effect was seen at 5 and 10 µg/ml concentration, when nearly all cells were dead after treatment with Amph-2 for 96 h.
Figure 1.
Growth and viability of HL-60 cells in absence and presence of Amph-1. Exponentially growing HL-60 cells were left untreated (Ctrl) or were treated with 1-10 µg/ml of Amph-2. The number and percent of cells excluding trypan blue was estimated at 0, 24, 48, 72 and 96 h after administration of Amph-1, and plotted as shown.
The effect of cell treatment with Amph-1 on cell cycle distribution and apoptosis is shown in Figure 2. Due to extensive DNA fragmentation during apoptosis the apoptotic cells (Ap) can be identified as the cells with fractional DNA content (“sub-G1” cell population).25,30 The data show that frequency of Ap cells was markedly increased in cultures containing Amph-1 and was progressively higher at higher Amph-1 concentration. However, even at the lowest (0.1 µg/ml; 8 nM) Amph-1 concentration there were 17% cells with fractional DNA content compared to 1% of such cells in control. The data also show a marked increase in percentage of G1 cells concomitant with a decrease in S and G2M-phase cells among the nonapoptotic cell population upon treatment with Amph-1 (Fig. 2). Analogous data showing induction of apoptosis, accumulation of cells in G1 and reduction in percentage of S- and G2M- cells of similar degree as for Amph-1, were induced by Amph-2, Amph-3 and Amph-4 (data not shown). The response of HL-60 cells to Amph-1-4 was essentially similar to that of U-937 although HL-cells appeared to be somewhat more sensitive (not shown).
Figure 2.
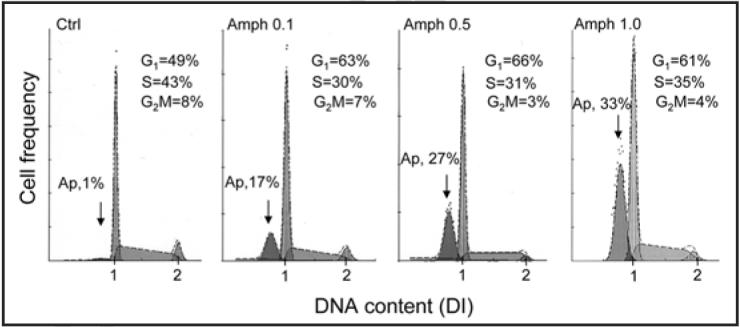
Effect of Amph-1 on cell cycle distribution and apoptosis of U-937 cells. Exponentially growing U-937 cells were untreated (Ctrl) or treated with 0.1, 0.5 or 1.0 µg/ml of Amph-1 for 72 h then fixed, their DNA stained with DAPI, and cellular fluorescence intensity measured by flow cytometry as described in references 25 and 30. The percent of apoptotic cells (Ap) with fractional DNA content (“sub-G1 cell population”) as well as the percent of cells in particular phases of the cell cycle, among the nonapoptotic cell population, is presented in each panel.
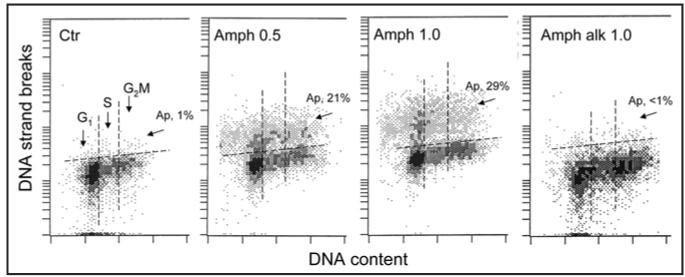
The induction of apoptosis characterized by DNA fragmentation by Amph-2 was confirmed using Jurkat cell line and identifying apoptotic cells by labeling DNA strand breaks in the TUNEL26 assay (Fig. 3). The increase in percentage of apoptotic cells from 1% to 21 and 29% was seen after cell treatment 0.5 or 1.0 µg/ml concentration of Amph-2 for 72 h, respectively. In contrast, the alkylated form of Amph-2, whose ribonuclease activity was reduced by over 98%,23 essentially was without effect on induction of apoptosis (Fig. 3D).
Figure 3.
Induction of apoptosis of Jurkat cell by Amph-2. The cells were untreated (Ctrl) or treated with 0.5 or 1.0 µg/ml of Amph-2, or with 1.0 µg/ml of the alkylated form of Amph-2 (Amph alk) for 72 h, then fixed and the presence of DNA strand breaks was detected by the TUNEL assay,26,32 DNA was counterstained with DAPI. The cells with DNA strand breaks above the threshold marked by the dashed line were classified as apoptotic and their percent is shown (Ap).
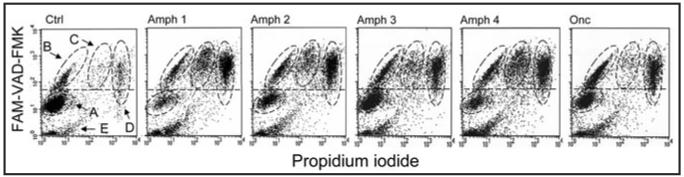
Figure 4 illustrates the effect of Amph-1 4 variants and Onc on caspases activation in HL-60 cells. It is quite evident that each of the four Amph variants as well as Onc strongly induced apoptosis characterized by caspases activation and consecutively, by the loss of plasma membrane integrity. Note that even that Onc was administered at five-fold higher (5 µg/ml) concentration than Amph (1 µg/ml) the induction of apoptosis in Amph and Onc treated cultures was seen at approximately similar frequency. The Onc effect was distinctly lower at 1 µg/ml concentration (not shown).
Figure 4.
Induction of apoptosis (caspase activation) of HL-60 cells by Amph 1-4 variants and by Onc. Exponentially growing cells were untreated (Ctrl) or treated with 1 µg/ml of Amph 1-4 or 5 µg/ml of Onc for 72 h, then incubated with the fluorochrome—(carboxyfluoresceine, FAM)—tagged inhibitor-ligand of activated caspases (FAM-VAD-FMK; pan-caspase ligand) and with propidium iodide (PI); their green (FAM) and red (PI) fluorescence was measured by flow cytometry as described in reference 27. Activation of caspases is characterized by increased binding of FAM-VAD-FMK above the background level marked by horizontal dashed line.31 Four cell subpopulations (A-D) can be distinguished based on differences in binding of FAM-VAD-FMK and degree of exclusion of PI.27,31 Population “A” represents live cells that did not activate caspases and exclude PI; “B” cell population represents very early apoptotic cells that show caspases activation but yet exclude PI; “C” cells are more advanced in apoptosis: have activated caspases and start to loose integrity of plasma membrane, partially excluding PI; “D” cells are late apoptotic cells, activated caspase-positive, that lost plasma membrane integrity and do not exclude, and thus strongly stain, with PI. The events marked “E” are cell debris.27,31
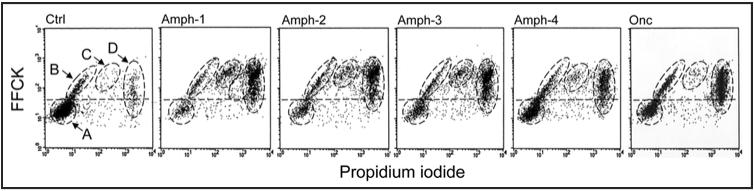
Similar pattern of induction of apoptosis by Amph 1-4 variants and by Onc was apparent when the cells were assessed for activation of serine proteases and exclusion of PI (Fig. 5). The probe, FFCK is a fluorescent analog of TPCK, the inhibitor and irreversible ligand of chymotrypsin-like proteases, which binds to such enzymes during their activation in apoptotic cells.28 The data show the strong response of HL-60 cells to all four variants of Amph-1 as well as to Onc.
Figure 5.
Induction of apoptosis (activation of serine proteases) of HL-60 cells treated with Amph 1-4 and by Onc. The cells were untreated (Ctrl) or treated with 1 µg/ml of Amph 1-4 or 5 µg/ml of Onc for 72 h, then incubated with the fluorochrome—(F; carboxyfluorescein)—tagged inhibitor and active center ligand of chymotrypsin-like proteases (FFCK) and with PI, and their green and red fluorescence was measured by flow cytometry as described in reference 28. Activation of serine proteases is detected by binding of FFCK above the background level marked by horizontal dashed line.28 As in Figure 4, four cell subpopulations, representing live cells (A) and cells at various stages of apoptosis (B-D), can be discriminated based on binding FFCK and exclusion of PI. The events representing cell debris (Fig. 4,”E”) were gated out.
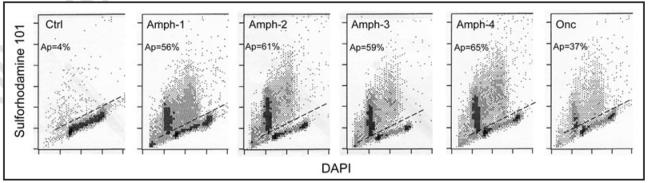
We have also tested effects of Amph 1-4 variants and Onc on activation of tTG in HL-60 cells (Fig. 6). This assay is based on the detection intra- and inter-molecular crosslinking of cytoplasmic proteins by the activated tTG which confers the resistance of cell to lysis by detergents.29 Thus, treatment of nonapoptotic cells with detergent leads to lysis of their plasma membrane and consequently leakage of cytosol and nuclear isolation. However, in apoptotic cells cytoplasmic proteins are insoluble and remain crosslinked to nuclei. The protein-fluorochrome sulforhodamine 101 used in this assay strongly stains apoptotic cells while isolated nuclei of nonapoptotic cells show markedly reduced stainability.29 Their DNA stainability is additionally reduced due to DNA fragmentation and loss of highly fragmented DNA sections during the staining procedure.25,31 The data (Fig. 6) demonstrate that all four Amph variants as well as Onc induced activation of tTG, still another marker of apoptosis, in a large number of HL-60 cells.
Figure 6.
Activation of tTG during apoptosis of U-937 cells induced by Amph variants 1-4 and by Onc. The cells, incubated for 72 h in the absence (Ctrl) or presence of Amph 1-4 variants at 1 µg/ml concentration, or with 5 µg/ml Onc, were treated with a solution of nonionic detergent Triton X-100 containing the DNA fluorochrome DAPI and the protein fluorochrome sulforhodamine 101. The nonapoptotic cells were lysed under these conditions, releasing isolated nuclei that stain minimally with sulforhodamine 101. In contrast, apoptotic cells (Ap) with activated tTG having crosslinked proteins, withstand the detergent treatment, their cytoplasmic proteins stain strongly with the sulforhodamine 101 while DNA, due to extensive fragmentation and chromatin condensation, stains at lower intensity compared to nonapoptotic cells.25,29
DISCUSSION
The present data demonstrate cytostatic and cytotoxic effects of all four Amph variants. The cytostatic effects manifest as accumulation of cells in G1 phase of the cell cycle and reduction of cells in S and G2M. This effect was most apparent at low concentration of Amph (0.1 µg/ml; Fig. 1), when the cytotoxicity was relatively low. Although the cell cycle data are presented with respect to HL-60 cells only, we observed similar accumulation of cells in G1 studying U-937 and Jurkat cells in the instances when the cytotoxic effects of Amph were not much pronounced [at low (0.1-1.0 µg/ml) Amph concentration and during the initial 24 - 48 h of the treatment; data not shown]. This cytostatic effect, which reflects prolongation of G1 phase of the cell cycle, is very much similar to the effect of Onc which was shown to partially arrest cells in G1,9 most likely by the mechanism involving suppression of cyclin D3 expression, upregulation of p16INK4A, p21WAF1/CIP1 and p27KIP and decreased pRb phosphorylation.11
The cytotoxic effect manifested as classical apoptosis, which involved morphological changes characterized by cell shrinkage, chromatin condensation and nuclear fragmentation (not shown). Also, extensive DNA fragmentation was apparent, revealed by the presence of the sub-G1 cell population with fractional DNA content (Fig. 2) and the presence of DNA strand breaks (Fig. 3) that were labeled with the fluorochrome tagged deoxynucleotide in the TUNEL assay utilizing exogenous terminal deoxynucleotidyl transferase to attach the fluorochrome-tagged deoxynucleotides to the 3′-OH ends of the breaks.26,32 Activation of caspases (Fig. 4), serine proteases (Fig. 5) and tTG (Fig. 6) was also detected. Activation of these enzymes was evident in the cells treated with each of the four Amph variants as well as with Onc. These findings are consistent with our earlier observations reporting pro-apoptotic effects of Onc on different cell lines when we observed sequential activation of caspases, serine proteases and tTG.27-30
The cytostatic and cytotoxic effectiveness of Amph, similar as that of Onc9-11 required its enzymatic ribonuclease activity. The alkylated form of Amph, which is enzymatically inactive but otherwise shows similar conformational structure as native Amph,23 neither showed evidence of cytotoxicity (Fig. 3D) nor of cytostatic effects (not shown). This is consistent with the hypothesis that the intercellular target of both Onc and Amph is intracellular RNA.
It has been claimed that the actual substrate of Onc within the cell is tRNA and its destruction by this drug, which leads to suppression of translation, is responsible for its cytostatic/cytotoxic effects.10 However, we observed that transcription of several genes is actually upregulated in the cells treated with Onc.11 This observation is incompatible with the mechanism in which Onc, by solely targeting tRNA, would lead to indiscriminate overall suppression of translation. Furthermore, the distinct specificity of Onc towards cancer cells can not be explained by targeting tRNA alone. We proposed,33 therefore, that one of the targets of Onc and Amph is the noncoding (micro; mi) RNA that is involved in regulation of gene expression through RNA interference (RNAi).34-36 Targeting RNAi may be the mechanism responsible for higher effectiveness of Onc towards tumor as compared to normal cells. Namely, it was recently reported that development of many tumors is associated with early alterations at the level of miRNA genes.37-40 The miRNA genes are often located at the genome hot spots associated with cancer. There is growing body of evidence that miRNAs are extensively involved in pathogenesis not only of leukemias or lymphomas but also of solid tumors and they promote neoplastic growth by controlling the expression of protein-coding tumor suppressors and oncogenes.38 By targeting RNAi, thus, ONC may be more effective in suppressing growth of tumor- rather than normal- cells.
Onc and Amph are in abundant quantity in frog eggs or developing embryos and its physiological function during embryogenesis is unknown. However, since RNAi plays critical role in gene regulation during development36,41-45 the presence of Onc and different variants of Amph in developing embryo would be consistent with their possible role in RNAi-mediated gene regulation during embryogenesis.
The initial step for the cytosolic internalization of Onc, and most likely of Amph, is binding to the cell surface, followed by a dynamin-independent endocytic pathway.46,47 The evidence with regard to the presence of cell surface receptors to Onc in tumor cells is contradictory.10,47 Onc and Amph, however, are strongly cationic molecules.7,22,23 The cell surface of most tumor types cells is known to have distinctly higher negative charge compared to their normal counterparts.47-49 In fact, cancer cells with high metastatic potential have still higher negative membrane charge compared to the cells with low metastatic ability.48 It is likely, therefore, that the electro-static interactions may play a role in preferential binding of Onc and Amph to cancer cells, which could be an additional factor contributing to their higher sensitivity to these RNases, as suggested by Lee and Raines.6 Consistent with this mechanism is also observation of Ilianskaya et al., that the bacterial RNase binase that is preferentially cytotoxic to tumor cells is strongly cationic.51
Although the comparable effects of Amph were seen at about five-fold lower its molar concentration compared with Onc, there was similarity between Onc and Amph in terms of mechanism of action, such as involvement of ribonucleolytic activity, induction of cytostasis (G1 arrest), and apoptosis, which manifested by morphological changes, caspase and serine protease activation, DNA fragmentation, and protein crosslinking by activated Tgase. As mentioned in the Introduction, sequence identities among the Amph variants are between 86.8 and 99.1 % while identities of the Amph variants with respect to Onc are only between 38.2 and 40.0 %. Thus, despite similarity of mechanism of action, in light of the differences in molecular structure of there RNases, it is likely that different tumor types may show preferential sensitivity to either Onc or Amph, or even to different Amph variants. Further studies involving in vivo testing of Amph to explore its potential in clinical application are warranted.
ACKNOWLEDGEMENTS
Supported in part by NCI CA RO1 28 704.
ABBREVIATIONS
- Amph
amphinase
- DAPI
diamidino-2-phenylindole
- FFCK
carboxyfluorescein phenylalanyl chloromethyl ketone
- Onc
onconase
- PI
propidium iodide
- TPCK
tosyl phenylalanyl chloromethyl ketone
- tTG
tissue transglutaminase (EC 2.3.2.13)
- TUNEL
terminal transferase dUTP nick end labeling
References
- 1.Leland PA, Raines RT. Cancer chemotherapy - Ribonucleases to the rescue. Chem Biol. 2001;8:405–13. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matoušek J. Ribonucleases and their antitumor activity. Comp Biochem Physiol. 2001;129C:175–91. doi: 10.1016/s1532-0456(01)90202-9. [DOI] [PubMed] [Google Scholar]
- 3.Benito A, Ribó M, Vilanova M. On the track of antitumour ribonucleases. Mol Biosyst. 2005;1:294–302. doi: 10.1039/b502847g. [DOI] [PubMed] [Google Scholar]
- 4.Costanzi J, Sidransky D, Navon A, Goldsweig H. Ribonucleases as a novel pro-apoptotic anticancer strategy: Review of the preclinical and clinical data for Ranpirnase. Cancer Invest. 2005;23:643–50. doi: 10.1080/07357900500283143. [DOI] [PubMed] [Google Scholar]
- 5.Arnold U, Ulbrich-Hofmann R. Natural and engineered ribonucleases as potential cancer therapeutics. Biotechnol Lett. 2006;28:1615–22. doi: 10.1007/s10529-006-9145-0. [DOI] [PubMed] [Google Scholar]
- 6.Lee JE, Raines RT. Ribonucleases as novel chemotherapeutics: The ranpirnase example. BioDrugs. 2007 doi: 10.2165/00063030-200822010-00006. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ardelt W, Mikulski SM, Shogen K. Amino acid sequence of an anti-tumor protein from Rana pipiens oocytes and early embryos. J Biol Chem. 1991;266:245–51. [PubMed] [Google Scholar]
- 8.Mosimann SC, Ardelt W, James MNG. Refined 1.7 Å X-ray crystallographic structure of P-30 protein, an amphibian ribonuclease with anti-tumor activity. J Mol Biol. 1994;236:1141–53. doi: 10.1016/0022-2836(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 9.Darzynkiewicz Z, Carter SP, Mikulski SM, Ardelt WJ, Shogen K. Cytostatic and cytotoxic effect of Pannon (P-30 Protein), a novel anticancer agent. Cell Tissue Kinet. 1988;21:169–82. doi: 10.1111/j.1365-2184.1988.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Mikulski SM, Ardelt W, Rybak SM, Youle RJ. A cytotoxic ribonuclease: Study of the mechanism of onconase cytotoxicity. J Biol Chem. 1993;268:10686–93. [PubMed] [Google Scholar]
- 11.Juan G, Ardelt B, Li X, Mikulski SM, Shogen K, Ardelt W, Mittelman A, Darzynkiewicz Z. G1 arrest of U937 cells by onconase is associated with suppression of cyclin D3 expression, induction of p16INK4A, p21WAF1/CIP1 and p27KIP and decreased pRb phosphorylation. Leukemia. 1998;12:1241–8. doi: 10.1038/sj.leu.2401100. [DOI] [PubMed] [Google Scholar]
- 12.Mikulski SM, Ardelt W, Shogen K, Bernstein EH, Menduke H. Striking increase of survival of mice bearing M109 Madison carcinoma treated with a novel protein from amphibian embryos. J Natl Cancer Inst. 1990;82:151–3. doi: 10.1093/jnci/82.2.151-a. [DOI] [PubMed] [Google Scholar]
- 13.Lee I, Lee YH, Mikulski SM, Shogen K. Effect of ONCONASE +/− tamoxifen on ASPC-1 human pancreatic tumors in nude mice. Adv Exp Med Biol. 2003;530:187–96. doi: 10.1007/978-1-4615-0075-9_18. [DOI] [PubMed] [Google Scholar]
- 14.Lee I, Kalota A, Gewirtz AM, Shogen K. Antitumor efficacy of the cytotoxic RNase, ranpirnase, on A549 human lung cancer xenografts of nude mice. Anticancer Res. 2007;27:299–307. [PubMed] [Google Scholar]
- 15.Mikulski SM, Viera A, Ardelt W, Menduke H, Shogen K. Tamoxifen and trifluoroperazine (Stelazine) potentiate cytostatic/cytotoxic effects of P-30 protein, a novel protein possessing anti-tumor activity. Cell Tissue Kinet. 1990;23:237–46. doi: 10.1111/j.1365-2184.1990.tb01119.x. [DOI] [PubMed] [Google Scholar]
- 16.Mikulski SM, Viera A, Darzynkiewicz Z, Shogen K. Synergism between a novel amphibian oocyte ribonuclease and lovastatin in inducing cytostatic and cytotoxic effects inhuman lung and pancreatic carcinoma cell lines. Br J Cancer. 1992;66:304–10. doi: 10.1038/bjc.1992.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deptala A, Halicka HD, Ardelt B, Ardelt W, Mikulski SM, Shogen K, Darzynkiewicz Z. Potentiation of tumor necrosis factor induced apoptosis by onconase. Int J Oncol. 1998;13:11–6. doi: 10.3892/ijo.13.1.11. [DOI] [PubMed] [Google Scholar]
- 18.Halicka HD, Murakami T, Papageorgio CN, Mittelman A, Mikulski SM, Shogen K, Darzynkiewicz Z. Induction of differentiation of leukaemic (HL-60) or prostate cancer (LNCaP, JCA-1) cells potentiates apoptosis triggered by onconase. Cell Prolif. 2000;33:407–17. doi: 10.1046/j.1365-2184.2000.00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halicka HD, Ardelt B, Shogen K, Darzynkiewicz Z. Mild hyperthermia predisposes tumor cells to undergo apoptosis upon treatment with onconase. Int J Oncol. 2007;30:841–7. [PubMed] [Google Scholar]
- 20.Costanzi J, Sidransky D, Navon A, Goldsweig H. Ribonucleases as a novel pro- apoptotic anticancer strategy: Review of the preclinical and clinical data for ranpirnase. Cancer Invest. 2005;23:643–50. doi: 10.1080/07357900500283143. [DOI] [PubMed] [Google Scholar]
- 21.Pavlakis N, Vogelzang NJ. Ranpirnase-an antitumour ribonuclease: Its potential role in malignant mesothelioma. Expert Opin Biol Ther. 2006;6:391–9. doi: 10.1517/14712598.6.4.391. [DOI] [PubMed] [Google Scholar]
- 22.Ardelt W, Vidunas E, Saxena S, Lee HS, Saxena A, Viera A, Shogen K. A novel cytotoxic ribonuclease from amphibian oocytes; Proceedings of 6th International Conference on Ribonucleases; Bath, UK. 2002. pp. 19–23. L-27. [Google Scholar]
- 23.Singh UP, Ardelt W, Saxena SK, Holloway DE, Vidunas E, Lee HS, Saxena A, Shogen K, Acharya KR. Enzymatic and structural characterisation of amphinase, a novel cytotoxic ribonuclease from Rana pipiens Oocytes. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.04.071. (Epub) [DOI] [PubMed] [Google Scholar]
- 24.Crestfield AM, Stein WH, Moore S. Alkylation and identification of the histidine residues at the active site of ribonuclease. J Biol Chem. 1963;238:2413–20. [PubMed] [Google Scholar]
- 25.Kajstura M, Halicka HD, Pryjma J, Darzynkiewicz Z. Discontinuous fragmentation of nuclear DNA during apoptosis revealed by discrete “sub-G1” peaks on DNA content histograms. Cytometry A. 2007;71A:125–31. doi: 10.1002/cyto.a.20357. [DOI] [PubMed] [Google Scholar]
- 26.Gorczyca W, Bruno S, Darzynkiewicz RJ, Gong J, Darzynkiewicz Z. DNA strand breaks occurring during apoptosis: Their early in situ detection by the terminal deoxynucleotidyl transferase and nick translation assays and prevention by serine protease inhibitors. Int J Onc. 1992;1:639–48. doi: 10.3892/ijo.1.6.639. [DOI] [PubMed] [Google Scholar]
- 27.Grabarek J, Du L, Johnson GL, Lee B, Phelps DJ, Darzynkiewicz Z. Sequential activation caspases and serine proteases (serpases) during apoptosis. Cell Cycle. 2002;1:124–31. [PubMed] [Google Scholar]
- 28.Grabarek J, Ardelt B, Du L, Darzynkiewicz Z. Activation of caspases and serine proteases during apoptosis induced by onconase (Ranpirnase) Exp Cell Res. 2002;278:61–71. doi: 10.1006/excr.2002.5568. [DOI] [PubMed] [Google Scholar]
- 29.Grabarek J, Ardelt B, Kunicki J, Darzynkiewicz Z. Detection of in situ activation of transglutaminase during apoptosis: Correlation with the cell cycle phase by multiparameter flow- and laser scanning- cytometry. Cytometry. 2002;49:83–89. doi: 10.1002/cyto.10150. [DOI] [PubMed] [Google Scholar]
- 30.Darzynkiewicz Z, Juan G, Li X, Gorczyca W, Murakami T, Traganos F. Cytometry in cell necrobiology: Analysis of apoptosis and accidental cell death (necrosis) Cytometry. 1997;27:1–20. [PubMed] [Google Scholar]
- 31.Pozarowski P, Huang X, Halicka DH, Lee B, Johnson G, Darzynkiewicz Z. Interactions of fluorochrome-labeled caspase inhibitors with apoptotic cells: A caution in data interpretation. Cytometry. 2003;55A:50–60. doi: 10.1002/cyto.a.10074. [DOI] [PubMed] [Google Scholar]
- 32.Gorczyca W, Gong J, Darzynkiewicz Z. Detection of DNA strand breaks in individual apoptotic cells by the in situ terminal deoxynucleotidyl transferase and nick translation assays. Cancer Res. 1993;53:1945–51. [PubMed] [Google Scholar]
- 33.Ardelt B, Ardelt W, Darzynkiewicz Z. Cytotoxic ribonucleases and RNA interference (RNAi) Cell Cycle. 2003;2:22–4. doi: 10.4161/cc.2.1.232. [DOI] [PubMed] [Google Scholar]
- 34.Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210:1526–47. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 35.Mattick JS, Makunin IV. Noncoding RNA. Hum Mol Genet. 2006;15(Spec No1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 36.Mattick JS. RNA regulation: A new genetics. Nat Rev Genet. 2004;5:316–23. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 37.Calin GA, Croce CM. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin Oncol. 2006;33:163–173. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yunaihara N, Lanza G, Scarpa A, Veccione A, Negrini M, Harris CC, Croce CM. A microRNAs expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernando E. microRNAs and cancer: Role in tumorigenesis, patient classification and therapy. Clin Transl Oncol. 2007;9:155–60. doi: 10.1007/s12094-007-0029-0. [DOI] [PubMed] [Google Scholar]
- 40.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing. miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;13:1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 41.Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: A paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24:119–29. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- 42.Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–42. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song L, Tuan RS. MicroRNAs and cell differentiation in mammalian development. Birth Defects Res C Embryo Today. 2006;78:140–9. doi: 10.1002/bdrc.20070. [DOI] [PubMed] [Google Scholar]
- 44.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–5. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 45.Looijenga LHJ, Gillis AJM, Stoop H, Hersmus R, Oosterhuis JW. Relevance of microRNAs in normal and malignant development, including human testicular germ cell tumors. Int J Androl. 2007;30:304–15. doi: 10.1111/j.1365-2605.2007.00765.x. [DOI] [PubMed] [Google Scholar]
- 46.Haigis MC, Raines RT. Secretory ribonucleases are internalized by a dynamin-independent endocytic pathway. J Cell Sci. 2003;116:313–24. doi: 10.1242/jcs.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez M, Torrent G, Bosch M, Rayne F, Dubremetz JF, Ribó M, Benito A, Vilanova M, Beaumelle B. Intracellular pathway of Onconase that enables its delivery to the cytosol. J Cell Sci. 2007;120:1405–11. doi: 10.1242/jcs.03427. [DOI] [PubMed] [Google Scholar]
- 48.James AM, Ambrose EJ, Lowick JH. Differences between the electrical charge carried by normal and homologous tumour cells. Nature. 1956;177:576–7. doi: 10.1038/177576a0. [DOI] [PubMed] [Google Scholar]
- 49.Márquez M, Nilsson S, Lennartsson L, Liu Z, Tammela T, Raitanen M, Holmberg AR. Charge-dependent targeting: Results in six tumor cell lines. Anticancer Res. 2004;24:1347–51. [PubMed] [Google Scholar]
- 50.Carter HB, Partin AW, Coffey DS. Prediction of metastatic potential in an animal model of prostate cancer: Flow cytometric quantification of cell surface charge. J Urol. 1989;142:13338–48. doi: 10.1016/s0022-5347(17)39093-6. [DOI] [PubMed] [Google Scholar]
- 51.Ilinskaya ON, Zelenikhin PV, Petrushanko IY, Mitkevich VA, Prassolov VS, Makarov AA. Binase introduces apoptosis of transformed myeloid cells and does not induce T-cell immune response. Biochem Biophys Res Commun. 2007 doi: 10.1016/j.bbrc.2007.07.143. In press. [DOI] [PubMed] [Google Scholar]