Abstract
The rate limiting step in biophysical characterization of membrane proteins is often the availability of suitable amounts of protein material. It was therefore of interest to demonstrate that micro-coil nuclear magnetic resonance (NMR) technology can be used to screen microscale quantities of membrane proteins for proper folding in samples destined for structural studies. Micoscale NMR was then used to screen a series of newly designed zwitterionic phosphocholine detergents for their ability to reconstitute membrane proteins, using the previously well characterized β-barrel E.coli outer membrane protein OmpX as a test case. Fold screening was thus achieved with μg-amounts of uniformly 2H,15N-labeld OmpX and affordable amounts of the detergents, and prescreening with SDS-gel electrophoresis ensured efficient selection of the targets for NMR studies. A systematic approach to optimize the phosphocholine motif for membrane protein refolding led to the identification of two new detergents, 138-Fos and 179-Fos, that yield 2D [15N,1H]-TROSY correlation NMR spectra of natively folded reconstituted OmpX.
Keywords: Detergent synthesis, fold screening, integral membrane proteins, micro-coil NMR, structural biology
Introduction
Integral membrane proteins (IMP) are embedded in lipid bilayers, performing a wide variety of essential functions in cellular biochemistry and cell-cell communication. IMPs are encoded by about one-third of a typical mammalian genome and comprise more than half of all current human drug targets.1-3 The structural characterization of IMPs has been an important challenge for decades, which has only recently been addressed with increasing success, primarily by X-ray crystallography. The use of detergents to extract and purify IMPs from their natural membrane environments is crucial for X-ray crystallography, which further requires the additional challenging step of making high-quality crystals containing the IMP, detergent and possibly other additives.4,5 In large part due to the difficulties of these operations, only a relatively small number of IMP structures have been solved to date.6
Solution NMR spectroscopy has found broad application as a complementary technique to X-ray crystallography in the structural characterization of biological macromolecules.7,8 However, its use for the study of IMPs is still limited,9-12 for three major reasons: 1) Reconstitution of IMPs with amphiphilic molecules gives rise to a substantial increase in molecular mass of the NMR target. 2) IMPs tend to be unstable in non-native environments. 3) Sample preparation is demanding due to low yields in membrane protein production and the requirement of stable-isotope labeling. While significant progress has been made in using solution NMR for larger particles,13 the difficulties of sample preparation must so far be addressed on a case-by-case basis.
Integral membrane proteins have a pronounced tendency to aggregate or fold incorrectly in the detergents used to extract them from their native membrane lipids; this is a critical problem for their structural, biochemical and biophysical characterization. Reconstitution of IMPs into detergent micelles by refolding is beneficial, because it allows the investigator to take advantage of more efficient synthesis of isotope-labeled proteins in aggregated form or possibly also by cell-free methods.12,14-20 A number of IMPs, mostly β-barrel proteins, have been successfully refolded into detergents, lipids or detergent-lipid mixtures, and the available comparisons have shown the refolded structures to exhibit nearly identical properties to the native proteins.15,21-26 In general, however, our knowledge of how to correctly fold a large pool of IMPs is still very limited, lagging far behind that for soluble proteins.27 It is therefore of great value to further explore appropriate folding conditions for IMP targets of biochemical and structural interest. Synthetic chemistry has a role to play in this endeavor, since the properties of small-molecule amphiphiles contribute in many ways to the isolation, stabilization, and characterization of IMPs.
In solution NMR studies, detergent micelles, as the smallest membrane-replacing agents, are the prime choice for IMP reconstitution.9-11,15-17,28 The number of detergents suitable for NMR analysis of IMPs is very limited, and only a few have been characterized for potential general use, including β-D-octylpyranoglucoside (OG), n-dodecylphosphocholine (DPC), 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC), and single-chain lysophopholipid analogues (e.g. LPPG, 1-palmitoyl-2-hydroxy-sn-glycero-3-[phosphor-RAC-(1-glycerol); LMPC, 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine).9,29-31
Fold screening of soluble proteins has been performed using micro-coil 1D 1H NMR spectroscopy.32,33 Membrane protein–detergent mixed micelles are more challenging targets. The detergents used to reconstitute and solubilize the proteins introduce a background that interferes with a straightforward interpretation of the protein signals in 1D 1H NMR spectra, and the molecular weight of the protein–detergent micelles is significantly larger than for the proteins alone, typically 50−100 kDa even for small membrane proteins.12,16,21 As a result, it is necessary to use isotope-labeled IMP–detergent mixed micelles for screening by NMR.
Selection of an appropriate detergent for NMR is a significant challenge. Experience from IMP crystallization so far provides little help: as might be expected given the different requirements of intermolecular association in the two situations, a detergent that is successful for crystallizing a specific IMP is often unsuitable for its NMR characterization. In a previous study,30 a selection of commercially available detergents were fold-screened with 2D [15N,1H]-HSQC experiments, using a conventional 5 mm NMR probe, and the single-chain lipid-like LPPG was singled out as a good detergent for selected α-helical membrane proteins. However, with conventional NMR probe technology, milligram amounts of protein are generally required for each experiment, which makes serial detergent screening impractical. We report here the screening of both commercial and newly synthesized detergents for successful NMR sample preparation of a well-characterized IMP with the use of an NMR microprobe, which requires only microgram quantities of protein for each measurement.
Methods
Detergent synthesis and purification
The synthesis of phosphocholine detergents followed the standard procedures.34 Details of the synthesis and characterization of new detergents may be found in the Supporting Information. Briefly, phosphocholines with branched alkyl chains were synthesized directly from the respective commercially available secondary alcohols. The ester-linked detergents were made by coupling of the hydroxyl group of 1-hydroxylethylphosphocholine (for the preparation of single chain detergents) or 138-Fos (for dialkyl chain detergents) with individual carboxylic acids. The amide-linked detergents were prepared by coupling the amine group of 1-aminoethylphosphocholine (for single-chain detergents) or 168-Fos (for dialkyl chain detergents) with the respective carboxylic acid chloride. The coupling procedure is described in the Supporting Information. Each detergent used for refolding and NMR experiments was prepared in 100 milligram batches, purified by reverse-phase HPLC (after silica-gel column chromatography) to > 99% purity, and its structure was confirmed by NMR and by mass spectrometry (Supporting Information).
Determination of critical micelle concentration (CMC)
CMC values of detergents were determined by monitoring the fluorescence of the ammonium salt of 8-anilino-1-naphtalenesulfonic acid (ANS), which becomes highly fluourescent (λex = 405 nm; λem = 465 nm) when incorporated into the hydrophobic micellar environment.35 Solutions containing 10 μM ANS and a range of concentrations for each detergent were examined on a DXT880 multiplate spectrofluorimeter (Beckman Coulter). The CMC is defined as the breakpoint in the plot of fluorescence intensity vs. concentration.
Expression and Purification of OmpX in D2O medium
OmpX was pre-cloned in the pET 3b plasmid and transformed into E. coli BL-21 (DE3) pLysS (Stratagene) competent cells for expression. One colony was used to inoculate a culture flask containing 20 mL of LB broth with the necessary antibiotics and shaken at 37 °C overnight. The cell culture was adapted to D2O by inoculating 4 mL of standard M9 minimal medium containing 33% v/v D2O with 40 μL of this LB culture and shaken at 37 °C overnight. 100 μL of the 33% D2O culture was then used to inoculate 10 mL of M9 minimal medium with 66% v/v D2O and similarly shaken overnight. A uniformly 2H,15N-labeled OmpX sample was prepared by inoculating 1 L of standard M9 minimal medium containing 99% D2O (Spectra Gases) and 1 g/L of [15N]-ammonium-chloride (CIL) with the resulting 10 mL of D2O-adapted culture. This culture was shaken at 37 °C until the cell density reached an OD600 of approximately 1.0, and protein expression was then induced with 1 mM isopropyl-α-D-thiogalactopyranoside. Upon reaching the stationary growth phase after approximately 4 hours, the cells were harvested at 5,000 g for 10 minutes. TE buffer (20 mM Tris-HCl at pH = 8.0, 5 mM EDTA) was used to resuspend the cell pellet in a volume corresponding to approximately 10 times the wet cell mass in grams, and sonicated for 25 minutes using a Misonix 3000 sonicator. The solution was pelleted at 4,300 g for 1 hour and the supernatant was discarded. The remaining insoluble pellet was resuspended in the same volume of TE buffer plus 2% (v/v) Triton X-100 at room temperature, pelleted at 4,300 g for 30 minutes, and the supernatant was discarded. Triton was then removed by repeating the same procedure with TE buffer. OmpX was recovered from the inclusion bodies by resuspending the cell debris in TE buffer plus 6 M urea for 2 hours at room temperature. Remaining cellular debris were then pelleted at 48,000 g for 20 minutes at 4 °C, and the unfolded OmpX in the supernatant was recovered for ion exchange purification.
Purification steps were performed using an ÄKTA purifier (Amersham/GE) with a 5 mL HiTrap Q HP anion exchange column (Amersham/GE). OmpX was bound to the column using buffer A (20 mM Tris–HCl at pH = 8.5, 6 M urea) and eluted with a linear NaCl gradient of 0 − 1 M at a rate of 33.3 mM per minute. All impure fractions were exchanged into buffer A and repurified, using the previously mentioned steps. The pooled pure OmpX fractions were concentrated to 1 mL using Vivaspin 2 (Sartorius) concentrators with a 3 kDa molecular weight cutoff. The concentrated sample was desalted with buffer A using a 5 mL HiTrap desalting column (Amersham/GE).
Reconstitution of OmpX into Detergent Micelles for SDS gel Electrophoresis
Unfolded OmpX (11.9 mg/mL, 20 mM Tris at pH 8.5, 6 M urea) was diluted 30-fold with 1% (wt/vol) detergent solution at 25 °C (1% detergent concentration is well above the CMC for each of the commercial or newly synthesized detergents used). The detergents were pre-dissolved in either of two buffers, 50 mM citrate with 1 mM EDTA for the pH-range 3.4−6.0 or 50 mM Tris-HCl with 1 mM EDTA for the pH-range 6.0−9.8. The solution was incubated at 25 °C for 3 hours before running the SDS-gel electrophoresis (using 15% polyacrylamide gels and without boiling the samples). The gels were stained with Coomassie blue and the fractions of refolded protein were estimated with densitometry.
Reconstitution of OmpX into Detergent Micelles for NMR spectroscopy
A previously established protocol16 for OmpX reconstitution in DHPC (Avanti Polar Lipids) detergent micelles was modified for micro-scale experiments with a variety of widely different detergents (Figure 1). At 4 °C, 100 μL of unfolded OmpX at a concentration of 10 mg/mL was added to 600 μL of refolding buffer (20 mM Tris–HCl at pH = 8.5, 5 mM EDTA, 600 mM L-Arg, and 2% (w/v) detergent) over a period of 4 hours with vigorous stirring. The resulting OmpX solution was vigorously stirred at 4 °C for about 16 hours. The detergent-refolded OmpX was then collected and exchanged into NMR buffer (20 mM sodium phosphate at pH = 6.8, 100 mM NaCl, 0.3% NaN3, 10 % D2O, detergent) by way of concentration and dilution using Vivaspin 500 concentrators (10,000 molecular weight cut-off), alternatively using NMR buffer with and without 2% (w/v) of detergent. The final sample was concentrated to 50 μL.
Figure 1.
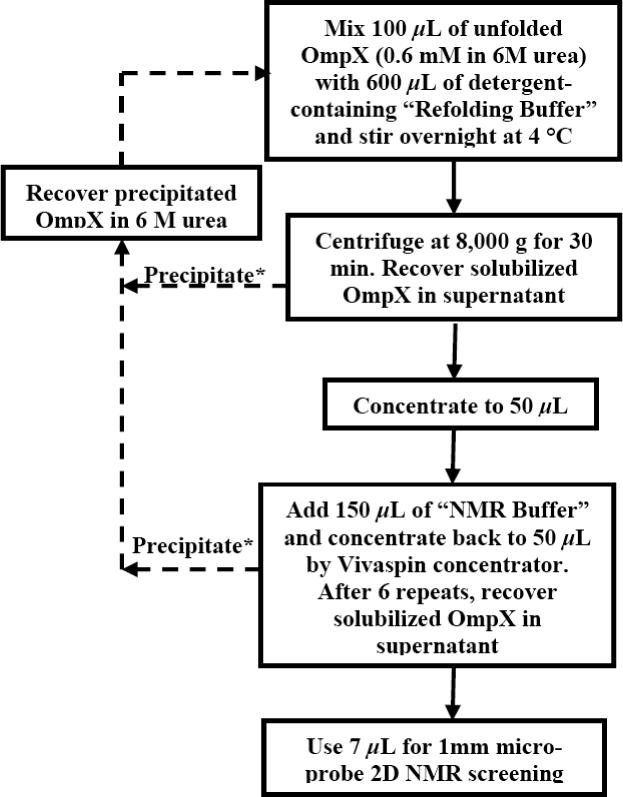
Flow chart for micro-scale OmpX reconstitution in detergent micelles. Solid lines indicate the route to reconstituted OmpX for structural studies, and dashed lines indicate the salvage pathways for precipitated OmpX. The buffers are described in the Methods section. Asterisks indicate procedural steps that were decisively improved with the use of the new detergents (see Table 3).
NMR Spectroscopy
All NMR experiments were recorded at 25 °C on a Bruker DRX-700 spectrometer (BrukerBiospin, Billerica, MA) equipped with a 1 mm TXI microprobe. The 1 mm NMR capillaries were filled with 7 μL of the solution containing the mixed OmpX–detergent micelles, using a 10 μL Gilson Syringe (Gilson Co., Reno, NV). 2D [15N,1H]-TROSY correlation experiments were recorded as described previously.16,36 Details of the parameter settings are given in the legend of Figure 4. Data processing and analysis were carried out using TOPSPIN 1.3 (Bruker) and XEASY,37 respectively.
Figure 4.
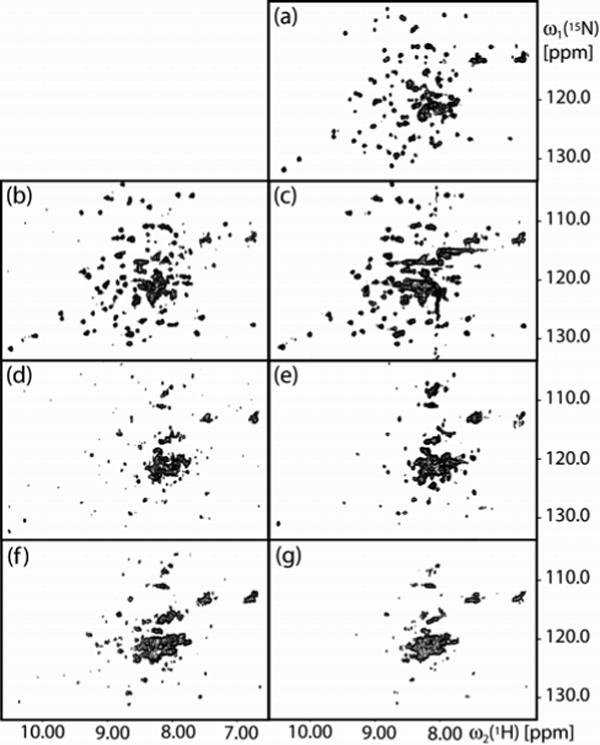
2D [15N,1H]-TROSY correlation NMR spectra of uniformly [2H,15N]-labeled OmpX reconstituted in mixed micelles with different detergents: (a) DHPC. (b) 138-Fos. (c) 179-Fos, where the vertical band of peaks near 8.0 ppm represents t1-noise from the signal of the amide proton of 179-Fos. (d) 115-Fos. (e) TPC. (f) 34-Fos. (g) 185-Fos. The spectra were collected with the following parameters: data size 50 (t1) × 1024 (t2) complex points; t1max = 25 ms; t2max = 86 ms; 300 scans per t1 increment, overall measurement time = 9 hours per experiment. Before Fourier transformation, the data matrices were multiplied with an exponential window function in the acquisition dimension, and with a 75°-shifted sine bell window46 in the indirect dimension.
Results and Discussion
OmpX is a 148-residue, 16.5 kD outer membrane protein with an 8-stranded β-barrel structure and structure-grade NMR spectra available for comparison.16,21 In general, the transmembrane domains of β-barrel proteins are less hydrophobic than those of helical IMPs, making the former highly soluble in chaotropic reagents as a starting point for refolding studies.
Twenty-three commercially available detergents (Table 1) were selected for OmpX reconstitution, which included the most popular representatives of the sugar (glucoside and maltoside), zwitterionic (Fos-choline series and lauryldimethylamine-N-oxide), cholate (sodium cholate, CHAPS, and CHAPSO), and amphipol (PMAL series: amphiphilic polymer detergents) classes; selected structures are shown in Supporting Information (Figure S1).
Table 1.
List of commercial detergents used (Anatrace, Inc.) and estimated OmpX refolding yields (± 5% error) by SDS-PAGE assay.
| Detergent | Yield (%) | Detergent | Yield (%) | Detergent | Yield (%) |
|---|---|---|---|---|---|
| n-Decyl-β-D-maltopyranoside | 40 | Triton X-100 | 20 | Sodium cholate | <5 |
| n-Undecyl-β-D-maltopyranoside | 55 | Fos-choline-10 | >90 | CHAPS | <5 |
| n-Dodecyl-β-D-maltopyranoside | 55 | Fos-choline-11 | >90 | CHAPSO | <5 |
| n-Tridecyl-β-D-maltopyranoside | 60 | Fos-choline-12 | >90 | PMAL-C10 | <5 |
| CYMAL-6 | 55 | Fos-choline-13 | >90 | PMAL-C12 | <5 |
| CYMAL-7 | 60 | Fos-choline-14 | >90 | PMAL-C16 | 10 |
| C-HEGA-11 | 35 | n-Octyl-β-D-gluopyranoside | <5 | Lauryldimethylamine-N-oxide | 80 |
| MEGA-8 | <5% | n-Nonyl-β-D-gluopyranoside | 10 |
OmpX is resistant to denaturation by SDS (as are many β-barrel membrane proteins38-43), yet this detergent inhibits refolding of the protein once it is denatured. Thus, folded and unfolded OmpX can be easily distinguished by SDS gel electrophoresis, migrating at 18 and 16 kD, respectively, on the standard Laemmli gels (15% polyacrylamide, Figure 2a). Interestingly, the folded and unfolded OmpX migrated at a reversed order (12 and 19 kD, respectively) on Bis-Tris gels (4 − 12% gradient polyacrylamide). This observation of the inverted running sequence of the folded and unfolded forms of β-barrel membrane proteins on different SDS gel matrices has not been recorded before to the best of our knowledge. The readout by SDS-PAGE was used to determine the OmpX folding efficiency in the presence of two well-known detergents, n-dodecyl-β-D-maltopyranoside (DDM) and Fos-choline-13 (TPC), as a function of pH (Figure 2b). OmpX refolding was thus found to be more efficient in TPC than in DDM in general, and was increasingly favored at higher pH, with > 90% folded protein achieved at pH 8.0 in TPC. The addition of folding additives, such as l-arginine and trimethylamine oxide, had no observable effect (data not shown).
Figure 2.
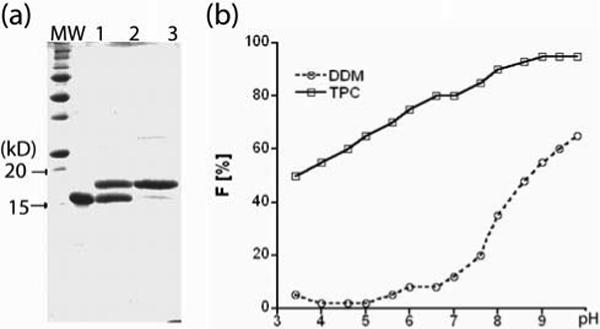
(a) SDS-gel electrophoresis of OmpX samples at pH 8.5. Lane 1: unfolded OmpX in 6 M urea; Lane 2: OmpX refolded in DDM; Lane 3: OmpX refolded in TPC. (b) pH effect on OmpX refolding. F is the fraction of the OmpX that was refolded. The protein (0.4 mg/mL), urea (0.2 M), and detergent (1% wt/vol) were incubated at room temperature for 3 hours, followed by quick analysis by SDS-PAGE. The fraction of folded OmpX was determined by densitometric analysis of the Coomassie stained gels (± 5% error). The relatively high concentration ensured that each detergent was present above CMC; the long incubation time ensured completion of the refolding process before the start of each measurement.44
Standardized folding conditions (pH 8.5 and 1% detergent concentration) were then used to screen the 23 aforementioned commercial detergents. As summarized in Table 1, the Fos-choline series (hydrophobic chain length 10−14 carbons) were the only candidates to support almost complete refolding of OmpX. In contrast, detergents with only glucose as the polar head (including octyl- and nonylglucoside), cholate-based detergents and amphipols were least efficient, with < 5% refolding in most of the cases. The larger maltose head group provided some improvement in refolding yield (40−60%), and lauryldimethylamine-N-oxide (LDAO) also gave a good yield (80%).
A majority of known favorable NMR detergents also fall into the phosphocholine class.9,29-31 In order to improve upon the phosphocholine theme, we introduced two types of modifications to make the detergents more closely resemble natural phospholipids (Figure 3). First, a spacer group to mimic the glycerol motif in lipids was inserted between the charged, potentially denaturing phosphocholine head group and the alkyl chain (Figure 3a). Second, short branches were added to mimic the dialkyl chain structure of lipids while maintaining their solubility (Figure 3b). Lysophospholipids and DHPC represent the former and latter patterns, respectively, and are generally regarded as mild phosphodetergents.
Figure 3.
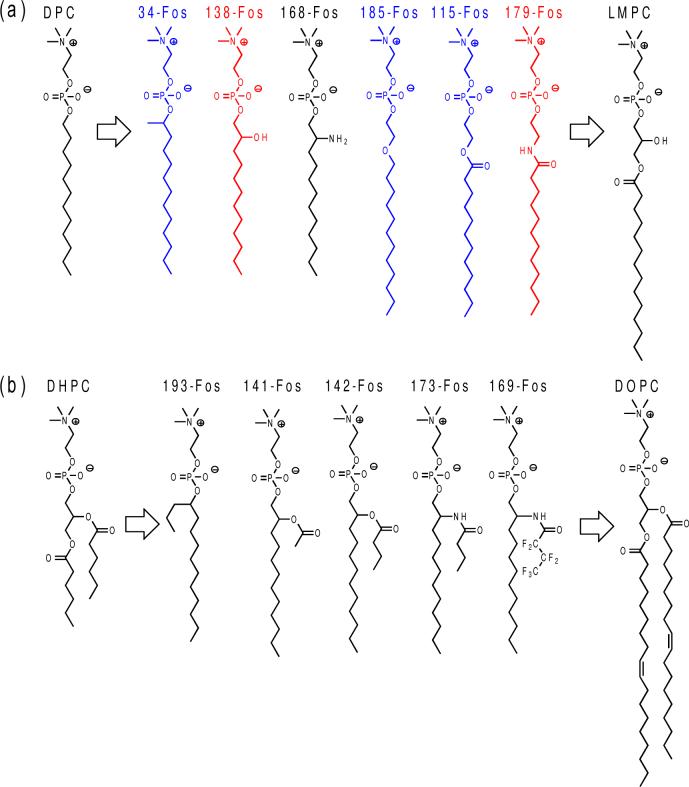
Representative examples of designed phosphocholine structures. (a) Single chain detergents. (b) Dialkyl-chain phospholipid (DOPC) and lipid-like detergents. The branched alkyl chain-containing detergents (34-Fos and 193-Fos) were assigned to both classes. Detergents examined by NMR are shown in blue and red, with the latter exhibiting the best performance.
We synthesized 42 new phosphocholine detergents (Figures 3 and S2) to test these design principles. The polar spacer groups inserted between the zwitterionic phosphocholine and the alkyl chain include secondary hydroxyl and amine groups (138-Fos and 168-Fos), ethylene oxide (185-Fos), ester (115-Fos, 141-Fos and 142-Fos), amide (179-Fos, 173-Fos and 169-Fos), and others (see Figure S2). For dialkyl chain detergents, the short chain was conveniently appended through an ester or amide bond or preinstalled on a secondary alcohol (branched detergents). Partially fluorinated detergents (e.g., 169-Fos) were also synthesized for their reportedly milder action.45
Similar refolding experimentations to Figure 2b yielded the data summarized in Table 2. Quite a number of the newly synthesized phosphocholine detergents could refold OmpX as efficiently as commercial ones. We have synthesized and evaluated other classes of new detergents, and the present study now corroborates that the phosphocholine series is among the top performers among all our new detergent structures for refolding OmpX. From the results shown in Table 2, it is apparent that detergents bearing dialkyl chains (branched through either an ester or amide linkage) are much less effective in refolding OmpX than analogues with single straight alkyl chains. Almost all the best candidates, with > 90% refolding, are either single alkyl chain detergents or those with a single methyl group on the branch (30-Fos, 31-Fos, 34-Fos and 38-Fos).
Table 2.
List of newly synthesized phosphocholine detergents and estimated OmpX refolding yields (± 5% error) by SDS-PAGE assay.
| Detergenta | Yield (%) | Detergenta | Yield (%) | Detergenta | Yield (%) |
|---|---|---|---|---|---|
| 30-Fos | > 90 | 168-Fos | > 90 | 171-Fos | 55 |
| 31-Fos | > 90 | 167-Fos | 84 | 142-Fos | 52 |
| 34-Fos | > 90 | 165-Fos | 71 | 193-Fos | 50 |
| 35-Fos | > 90 | 140-Fos | 70 | 173-Fos | 49 |
| 38-Fos | > 90 | 172-Fos | 69 | 170-Fos | 46 |
| 115-Fos | > 90 | 117-Fos | 67 | 175-Fos | 45 |
| 116-Fos | > 90 | 37-Fos | 63 | 178-Fos | 44 |
| 180-Fos | > 90 | 190-Fos | 60 | 194-Fos | 44 |
| 179-Fos | > 90 | 177-Fos | 60 | 174-Fos | 43 |
| 185-Fos | > 90 | 192-Fos | 58 | 195-Fos | 30 |
| 228-Fos | > 90 | 141-Fos | 57 | 169-Fos | 28 |
| 229-Fos | > 90 | 137-Fos | 56 | 144-Fos | 28 |
| 182-Fos | > 90 | 176-Fos | 55 | 145-Fos | 25 |
| 138-Fos | > 90 | 33-Fos | 55 | 41-Fos | < 5 |
Detergent structures can be found in Figures 3 and S2.
To extend the electrophoresis assay of Figure 2b, we used uniform 2H- and 15N-labeling of the protein OmpX and TROSY-type 2D [15N,1H]-correlation NMR spectroscopy to assess the fold of OmpX in mixed micelles with different detergents. To accomplish this goal, a screening protocol using micro-coil NMR equipment was first tested with OmpX−DHPC mixed micelles (OmpX/DHPC). OmpX/DHPC has previously been studied in great detail, so that nearly complete polypeptide backbone assignments and a solution structure are available.16,21 The 2D [15N,1H]-TROSY correlation spectrum of OmpX/DHPC measured with the 1 mm TXI microprobe shows a wide dispersion of cross peaks with roughly uniform intensities and line shapes (Figure 4a). Comparison with the previously determined chemical shift list (BMRB accession code 4936) showed that all cross peaks were visible, with the sole exception of the four peaks belonging to the polypeptide segment E32−S35. This shows that our screening protocol, which required only about 10 μL of protein solution (active volume in the NMR micro-coil = 7 μL), provided high-quality [15N,1H]-correlation maps that can be used as diagnostic fingerprints to assess the conformation of OmpX in mixed micelles with detergents.
Using the electrophoresis results to guide the selection of candidates, we examined [2H,15N]-labeled OmpX in mixed micelles with five newly synthesized detergents. The structures are representative of distinct classes (spacer groups) that fully refolded the protein (138-Fos, CMC 0.1%; 179-Fos, CMC 0.062%; 115-Fos, CMC 0.025%; 34-Fos, CMC 0.036%; 185-Fos, CMC 0.01%). A widely used commercial phosphocholine detergent (TPC, CMC 0.027%) was also used for comparison. OmpX was readily reconstituted into these detergent micelles at high concentrations (100 μL of 10 mg/mL unfolded OmpX was refolded into 600 μL detergent solution by slow addition); no protein precipitation was observed, and SDS-gel electrophoresis experiments confirmed complete solubilization.
The 2D [15N,1H]-TROSY correlation spectra of mixed OmpX/138-Fos and OmpX/179-Fos micelles (Figure 4, b and c) are both closely similar to the spectrum of OmpX/DHPC, with respect to the number of cross-peaks, their chemical shift dispersion and the peak intensity distribution, indicating that OmpX was homogeneously reconstituted and adopted a similar conformation in the new detergents as in DHPC. A decisive advance, which results in significant savings of detergent and working time, is that there was no precipitation during the micro-scale reconstitution in 138-Fos or in 179-Fos, whereas more than 50% of the protein precipitated during each cycle of reconstitution in DHPC (Table 3). Therefore, reconstitution in 138-Fos or in 179-Fos can efficiently be achieved, whereas with DHPC several recovery-cycles were needed in order to get a NMR sample (Figure 1).
Table 3.
Micro-scale OmpX reconstitution with different detergents.
| Detergent | OmpX precipitation in refolding buffera | OmpX precipitation in NMR bufferb | NMR Qualityc |
|---|---|---|---|
| DHPC | ∼30% | ∼40% | ++ |
| 138-Fos | 0% | 0% | ++ |
| 179-Fos | 0% | 0% | + |
| 115-Fos | 0% | 0% | − |
| TPC | 0% | 0% | − |
| 34-Fos | 0% | 0% | − |
| 185-Fos | 0% | 0% | − |
Recoverable OmpX loss due to precipitation during refolding.
Recoverable OmpX loss due to buffer exchange (Figure 1).
2D [15N,1H]-TROSY spectra (Figure 4) rated “++” for spectra suitable for a structure determination, “+” for spectra with detergent artifacts, and “-” for spectra showing that OmpX was solubilized, but not reconstituted.
The 2D [15N,1H]-TROSY correlation spectra of the mixed micelles with OmpX/115-Fos,OmpX/TPC, OmpX/34-Fos and OmpX/185-Fos (Figure 4, d–g) all show a cluster of broad lines in the center of the spectrum and variable numbers of additional, resolved cross peaks of unequal shapes and intensities, which indicates that these protein samples are not homogeneously folded, and probably form non-specific soluble aggregates with these detergents.
In conclusion, the combined use of preliminary screening of a library of candidate detergents for OmpX refolding using SDS-gel electrophoresis, and further characterization of the OmpX conformation in mixed micelles with selected detergents by 2D [15N,1H]-TROSY correlation NMR spectroscopy resulted in the identification of two new detergents, 138-Fos and 179-Fos, that afford reconstitution of OmpX in its native form more efficiently than DHPC. These two single-chain phosphocholine detergents are the structural mimics of the lysophospholipids that have been shown to retain the folded conformation of several α-helical membrane proteins in solution NMR studies.30 The use of micro-coil NMR technology was key to this success, reducing the cost of the protein as well as the novel amphiphiles, which are often initially made in small quantities.47-53
An important result is the observation that dialkyl chain lipid-like detergents are not as efficient as straight-chain detergents in refolding OmpX, which is consistent with the known difficulty of refolding β-barrel proteins into lipids with long double-alkyl chains.42,43 Presumably, detergents with two hydrophobic chains self-assemble into larger structures with smaller surface curvature than the single-chain analogues, and the smaller micelles formed by single-chain detergents tend to exhibit more rapid exchange with the detergent monomers. These properties might contribute to the high refolding efficiency for OmpX.54,55
Minor structural changes of the detergent chemical structures can have dramatic effects on their ability to complex with membrane proteins in solution. We and others have observed the same type of sensitivity to detergent modification in crystallization studies of IMPs.4 Thus, among the six phosphocholine detergents studied here by NMR spectroscopy, 115-Fos and 179-Fos differ only in the substitution of an ester bond by an amide linkage, but only 179-Fos was found to properly reconstitute OmpX. The two new molecules found to support full folding of OmpX under NMR conditions, 138-Fos and 179-Fos, are superior to the previously used DHPC and to lysophospholipids in terms of detergent stability, since the ester bonds in DHPC and in lysophospholipids are prone to hydrolysis at elevated temperature and basic pH.56-58 It is likely that different proteins will require different detergents to achieve similar results; toward this end, we will continue our amphiphile synthesis and characterization efforts with new classes of detergents and with other IMPs.
Acknowledgements
We thank Prof. G. Chang for providing some of the commercial detergents, and Prof. J. Kelly and P. Dawson for helpful discussions. This work was supported by the Joint Center for Innovative Membrane Protein Technologies (NIH Roadmap grant GM073197) and the Joint Center for Structural Genomics (NIH grant U54 GM074898). K.W. is the Cecil H. and Ida M. Green Professor of Structural Biology at The Scripps Research Institute.
Footnotes
Supporting Information Available: Structures of the commercial and newly-synthesized detergents; synthetic procedures and characterization data for representative detergents. Complete ref. 51. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Hopkins AL, Groom CR. Nat. Rev. Drug Discov. 2002;1:727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Lundstrom K. Curr. Protein Pept. Sci. 2006;7:465–70. doi: 10.2174/138920306778559403. [DOI] [PubMed] [Google Scholar]
- 3.Wallin E, von Heijne G. Protein Sci. 1998;7:1029–38. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunte C, von Jagow G, Schägger H, editors. Membrane protein purification and crystallization, a practical guide. 2 ed. Academic Press; San Deigo: 2003. [Google Scholar]
- 5.Iwata S, editor. Methods and Results in Crystallization of Membrane Proteins. International University Line; La Jolla: 2003. [Google Scholar]
- 6.Michel's H, White's S. summaries at http://www.mpibp-frankfurt.mpg.de/michel/public/memprotstruct.html and http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html.
- 7.Wüthrich K. Angew. Chem. Int. Ed. Engl. 2003;42:3340–63. doi: 10.1002/anie.200300595. [DOI] [PubMed] [Google Scholar]
- 8.Ferentz AE, Wagner G. Quat. Rev. Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 9.Sanders CR, Sönnichsen F. Magn. Reson. Chem. 2006;44:S24–40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 10.Sanders CR, Oxenoid K. Biochim. Biophys. Acta. 2000;1508:129–45. doi: 10.1016/s0005-2736(00)00308-4. [DOI] [PubMed] [Google Scholar]
- 11.Arora A, Tamm LK. Curr. Opin. Struct. Biol. 2001;11:540–7. doi: 10.1016/s0959-440x(00)00246-3. [DOI] [PubMed] [Google Scholar]
- 12.Fernández C, Wüthrich K. FEBS Lett. 2003;555:144–50. doi: 10.1016/s0014-5793(03)01155-4. [DOI] [PubMed] [Google Scholar]
- 13.The size limitations of solution NMR, exemplified in many early studies of small membrane-associated peptides or membrane protein fragments, have been remarkably improved by higher field instrumentation and new techniques such as uniform and selective isotope labeling and heteronuclear multidimensional NMR spectroscopy. For example, transverse relaxation-optimized spectroscopy (TROSY) Wüthrich K, Wider G. Encycl. Nucl. Magn. Reson. 2002;9:468–477. has pushed the size limit beyond 100 kDa also in applications to the study of IMPs.
- 14.Etezady-Esfarjani T, Hiller S, Villalba C, Wüthrich K. J. Biomol. NMR. 2007;39:229–238. doi: 10.1007/s10858-007-9188-0. [DOI] [PubMed] [Google Scholar]
- 15.Arora A, Abildgaard F, Bushweller JH, Tamm LK. Nat. Struct. Biol. 2001;8:334–8. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 16.Fernández C, Adeishvili K, Wüthrich K. Proc. Natl. Acad. Sci. USA. 2001;98:2358–63. doi: 10.1073/pnas.051629298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Privé GG, Bishop RE, Kay LE. Proc. Natl. Acad. Sci. USA. 2002;99:13560–5. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. Science. 2005;307:1317–21. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 19.Fernández C, Hilty C, Bonjour S, Adeishvili K, Pervushin K, Wüthrich K. FEBS Lett. 2001;504:173–8. doi: 10.1016/s0014-5793(01)02742-9. [DOI] [PubMed] [Google Scholar]
- 20.Koglin A, Klammt C, Trbovic N, Schwarz D, Schneider B, Schäfer B, Löhr F, Bernhard F, Dötsch V. Magn. Reson. Chem. 2006;44:S17–23. doi: 10.1002/mrc.1833. [DOI] [PubMed] [Google Scholar]
- 21.Fernández C, Hilty C, Wider G, Güntert P, Wüthrich K. J. Mol. Biol. 2004;336:1211–1221. doi: 10.1016/j.jmb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Pautsch A, Schulz GE. Nat. Struct. Biol. 1998;5:1013–7. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 23.Pautsch A, Schulz GE. J. Mol. Biol. 2000;298:273–82. doi: 10.1006/jmbi.2000.3671. [DOI] [PubMed] [Google Scholar]
- 24.Vogt J, Schulz GE. Structure. 1999;7:1301–9. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 25.Huang KS, Bayley H, Liao MJ, London E, Khorana HG. J. Biol. Chem. 1981;256:3802–9. [PubMed] [Google Scholar]
- 26.Popot JL, Trewhella J, Engelman DM. EMBO J. 1986;5:3039–44. doi: 10.1002/j.1460-2075.1986.tb04603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Booth PJ, Curnow P. Curr. Opin. Struct. Biol. 2006;16:480–8. doi: 10.1016/j.sbi.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Bicelles (a mixture of detergents and lipids) and amphipols (amphiphilic polymer detergents) have recently been used in the solution NMR study of native integral membrane proteins: Poget SF, Cahill SM, Girvin ME. J. Am. Chem. Soc. 2007;129:2432–3. doi: 10.1021/ja0679836..Zoonens M, Catoire LJ, Giusti F, Popot JL. Proc. Natl. Acad. Sci. USA. 2005;102:8893–8. doi: 10.1073/pnas.0503750102.
- 29.Vinogradova O, Sönnichsen F, Sanders CR. J. Biomol. NMR. 1998;11:381–6. doi: 10.1023/a:1008289624496. [DOI] [PubMed] [Google Scholar]
- 30.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. J. Biomol. NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 31.Brown LR, Bösch C, Wüthrich K. Biochim. Biophys. Acta. 1981;642:296–312. doi: 10.1016/0005-2736(81)90447-8. [DOI] [PubMed] [Google Scholar]
- 32.Peti W, Norcross J, Eldridge G, O'Neil-Johnson M. J. Am. Chem. Soc. 2004;126:5873–8. doi: 10.1021/ja039779d. [DOI] [PubMed] [Google Scholar]
- 33.Peti W, Page R, Moy K, O'Neil-Johnson M, Wilson IA, Stevens RC, Wüthrich K. J. Struct. Funct. Genom. 2005;6:259–67. doi: 10.1007/s10969-005-9000-x. [DOI] [PubMed] [Google Scholar]
- 34.Tegoulia VA, Rao WS, Kalambur AT, Rabolt JR, Cooper SL. Langmuir. 2001;17:4396–404. [Google Scholar]
- 35.De Vendittis E, Palumbo G, Parlato G, Bocchini V. Anal. Biochem. 1981;115:278–86. doi: 10.1016/0003-2697(81)90006-3. [DOI] [PubMed] [Google Scholar]
- 36.Pervushin K, Riek R, Wider G, Wüthrich K. Proc. Natl. Acad. Sci. USA. 1997;94:12366–71. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 38.Kleinschmidt JH. Chem. Phys. Lipids. 2006;141:30–47. doi: 10.1016/j.chemphyslip.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Tamm LK, Hong H, Liang B. Biochim. Biophys. Acta. 2004;1666:250–63. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 40.Ohnishi S, Kameyama K. Biochim. Biophys. Acta. 2001;1515:159–66. doi: 10.1016/s0005-2736(01)00410-2. [DOI] [PubMed] [Google Scholar]
- 41.Conlan S, Bayley H. Biochemistry. 2003;42:9453–65. doi: 10.1021/bi0344228. [DOI] [PubMed] [Google Scholar]
- 42.Pocanschi CL, Apell HJ, Puntervoll P, Hogh B, Jensen HB, Welte W, Kleinschmidt JH. J. Mol. Biol. 2006;355:548–61. doi: 10.1016/j.jmb.2005.10.060. [DOI] [PubMed] [Google Scholar]
- 43.Surrey T, Jähnig F. Proc. Natl. Acad. Sci. USA. 1992;89:7457–61. doi: 10.1073/pnas.89.16.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinschmidt JH, Wiener MC, Tamm LK. Protein Sci. 1999;8:2065–71. doi: 10.1110/ps.8.10.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaudier Y, Zito F, Barthélémy P, Stroebel D, Améduri B, Popot JL, Pucci B. Bioorg. Med. Chem. Lett. 2002;12:1587–90. doi: 10.1016/s0960-894x(02)00242-1. [DOI] [PubMed] [Google Scholar]
- 46.DeMarco A, Wüthrich K. J. Magn. Reson. 1976;24:201–204. [Google Scholar]
- 47.Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G. Angew. Chem. Int. Ed. Engl. 2007;46:7023–5. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 48.Kiley P, Zhao X, Vaughn M, Baldo MA, Bruce BD, Zhang S. PLoS Biol. 2005;3:e230. doi: 10.1371/journal.pbio.0030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGregor CL, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Privé GG. Nat. Biotechnol. 2003;21:171–6. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]
- 50.McQuade DT, Quinn MA, Yu SM, Polans AS, Krebs MP, Gellman SH. Angew. Chem. Int. Ed. Engl. 2000;39:758–761. [PubMed] [Google Scholar]
- 51.Popot JL, et al. Cell Mol. Life Sci. 2003;60:1559–74. doi: 10.1007/s00018-003-3169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schafmeister CE, Miercke LJ, Stroud RM. Science. 1993;262:734–8. doi: 10.1126/science.8235592. [DOI] [PubMed] [Google Scholar]
- 53.Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Proc. Natl. Acad. Sci. USA. 2006;103:17707–12. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pocanschi CL, Patel GJ, Marsh D, Kleinschmidt JH. Biophys. J. 2006;91:L75–7. doi: 10.1529/biophysj.106.091439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong H, Tamm LK. Proc. Natl. Acad. Sci. USA. 2004;101:4065–70. doi: 10.1073/pnas.0400358101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burns RA, Jr., Friedman JM, Roberts MF. Biochemistry. 1981;20:5945–50. doi: 10.1021/bi00524a004. [DOI] [PubMed] [Google Scholar]
- 57.Dannenberg A, Wong T, Zakim D, Eibl H. Anal. Biochem. 1990;191:183–6. doi: 10.1016/0003-2697(90)90406-y. [DOI] [PubMed] [Google Scholar]
- 58.Das AK, Hajra AK. J. Lipid Res. 1995;36:2459–68. [PubMed] [Google Scholar]